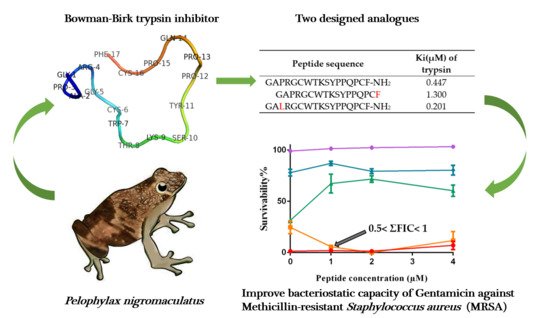
Ranacyclin-NF, a Novel Bowman–Birk Type Protease Inhibitor from the Skin Secretion of the East Asian Frog, Pelophylax nigromaculatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of Skin Secretion
2.2. “Shotgun” Cloning and Protein Bioinformatic Analyses
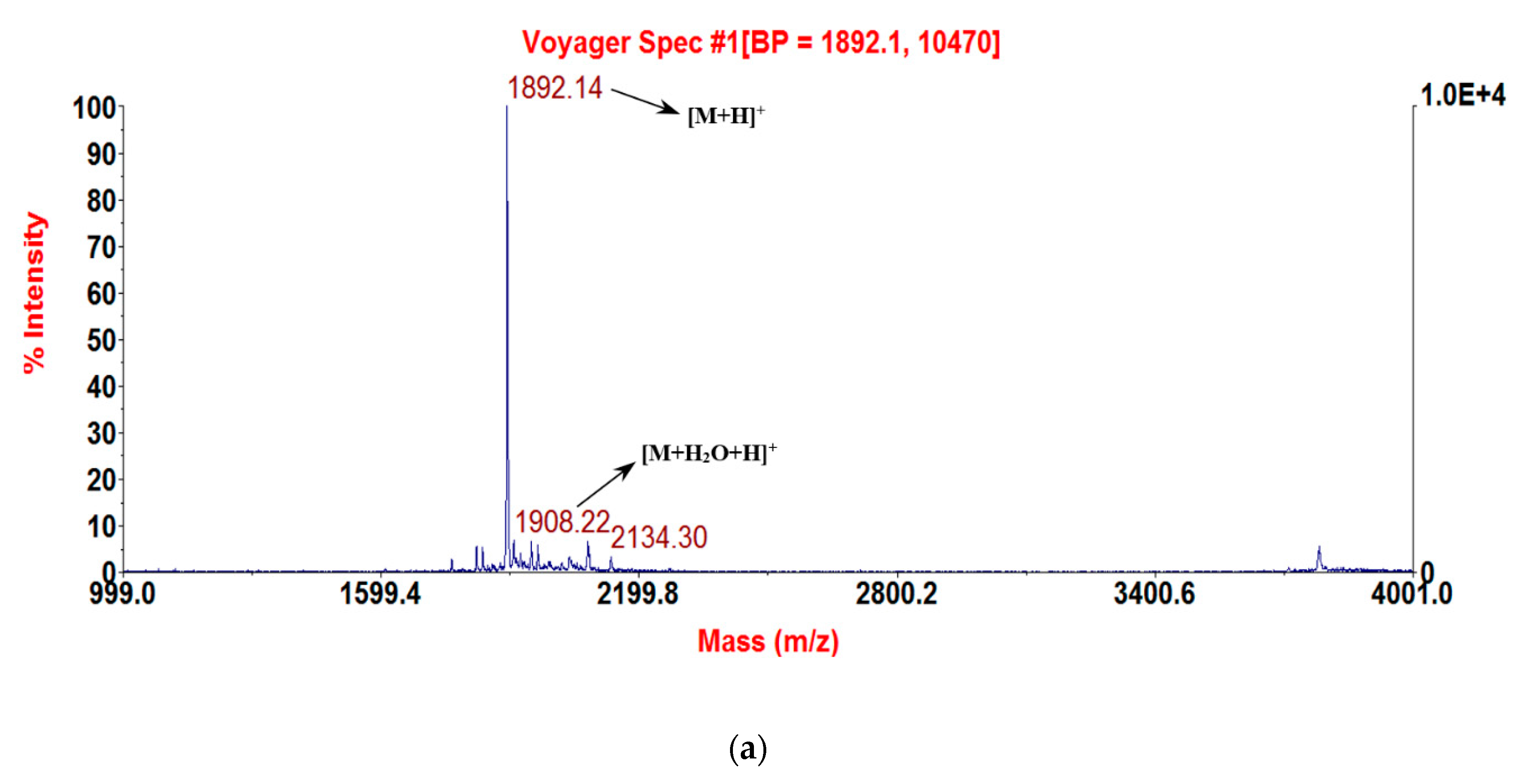
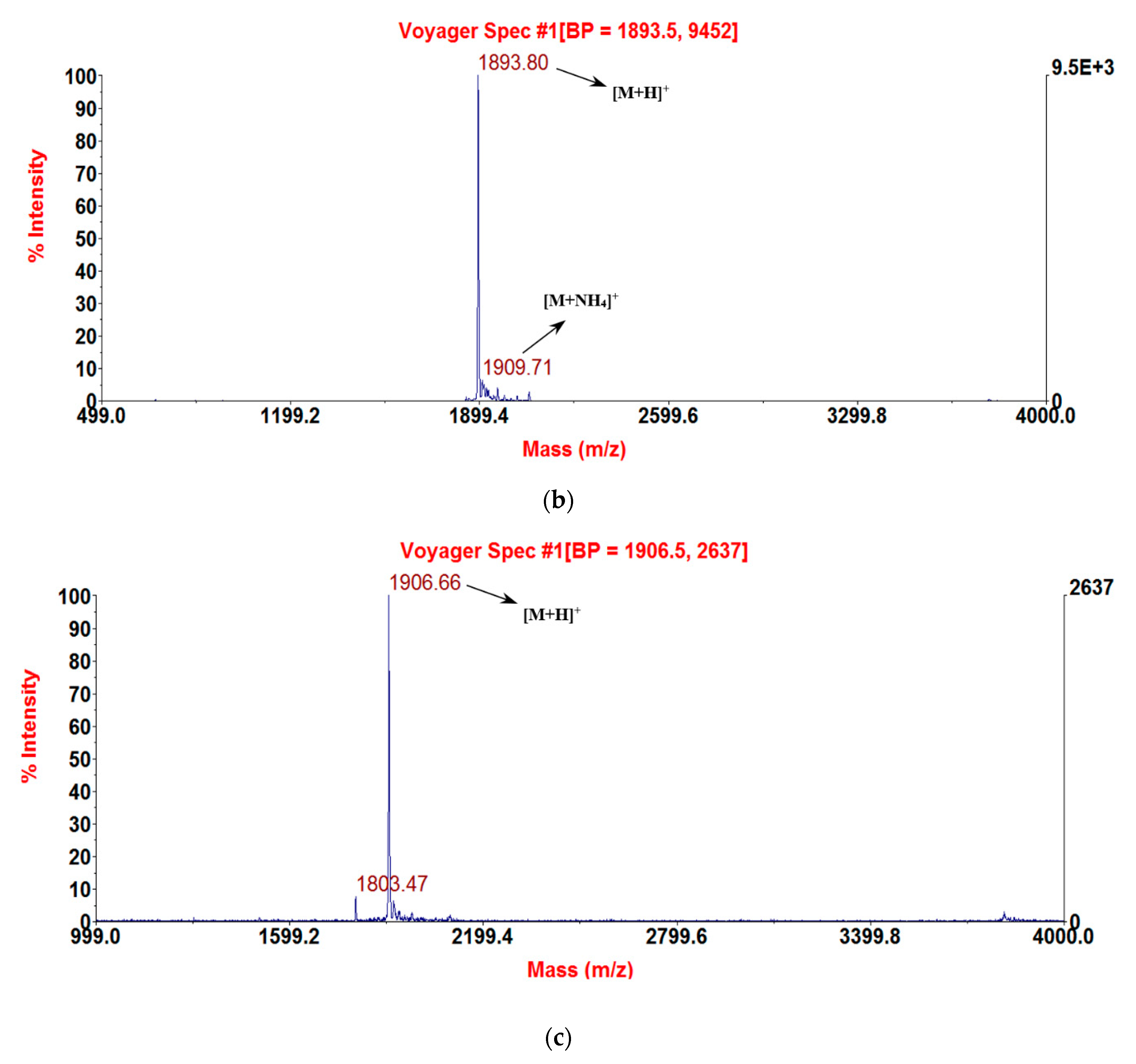
2.3. Solid-Phase Peptide Synthesis and Confirmation of Structure
2.4. Circular Dichroism (CD) Analysis
2.5. Trypsin, Chymotrypsin and Tryptase Inhibition Assays
2.6. Minimum Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC) Assays
2.7. Checkerboard Assays
2.8. Haemolysis and Cytotoxicity Assays
2.9. Statistical Analysis
3. Results
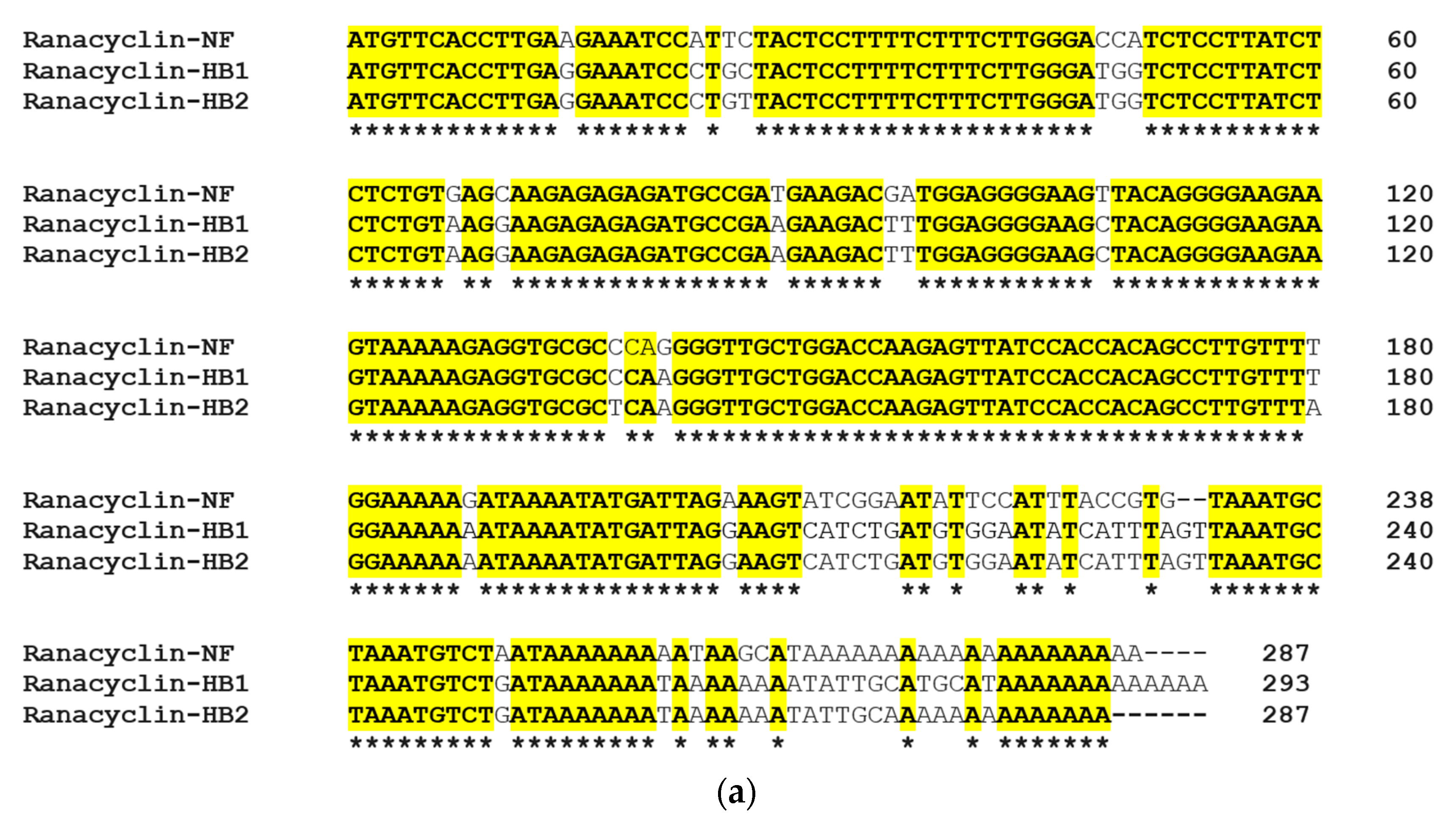
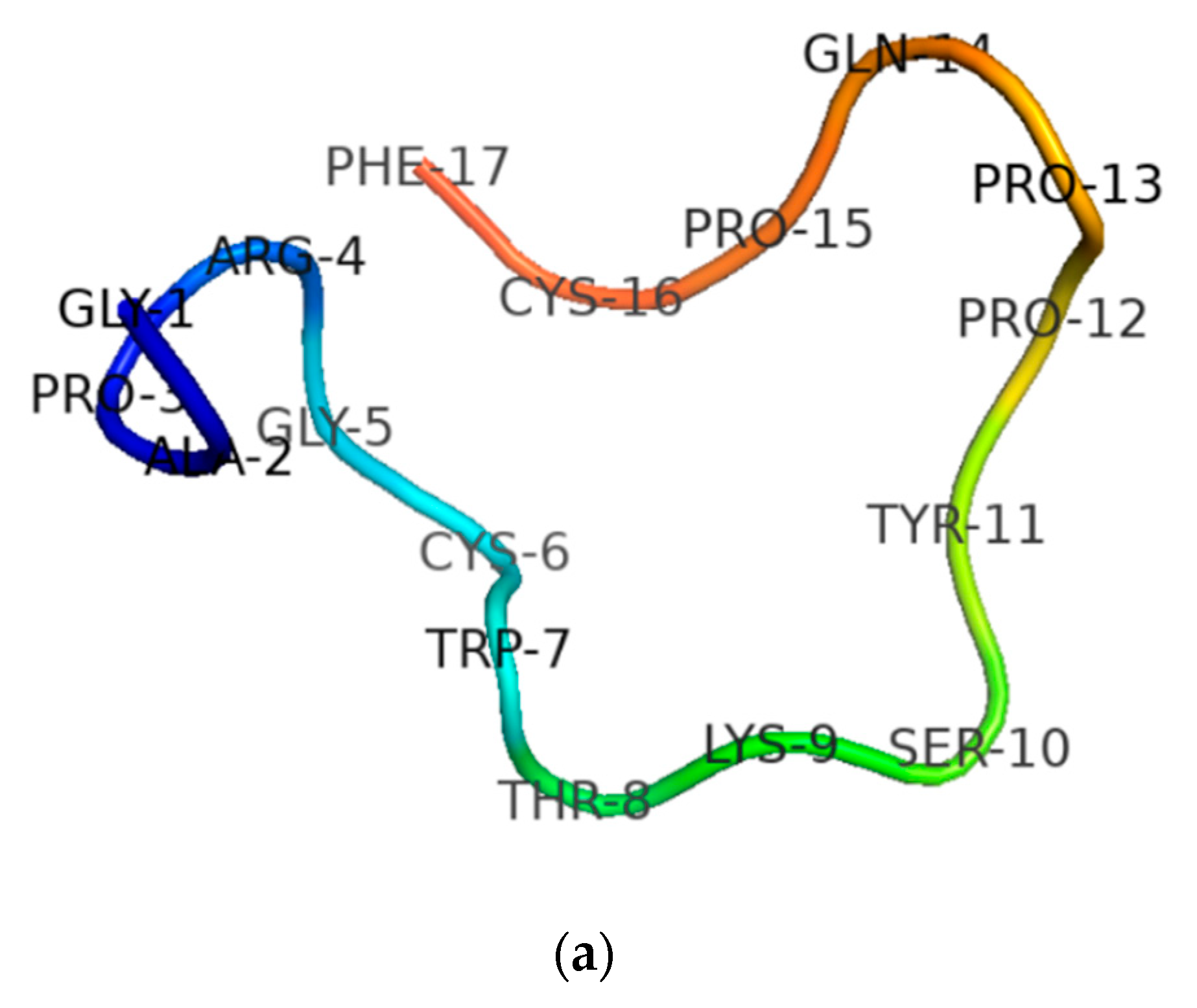
3.1. Molecular Cloning of the RNF Precursor-Encoding cDNA and Sequence Analysis
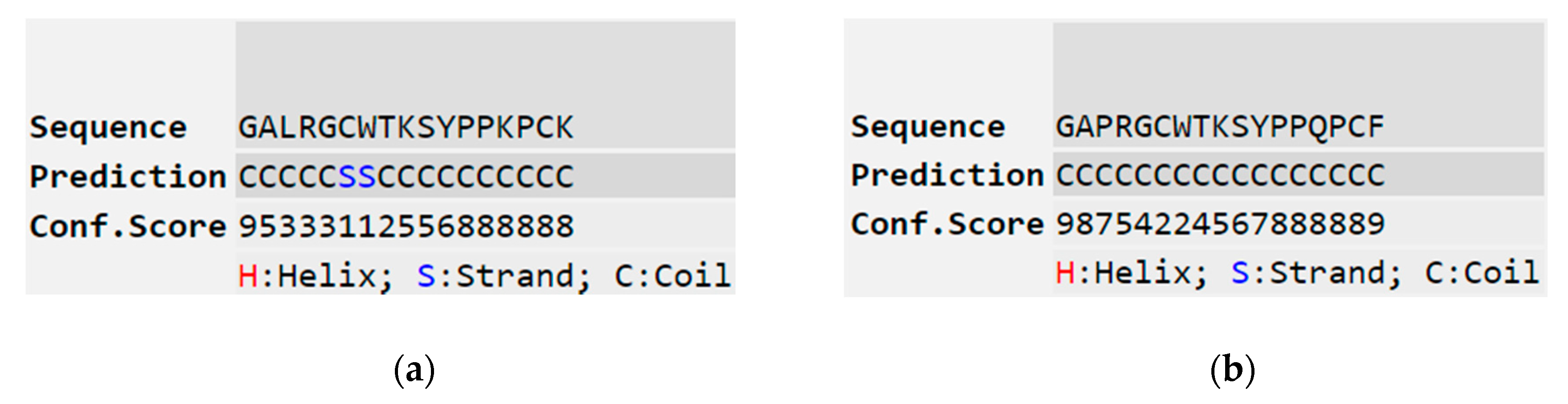
3.2. Secondary Structure Determinations
3.3. Protease Inhibitory Activity Assays
3.4. Antimicrobial Activity
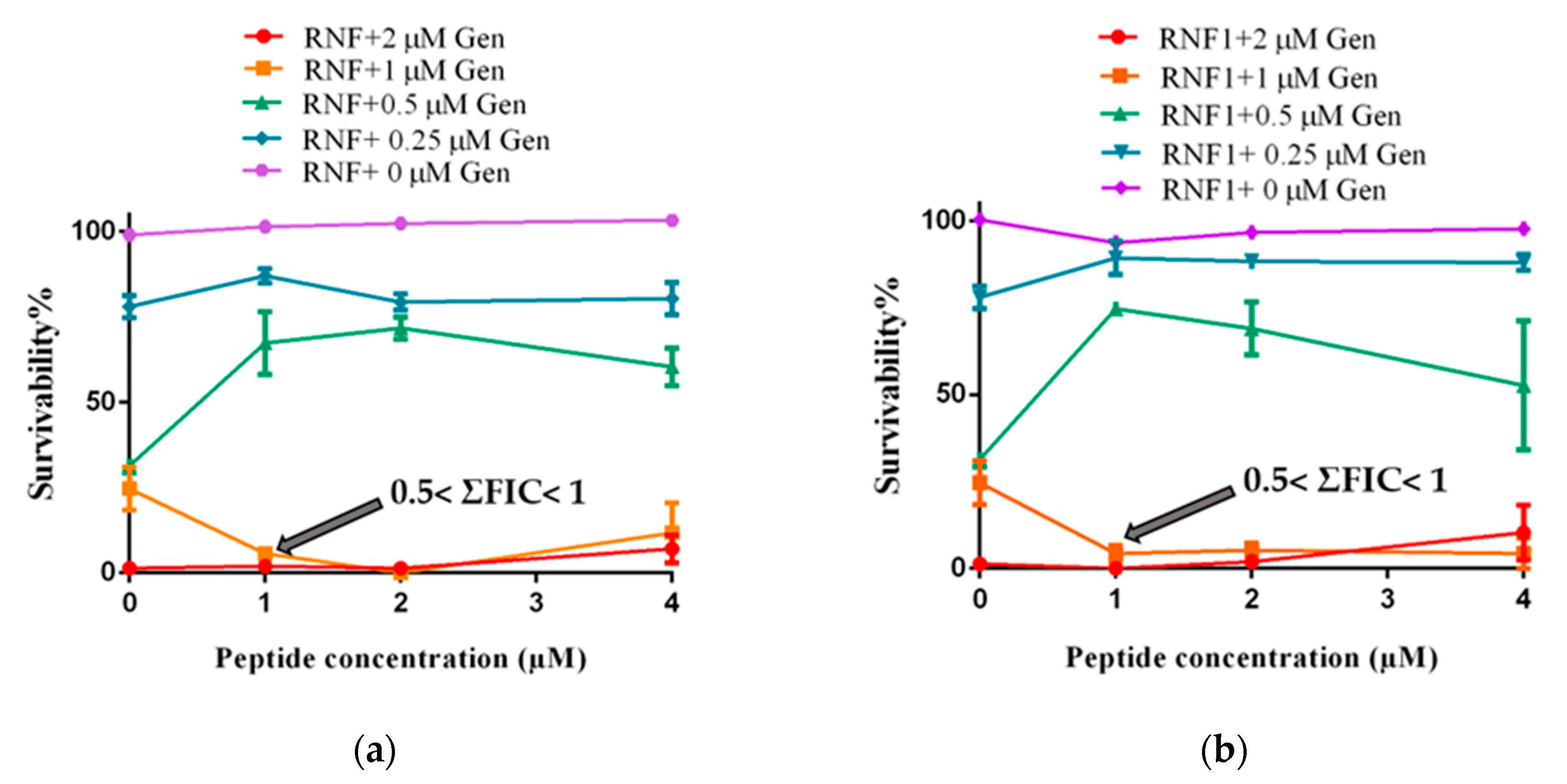
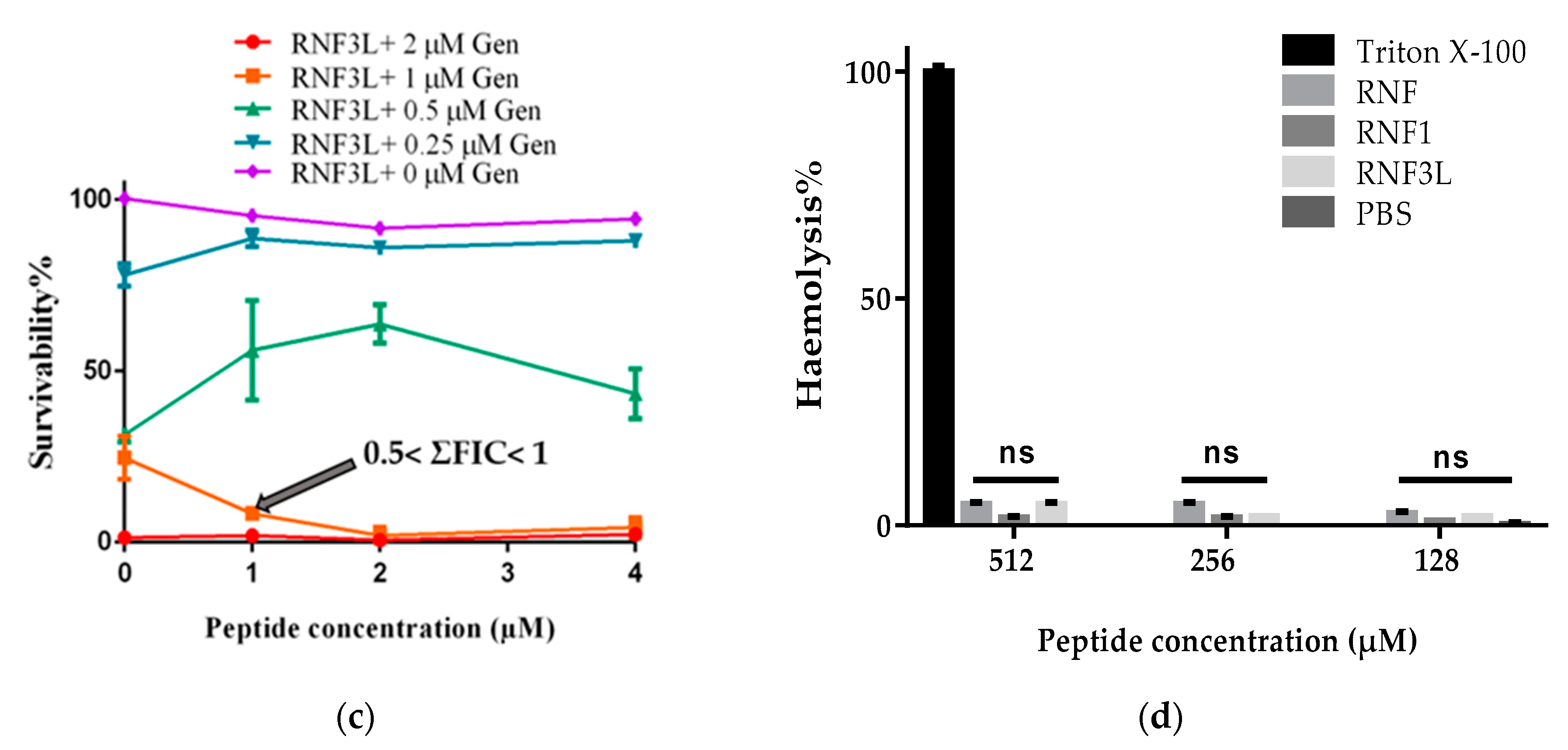
3.5. Additive Antimicrobial Effects with Gentamicin
3.6. Haemolysis and Cytotoxicity
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hedstrom, L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef] [PubMed]
- Bachovchin, D.; Cravatt, B. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discov. 2012, 11, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Drag, M.; Salvesen, G. