Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury
Abstract
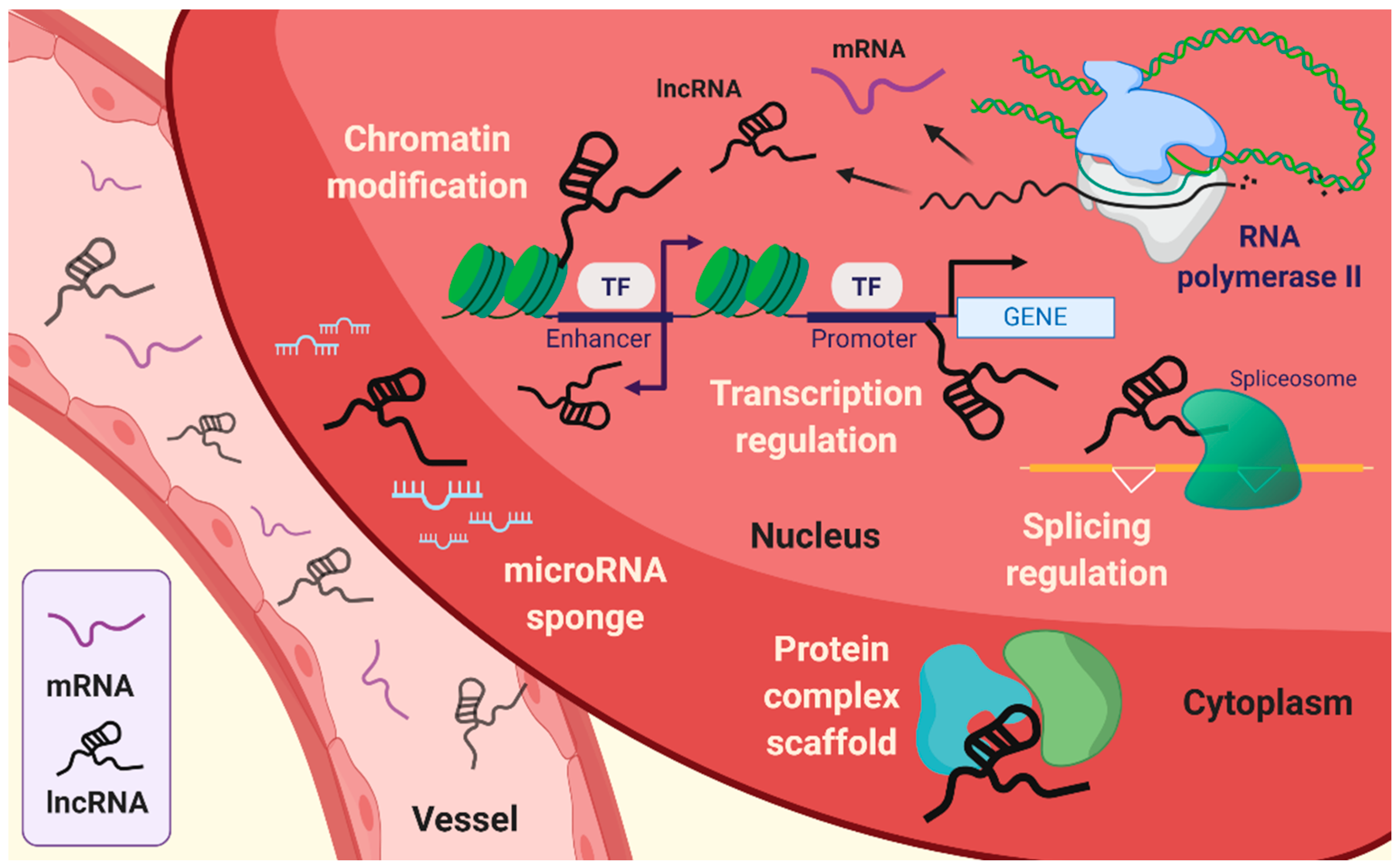
Simple Summary
Abstract
1. Introduction
2. Stroke
2.1. LncRNAs in TBI
2.2. Molecular Features of LncRNAs in Stroke
3. Hypoxia
3.1. Influence of LncRNAs in Neuronal Cell Fate
3.2. Possibilities of LncRNA for Targeted Therapy for TBI
4. Further Research Directions
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Yousefi, H.; Maheronnaghsh, M.; Molaei, F.; Mashouri, L.; Reza Aref, A.; Momeny, M.; Alahari, S.K. Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 2020, 39, 953–974. [Google Scholar] [CrossRef] [PubMed]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Schaukowitch, K.; Joo, J.Y.; Liu, X.; Watts, J.K.; Martinez, C.; Kim, T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 2014, 56, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.Y.; Schaukowitch, K.; Farbiak, L.; Kilaru, G.; Kim, T.K. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat. Neurosci. 2016, 19, 75–83. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Chillón, I.; Marcia, M. The molecular structure of long non-coding RNAs: Emerging patterns and functional implications. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 662–690. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, 6320. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef] [PubMed]
- Aillaud, M.; Schulte, L.N. Emerging Roles of Long Noncoding RNAs in the Cytoplasmic Milieu. NcRNA 2020, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chi, Y.J.; Lin, G.Q.; Xiao, L.F.; Su, G.L.; Yang, L.M. LncRNA MEG3 participates in neuronal cell injury induced by subarachnoid hemorrhage via inhibiting the Pi3k/Akt pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2824–2831. [Google Scholar] [PubMed]
- Zhou, X.B.; Lai, L.F.; Xie, G.B.; Ding, C.; Xu, X.; Wang, Y. LncRNAGAS5 sponges miRNA-221 to promote neurons apoptosis by up-regulated PUMA under hypoxia condition. Neurol. Res. 2020, 42, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Qiu, Y.; Lin, Y.; Medina, R.; Zhuang, S.; Rosenblum, J.S.; Cui, J.; Li, Z.; Zhang, X.; Guo, L. Blocking lncRNA H19-miR-19a-Id2 axis attenuates hypoxia/ischemia induced neuronal injury. Aging 2019, 11, 3585–3600. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. Cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 2012, 19, 1068–1075. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Tsai, P.F.; Dell’Orso, S.; Rodriguez, J.; Vivanco, K.O.; Ko, K.D.; Jiang, K.; Juan, A.H.; Sarshad, A.A.; Vian, L.; Tran, M.; et al. A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol. Cell 2018, 71, 129–141. [Google Scholar] [CrossRef]
