Multipath: An R Package to Generate Integrated Reproducible Pathway Models
Abstract
Simple Summary
Abstract
1. Introduction
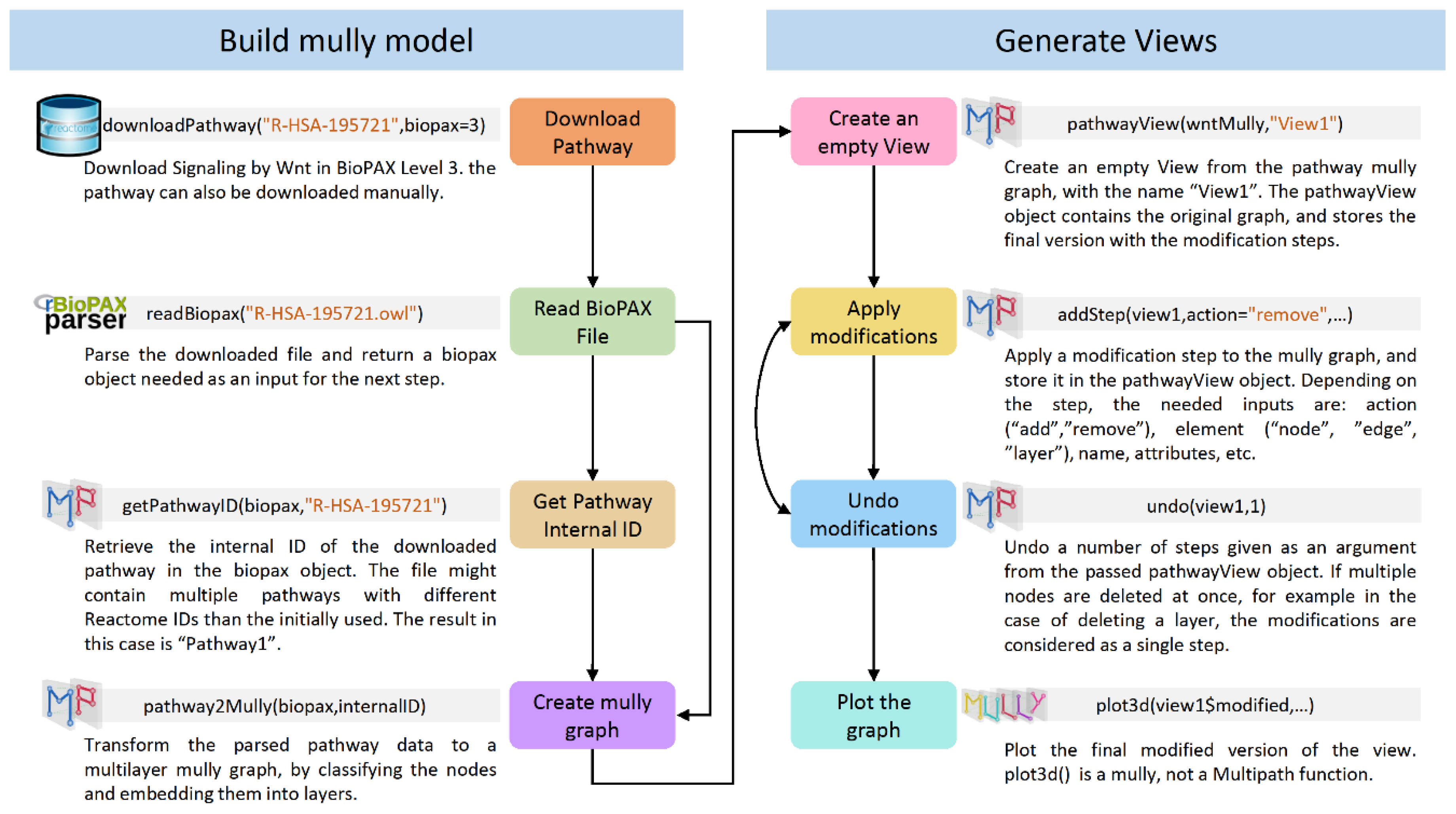
2. Materials and Methods
- Read the BioPAX file using the rBiopaxparser function readBiopax() and provide the path to the BioPAX file containing the pathway information as an argument. The file can be downloaded manually or using the function downloadPathway().
- Fetch the pathway’s internal ID using the function getPathwayID() from the BioPAX object returned in step 1 which might contain multiple pathways.
- Provide the pathway’s ID alongside the BioPAX object as arguments to the Multipath function pathway2Mully() which returns the mully graph created from the parsed pathway information in the BioPAX object.