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010, 9, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; de Veer, S.; White, A.; Chen, X.; Harris, J.; Swedberg, J.; Craik, D. Amino Acid Scanning at P5′ within the Bowman–Birk Inhibitory Loop Reveals Specificity Trends for Diverse Serine Proteases. J. Med. Chem. 2019, 62, 3696–3706. [Google Scholar] [CrossRef]
- Qi, R.; Song, Z.; Chi, C. Structural Features and Molecular Evolution of Bowman-Birk Protease Inhibitors and Their Potential Application. Acta Biochim. Biophys. Sin. 2005, 37, 283–292. [Google Scholar] [CrossRef]
- BIRK, Y. The Bowman-Birk inhibitor. Trypsin- and chymotrypsin-inhibitor from soybeans. Int. J. Pept. Protein Res. 1985, 25, 113–131. [Google Scholar] [CrossRef]
- McBride, J.; Leatherbrrow, R. Synthetic Peptide Mimics of the Bowman-Birk Inhibitor Protein. Curr. Med. Chem. 2001, 8, 909–917. [Google Scholar] [CrossRef]
- Lin, Y.; Hang, H.; Chen, T.; Zhou, M.; Wang, L.; Shaw, C. pLR-HL: A Novel Amphibian Bowman-Birk-type Trypsin Inhibitor from the Skin Secretion of the Broad-folded Frog, Hylarana latouchii. Chem. Biol. Drug Des. 2016, 87, 91–100. [Google Scholar] [CrossRef]
- Li, J.; Xu, X.; Xu, C.; Zhou, W.; Zhang, K.; Yu, H.; Zhang, Y.; Zheng, Y.; Rees, H.; Lai, R.; et al. Anti-infection Peptidomics of Amphibian Skin. Mol. Cell Proteom. 2007, 6, 882–894. [Google Scholar] [CrossRef]
- Rothemund, S.; Sönnichsen, F.; Polte, T. Therapeutic Potential of the Peptide Leucine Arginine as a New Nonplant Bowman–Birk-Like Serine Protease Inhibitor. J. Med. Chem. 2013, 56, 6732–6744. [Google Scholar] [CrossRef]