- Dolezel, S.; Filkuka, J.; Tomasek, V.; Vlasin, Z. Histochemical demonstration of 5-hydroxytryptamin in a malignant carcinoid of the small intestine. Neoplasma 1969, 16, 209–214. [Google Scholar]
- Sigova, A.A.; Abraham, B.J.; Ji, X.; Molinie, B.; Hannett, N.M.; Guo, Y.E.; Jangi, M.; Giallourakis, C.C.; Sharp, P.A.; Young, R.A. Transcription factor trapping by RNA in gene regulatory elements. Science 2015, 350, 978–981. [Google Scholar] [CrossRef]
- Schier, A.C.; Taatjes, D.J. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020, 34, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Mollayeva, T.; Mollayeva, S.; Colantonio, A. Traumatic brain injury: Sex, gender and intersecting vulnerabilities. Nat. Rev. Neurol. 2018, 14, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, F.A.; Thelin, E.P.; Donnelly, J.; Stevens, A.R.; Smielewski, P.; Czosnyka, M.; Hutchinson, P.J.; Menon, D.K. Genetic drivers of cerebral blood flow dysfunction in TBI: A speculative synthesis. Nat. Rev. Neurol. 2019, 15, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Mitsis, E.M.; Riggio, S.; Kostakoglu, L.; Dickstein, D.L.; Machac, J.; Delman, B.; Goldstein, M.; Jennings, D.; D’Antonio, E.; Martin, J.; et al. Tauopathy PET and amyloid PET in the diagnosis of chronic traumatic encephalopathies: Studies of a retired NFL player and of a man with FTD and a severe head injury. Transl. Psychiatry 2014, 4, e441. [Google Scholar] [CrossRef]
- Zeiler, F.A.; McFadyen, C.; Newcombe, V.F.J.; Synnot, A.; Donoghue, E.L.; Ripatti, S.; Steyerberg, E.W.; Gruen, R.L.; McAllister, T.W.; Rosand, J.; et al. Genetic Influences on Patient-Oriented Outcomes in Traumatic Brain Injury: A Living Systematic Review of Non-Apolipoprotein E Single-Nucleotide Polymorphisms. J. Neurotrauma 2019, 35, 1–17. [Google Scholar] [CrossRef]
- Hodge, R.D.; Bakken, T.E.; Miller, J.A.; Smith, K.A.; Barkan, E.R.; Graybuck, L.T.; Close, J.L.; Long, B.; Johansen, N.; Penn, O.; et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 2019, 573, 61–68. [Google Scholar] [CrossRef]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011, 12, 641–655. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.M.; Pellequer, J.L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982–995. [Google Scholar] [CrossRef]
- Kim, D.N.; Thiel, B.C.; Mrozowich, T.; Hennelly, S.P.; Hofacker, I.L.; Patel, T.R.; Sanbonmatsu, K.Y. Zinc-finger protein CNBP alters the 3-D structure of lncRNA Braveheart in solution. Nat. Commun. 2020, 11, 148. [Google Scholar] [CrossRef]
- Campbell, B.C.V.; Khatri, P. Stroke. Lancet 2020, 396, 129–142. [Google Scholar] [CrossRef]