3. Results
3.1. Pathway Data Integration
3.1.1. DrugBank
3.1.2. UniProt
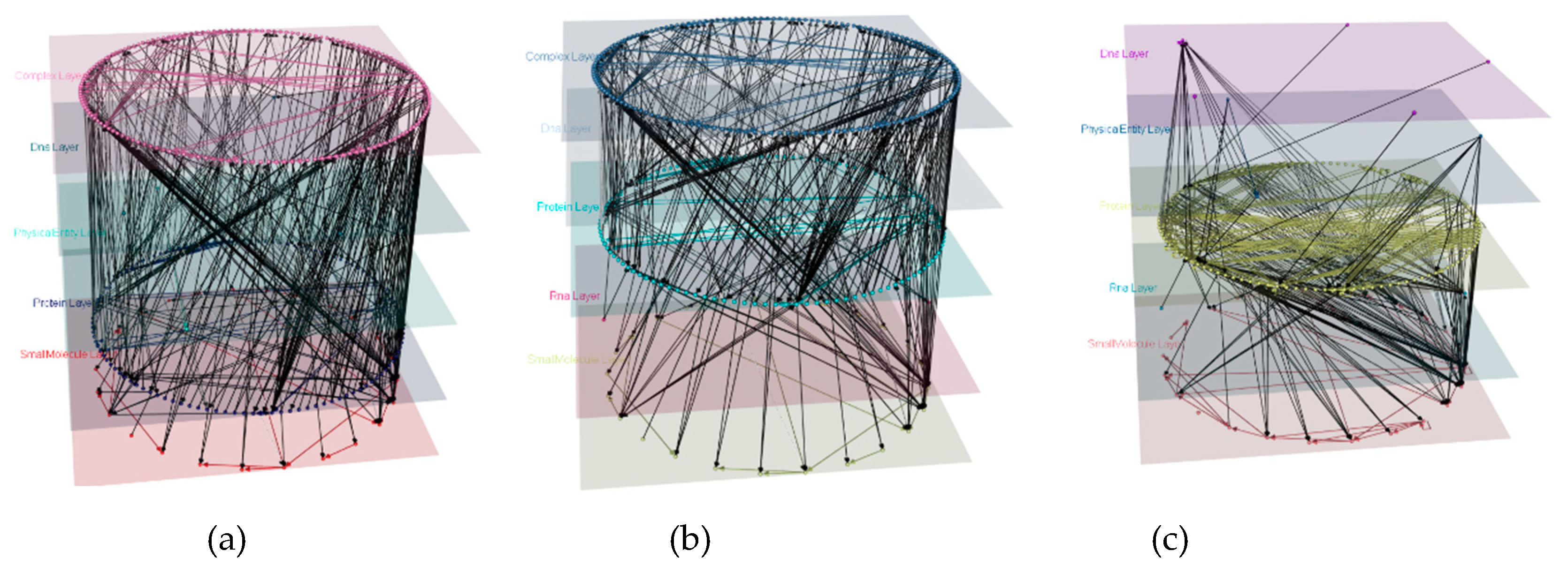
3.2. Pathway Models’ Reproducibility
4. Discussion
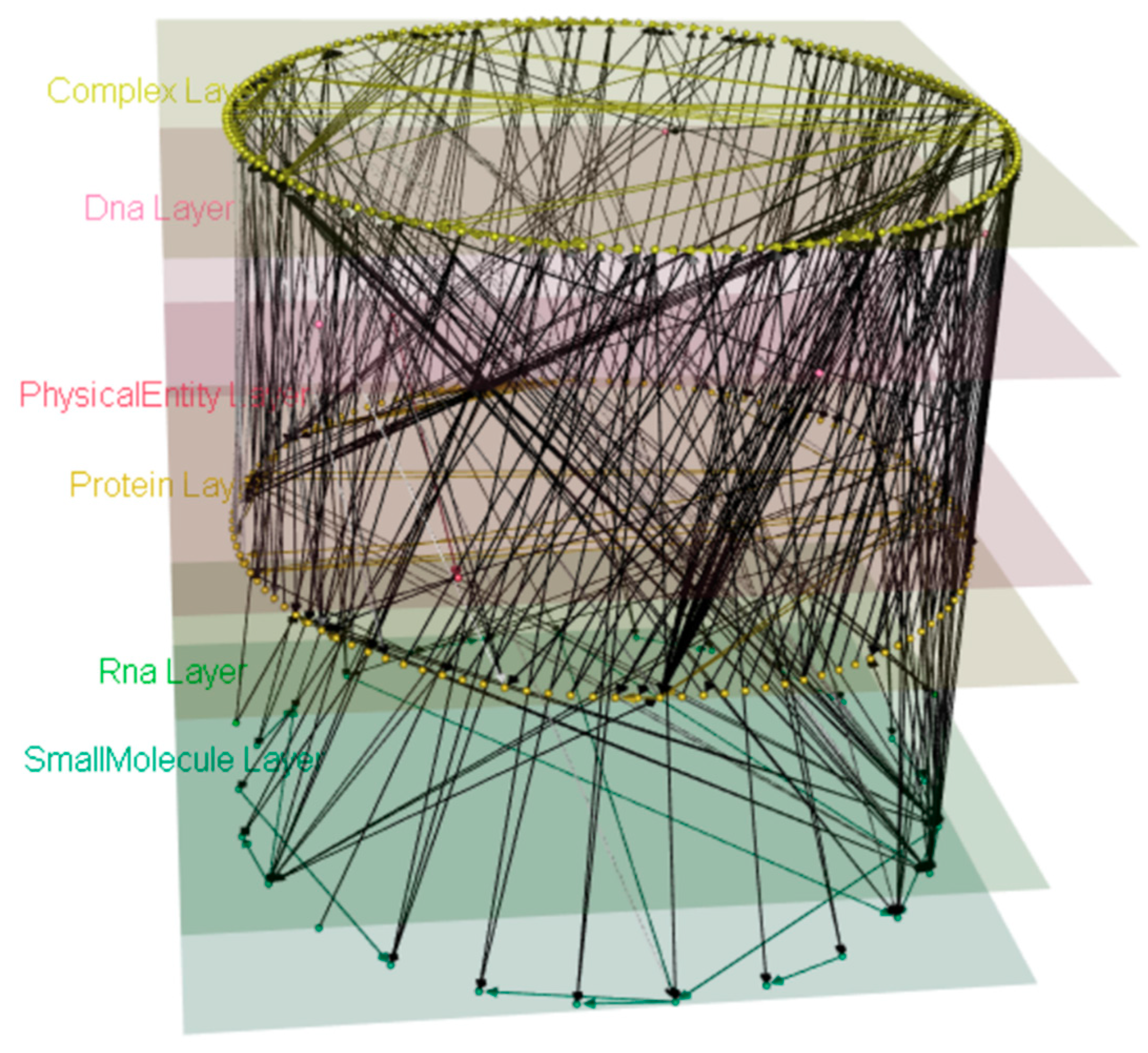
4.1. Wnt Multipath Model
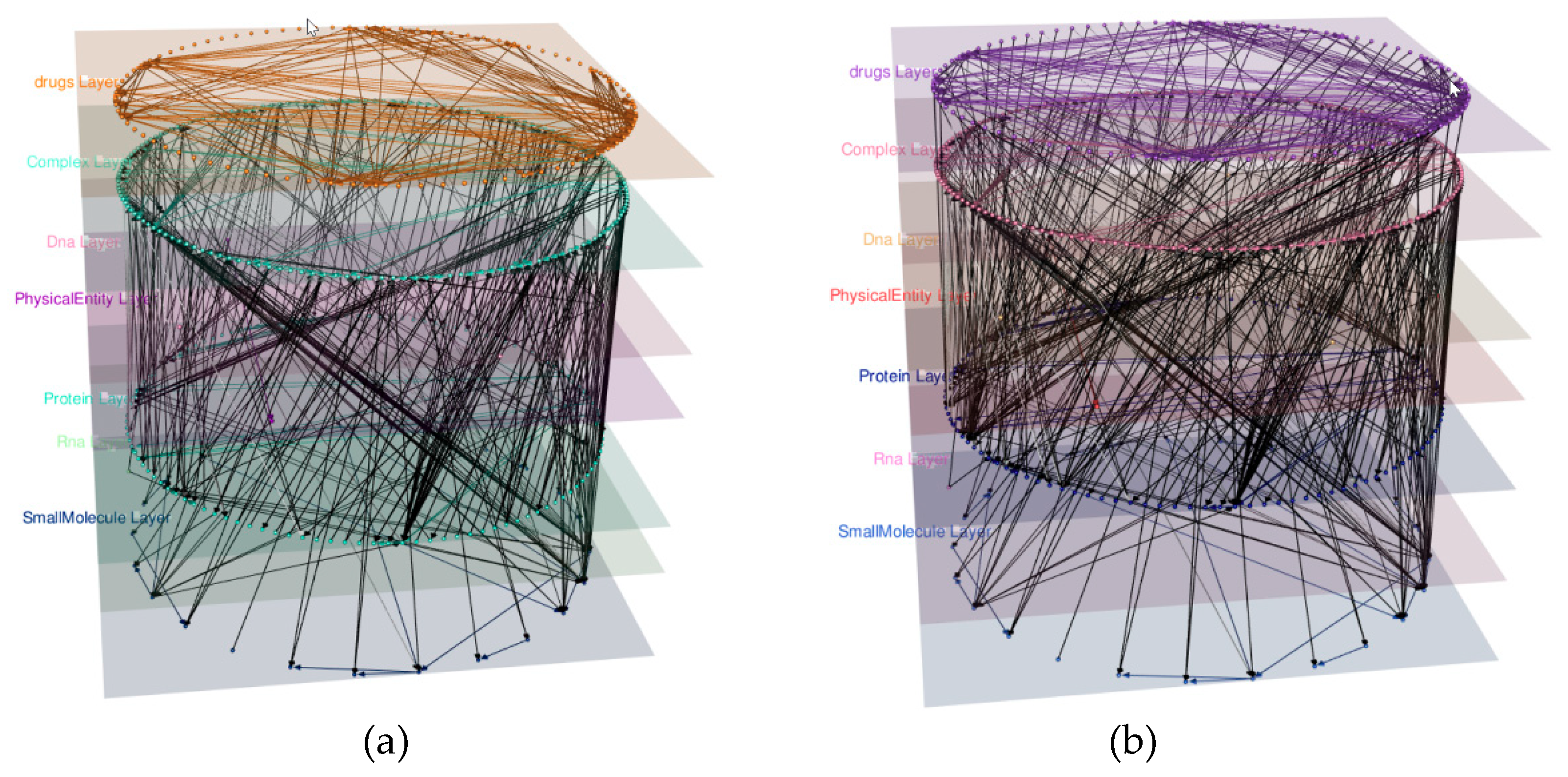
4.2. Adding a Drug Layer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Goldberg, R.N.; Cary, M.; Demir, E. BioPAX—A community standard for pathway data sharing. Nat. Biotechnol. 2010, 28, 935–942. [Google Scholar]
- Kramer, F.; Bayerlová, M.; Klemm, F.; Bleckmann, A.; Beissbarth, T. rBiopaxParser--an R package to parse, modify and visualize BioPAX data. Bioinformatics 2013, 29, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, Z.; Kramer, F. mully: An R Package to Create, Modify and Visualize Multilayered Graphs. Genes (Basel) 2018, 9, 519. [Google Scholar] [CrossRef] [PubMed]
- The Dbparser R Package. Available online: https://github.com/ropensci/dbparser (accessed on 29 September 2020).
- The UniProt.ws R Interface. Available online: https://bioconductor.org/packages/UniProt.ws/ (accessed on 29 September 2020).