- McBride, J.; Watson, E.; Brauer, A.; Jaulent, A.; Leatherbarrow, R. Peptide mimics of the Bowman-Birk inhibitor reactive site loop. Biopolymers 2002, 66, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Safavi, F.; Rostami, A. Role of serine proteases in inflammation: Bowman–Birk protease inhibitor (BBI) as a potential therapy for autoimmune diseases. Exp. Mol. Pathol. 2012, 93, 428–433. [Google Scholar] [CrossRef]
- Mangoni, M.; Papo, N.; Mignogna, G.; Andreu, D.; Shai, Y.; Barra, D.; Simmaco, M. Ranacyclins, a New Family of Short Cyclic Antimicrobial Peptides: Biological Function, Mode of Action, and Parameters Involved in Target Specificity. Biochemistry 2003, 42, 14023–14035. [Google Scholar] [CrossRef]
- Xu, X.; Lai, R. The chemistry and biological activities of peptides from amphibian skin secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.; Irvine, A.E.; McClean, S.; Richter, S.C.; Flatt, P.R.; Shaw, C. Peptide Tyrosine Arginine, a potent immunomodulatory peptide isolated and structurally characterized from the skin secretions of the dusky gopher frog, Rana sevosa. Peptides 2005, 26, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.L.; Cross, L.J.; Irvine, A.E.; Lappin, T.R.; Dathe, M.; Krause, G.; Canning, P.; Thim, L.; Beyermann, M.; Rothemund, S.; et al. Peptide leucine arginine, a potent immunomodulatory peptide isolated and structurally characterized from the skin of the Northern Leopard frog, Rana pipiens. J. Biol. Chem. 2001, 276, 10145–10152. [Google Scholar] [CrossRef]
- Hao, X.; Tang, X.; Luo, L.; Wang, Y.; Lai, R.; Lu, Q. A novel ranacyclin-like peptide with anti-platelet activity identified from skin secretions of the frog Amolops loloensis. Gene 2016, 576, 171–175. [Google Scholar] [CrossRef]