- Campbell, B.C.V.; De Silva, D.A.; Macleod, M.R.; Coutts, S.B.; Schwamm, L.H.; Davis, S.M.; Donnan, G.A. Ischaemic stroke. Nat. Rev. Dis. Primers 2019, 5, 70. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, M.T.; Zubair, A.C.; Meschia, J.F.; Freeman, W.D. Mesenchymal stem cells for hemorrhagic stroke: Status of preclinical and clinical research. NPJ Regen. Med. 2019, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Kang, J.H.; Lin, H.C. Patients with traumatic brain injury: Population-based study suggests increased risk of stroke. Stroke 2011, 42, 2733–2739. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, R.G.; Haarbauer-Krupa, J.K.; Bell, J.M.; Corrigan, J.D.; Hammond, F.M.; Torbey, M.T.; Hofmann, M.C.; Dams-O’Connor, K.; Miller, A.C.; Whiteneck, G.G. Acute Ischemic Stroke After Moderate to Severe Traumatic Brain Injury: Incidence and Impact on Outcome. Stroke 2017, 48, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Jiang, L.; Cheng, C.; Huang, Z.; Zhang, H.; Liu, H.; He, J.; Cao, F.; Peng, J.; Jiang, Y.; et al. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016, 1646, 589–600. [Google Scholar] [CrossRef]
- Ren, D.; Chen, W.; Cao, K.; Wang, Z.; Zheng, P. Expression Profiles of Long Non-coding RNA and Messenger RNA in Human Traumatic Brain Injury. Mol. Ther. Nucleic Acids 2020, 22, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, X.; Liu, K.; Hamblin, M.H.; Yin, K.J. Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J. Neurosci. 2017, 37, 1797–1806. [Google Scholar] [CrossRef]
- Patel, N.A.; Moss, L.D.; Lee, J.Y.; Tajiri, N.; Acosta, S.; Hudson, C.; Parag, S.; Cooper, D.R.; Borlongan, C.V.; Bickford, P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflammation 2018, 15, 204. [Google Scholar] [CrossRef]
- Shao, H.F.; Li, Z.Z.; Zheng, X.F.; Wang, H.J.; Wang, Y.G.; Ma, Z.L.; Liu, Z.B.; Han, Q.Y. Research on the correlation of changes in plasma lncRNA MEG3 with change in inflammatory factors and prognosis in patients with traumatic brain injury. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4341–4347. [Google Scholar]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An update on long intergenic noncoding RNA p21: A regulatory molecule with various significant functions in cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Cao, F.; Ran, Q.; Wang, F. Long non-coding RNA Gm4419 promotes trauma-induced astrocyte apoptosis by targeting tumor necrosis factor α. Biochem. Biophys. Res. Commun. 2017, 491, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Zhang, Y.; Chen, S.; Zhou, Y. LncRNA HOTAIR Participates in Microglia Activation and Inflammatory Factor Release by Regulating the Ubiquitination of MYD88 in Traumatic Brain Injury. J. Mol. Neurosci. 