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [CrossRef] [PubMed]
- Croft, D.; Mundo, A.F.; Haw, R.; Milacic, M.; Weiser, J.; Wu, G.; Caudy, M.; Garapati, P.; Gillespie, M.; Kamdar, M.R.; et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014, 42, D472–D477. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Cary, M.P.; Sander, C. Pathguide: A Pathway Resource List. Nucleic Acids Res. 2006, 34 (Suppl. 1), D504–D506. [Google Scholar] [CrossRef] [PubMed]
- Degtyarenko, K.; De Matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; McNaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008, 36, D344–D350. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Szöcs, E.; Stirling, T.; Scott, E.R.; Scharmüller, A.; Schäfer, R.B. webchem: An R Package to Retrieve Chemical Information from the Web. J. Stat. Softw. 2020, 93, 1–17. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The Ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef] [PubMed]
- The DrugBank Website. Available online: drugbank.ca (accessed on 29 September 2020).
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Signaling by Wnt. Available online: https://reactome.org/content/detail/R-HSA-195721 (accessed on 29 September 2020).
- The Multipath R Package. Available online: https://github.com/frankkramer-lab/Multipath (accessed on 29 September 2020).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammoud, Z.; Kramer, F. Multipath: An R Package to Generate Integrated Reproducible Pathway Models. Biology 2020, 9, 483. https://doi.org/10.3390/biology9120483
Hammoud Z, Kramer F. Multipath: An R Package to Generate Integrated Reproducible Pathway Models. Biology. 2020; 9(12):483. https://doi.org/10.3390/biology9120483
Chicago/Turabian StyleHammoud, Zaynab, and Frank Kramer. 2020. "Multipath: An R Package to Generate Integrated Reproducible Pathway Models" Biology 9, no. 12: 483. https://doi.org/10.3390/biology9120483
APA StyleHammoud, Z., & Kramer, F. (2020). Multipath: An R Package to Generate Integrated Reproducible Pathway Models. Biology, 9(12), 483. https://doi.org/10.3390/biology9120483