- Wang, X.; Ren, S.; Guo, C.; Zhang, W.; Zhang, X.; Zhang, B.; Li, S.; Ren, J.; Hu, Y.; Wang, H. Identification and functional analyses of novel antioxidant peptides and antimicrobial peptides from skin secretions of four East Asian frog species. Acta Biochim. Biophys. Sin. 2017, 49, 550–559. [Google Scholar] [CrossRef]
- Yang, H.; Wang, X.; Liu, X.; Wu, J.; Liu, C.; Gong, W.; Zhao, Z.; Hong, J.; Lin, D.; Wang, Y.; et al. Antioxidant peptidomics reveals novel skin antioxidant system. Mol. Cell Proteom. 2009, 8, 571–583. [Google Scholar] [CrossRef]
- Demori, I.; Rashed, Z.E.; Corradino, V.; Catalano, A.; Rovegno, L.; Queirolo, L.; Salvidio, S.; Biggi, E.; Zanotti-Russo, M.; Canesi, L.; et al. Peptides for Skin Protection and Healing in Amphibians. Molecules 2019, 24, 347. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Xu, X.; Wang, J.; Yu, H.; Lai, R.; Gong, W. Trypsin inhibitory loop is an excellent lead structure to design serine protease inhibitors and antimicrobial peptides. FASEB J. 2007, 21, 2466–2473. [Google Scholar] [CrossRef]
- Yuan, Y.; Zai, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Shaw, C.; Chen, T. A novel membrane-disruptive antimicrobial peptide from frog skin secretion against cystic fibrosis isolates and evaluation of anti-MRSA effect using Galleria mellonella model. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Louis-Jeune, C.; Andrade-Navarro, M.A.; Perez-Iratxeta, C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 2012, 80, 374–381. [Google Scholar] [CrossRef]
- Wu, Y.; Long, Q.; Xu, Y.; Guo, S.; Chen, T.; Wang, L.; Zhou, M.; Zhang, Y.; Shaw, C.; Walker, B. A structural and functional analogue of a Bowman–Birk-type protease inhibitor from Odorrana schmackeri. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xi, X.; Ma, C.; Chen, X.; Zhou, M.; Burrows, J.F.; Chen, T.; Wang, L. A Novel Dermaseptin Isolated from the Skin Secretion of Phyllomedusa tarsius and Its Cationicity-Enhanced Analogue Exhibiting Effective Antimicrobial and Anti-Proliferative Activities. Biomolecules 2019, 9, 628. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Zhou, M.; Wu, Y.; Chen, T.; Shaw, C.; Wang, L. The synergistic antimicrobial effects of novel bombinin and bombinin H peptides from the skin secretion of Bombina orientalis. Biosci. Rep. 2017, 37, BSR20170967. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]
- Hall, M.J.; Middleton, R.F.; Westmacott, D. The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 1983, 11, 427–433. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Y.; Wang, T.; Chen, X.; Zhou, M.; Ma, C.; Xi, X.; Zhang, Y.; Chen, T.; Shaw, C.; et al. Brevinin-1GHd: A novel Hylarana guentheri skin secretion-derived Brevinin-1 type peptide with antimicrobial and anticancer therapeutic potential. Biosci. Rep. 2020, 40, BSR20200019. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Chen, T.; Walker, B.; Zhou, M.; Sui, D.; Conlon, J.; Shaw, C. Identification and molecular cloning of a novel amphibian Bowman Birk-type trypsin inhibitor from the skin of the Hejiang Odorous Frog; Odorrana hejiangensis. Peptides 2012, 33, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Chen, G.; Xi, X.; Ma, C.; Wang, L.; Burrows, J.; Duan, J.; Zhou, M.; Chen, T. Discovery and Rational Design of a Novel Bowman-Birk Related Protease Inhibitor. Biomolecules 2019, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.; Rubenstein, E. Proline: The Distribution, Frequency, Positioning, and Common Functional Roles of Proline and Polyproline Sequences in the Human Proteome. PLoS ONE 2013, 8, e53785. [Google Scholar] [CrossRef] [PubMed]
- Groß, A.; Hashimoto, C.; Sticht, H.; Eichler, J. Synthetic Peptides as Protein Mimics. Front. Bioeng. Biotechnol. 2016, 3, 211. [Google Scholar] [CrossRef] [PubMed]
- Cristina Oliveira de Lima, V.; Piuvezam, G.; Leal Lima Maciel, B.; Heloneida de Araújo Morais, A. Trypsin inhibitors: Promising candidate satietogenic proteins as complementary treatment for obesity and metabolic disorders? J. Enzym. Inhib. Med. Chem. 2019, 34, 405–419. [Google Scholar] [CrossRef]
- Ware, J.H.; Wan, X.S.; Rubin, H.; Schechter, N.M.; Kennedy, A.R. Soybean Bowman-Birk protease inhibitor is a highly effective inhibitor of human mast cell chymase. Arch. Biochem. Biophys. 1997, 344, 133–138. [Google Scholar] [CrossRef]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from the skins of North American frogs. Biochim. Biophys. Acta 2009, 1788, 1556–1563. [Google Scholar] [CrossRef]
- Cancelarich, N.L.; Wilke, N.; Fanani, M.A.L.; Moreira, D.C.; Pérez, L.O.; Alves Barbosa, E.; Plácido, A.; Socodato, R.; Portugal, C.C.; Relvas, J.B.; et al. Somuncurins: Bioactive Peptides from the Skin of the Endangered Endemic Patagonian Frog Pleurodema somuncurense. J. Nat. Prod. 2020, 83, 972–984. [Google Scholar] [CrossRef]
- Smith, A.; Smith, D. Gentamicin: Adenine Mononucleotide Transferase: Partial Purification, Characterization, and Use in the Clinical Quantitation of Gentamicin. J. Infect. Dis. 1974, 129, 391–401. [Google Scholar] [CrossRef]
- Hariprasad, G.; Kumar, M.; Rani, K.; Kaur, P.; Srinivasan, A. Aminoglycoside induced nephrotoxicity: Molecular modeling studies of calreticulin-gentamicin complex. J. Mol. Model. 2012, 18, 2645–2652. [Google Scholar] [CrossRef]
- Smetana, S.; Khalef, S.; Bar-Khayim, Y.; Birk, Y. Antimicrobial Gentamicin Activity in the Presence of Exogenous Protease Inhibitor (Bowman-Birk Inhibitor) in Gentamicin-lnduced Nephrotoxicity in Rats. Nephron 1992, 61, 68–72. [Google Scholar] [CrossRef] [PubMed]




| Peptide | Peptide Sequence | Ki(µM) of Trypsin | Ki(µM) of Chymotrypsin | Ki(µM) of Tryptase |
|---|---|---|---|---|
| RNF | GAPRGCWTKSYPPQPCF-NH2 | 0.447 | N.I. | 6.774 |
| RNF1 | GAPRGCWTKSYPPQPCF | 1.300 | N.I. | 9.059 |
| RNF3L | GALRGCWTKSYPPQPCF-NH2 | 0.201 | N.I. | 12.5 |
| Ranacyclin-T [10] | GALRGCWTKSYPPKPCK | 0.116 | - | - |
| HJTI [31] | GAPKGCWTKSYPPQPCS-NH2 | 0.388 | - | - |
| OSTI [24] | AALKGCWTKSIPPKPCF-NH2 | 0.0003 | N.I. | 2.5 |
| PPF-BBI [32] | ALRGCWTKSIPPKPCP-NH2 | 0.17 | N.I. | 30.73 |
| MICs/MBCs (µM) | |||||||
|---|---|---|---|---|---|---|---|
| Peptide | S. aureus | MRSA | E. faecalis | E. coli | P. aeruginosa | K. pneumoniae | C. albicans |
| RNF | 512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 |
| RNF1 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 |
| RNF3L | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 | >512/>512 |
| Peptide | Peptide Sequence | Net Charge | Trypsin Inhibition Ki (nM) |
|---|---|---|---|
| RNF | GAPRGCWTKSYPPQPCF-NH2 | 3 | 447 |
| Ranacyclin-T [10] | GALRGCWTKSYPPKPCK-NH2 | 5 | 116 |
| HJTI [31] | GAPKGCWTKSYPPQPCS-NH2 | 3 | 388 |
| OSTI [24] | AALKGCWTKSIPPKPCF-NH2 | 4 | 0.3 |
| PPF-BBI [32] | ALRGCWTKSIPPKPCP-NH2 | 4 | 170 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Jiang, Y.; Chen, X.; Wang, L.; Ma, C.; Xi, X.; Zhang, Y.; Chen, T.; Shaw, C.; Zhou, M. Ranacyclin-NF, a Novel Bowman–Birk Type Protease Inhibitor from the Skin Secretion of the East Asian Frog, Pelophylax nigromaculatus. Biology 2020, 9, 149. https://doi.org/10.3390/biology9070149
Wang T, Jiang Y, Chen X, Wang L, Ma C, Xi X, Zhang Y, Chen T, Shaw C, Zhou M. Ranacyclin-NF, a Novel Bowman–Birk Type Protease Inhibitor from the Skin Secretion of the East Asian Frog, Pelophylax nigromaculatus. Biology. 2020; 9(7):149. https://doi.org/10.3390/biology9070149
Chicago/Turabian StyleWang, Tao, Yangyang Jiang, Xiaoling Chen, Lei Wang, Chengbang Ma, Xinping Xi, Yingqi Zhang, Tianbao Chen, Chris Shaw, and Mei Zhou. 2020. "Ranacyclin-NF, a Novel Bowman–Birk Type Protease Inhibitor from the Skin Secretion of the East Asian Frog, Pelophylax nigromaculatus" Biology 9, no. 7: 149. https://doi.org/10.3390/biology9070149
APA StyleWang, T., Jiang, Y., Chen, X., Wang, L., Ma, C., Xi, X., Zhang, Y., Chen, T., Shaw, C., & Zhou, M. (2020). Ranacyclin-NF, a Novel Bowman–Birk Type Protease Inhibitor from the Skin Secretion of the East Asian Frog, Pelophylax nigromaculatus. Biology, 9(7), 149. https://doi.org/10.3390/biology9070149