2020, 70, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dykstra-Aiello, C.; Jickling, G.C.; Ander, B.P.; Shroff, N.; Zhan, X.; Liu, D.; Hull, H.; Orantia, M.; Stamova, B.S.; Sharp, F.R. Altered Expression of Long Noncoding RNAs in Blood After Ischemic Stroke and Proximity to Putative Stroke Risk Loci. Stroke 2016, 47, 2896–2903. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.H.; Szeto, V.; Yang, B.B.; Zhu, S.Z.; Sun, H.S.; Feng, Z.P. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018, 9, 281. [Google Scholar] [CrossRef]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gabel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef]
- Feng, L.; Guo, J.; Ai, F. Circulating long noncoding RNA ANRIL downregulation correlates with increased risk, higher disease severity and elevated pro-inflammatory cytokines in patients with acute ischemic stroke. J. Clin. Lab. Anal. 2019, 33, e22629. [Google Scholar] [CrossRef]
- Wu, W.; Hu, Q.; Nie, E.; Yu, T.; Wu, Y.; Zhi, T.; Jiang, K.; Shen, F.; Wang, Y.; Zhang, J.; et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci. Rep. 2017, 7, 45029. [Google Scholar] [CrossRef]
- Wang, J.; Cao, B.; Han, D.; Sun, M.; Feng, J. Long Non-coding RNA H19 Induces Cerebral Ischemia Reperfusion Injury via Activation of Autophagy. Aging Dis. 2017, 8, 71–84. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, P.; Zuo, X.; Yu, N.; Qin, Y.; Xu, Q.; He, S.; Cen, B.; Liao, W.; Ji, A. LncRNA-N1LR Enhances Neuroprotection Against Ischemic Stroke Probably by Inhibiting p53 Phosphorylation. Mol. Neurobiol. 2017, 54, 7670–7685. [Google Scholar] [CrossRef]
- Xu, Q.; Deng, F.; Xing, Z.; Wu, Z.; Cen, B.; Xu, S.; Zhao, Z.; Nepomuceno, R.; Bhuiyan, M.I.; Sun, D.; et al. Long non-coding RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal survival through the NF-kappaB signaling pathway following cerebral ischemia. Cell Death Dis. 2016, 7, e2173. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhang, L.; Wang, E.; Zhang, C.; Li, X. LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem. Biophys. Res. Commun. 2018, 496, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, K.; Unterberg, A. Traumatic brain injury. Nervenarzt 2016, 87, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Unterberg, A.W.; Stover, J.; Kress, B.; Kiening, K.L. Edema and brain trauma. Neuroscience 2004, 129, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Dings, J.; Meixensberger, J.; Jager, A.; Roosen, K. Clinical experience with 118 brain tissue oxygen partial pressure catheter probes. Neurosurgery 1998, 43, 1082–1095. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, W.E.; Charbel, F.T.; Edelman, G. Brain tissue oxygen, carbon dioxide, and pH in neurosurgical patients at risk for ischemia. Anesth. Analg. 1996, 82, 582–586. [Google Scholar]
- Cervos-Navarro, J.; Diemer, N.H. Selective vulnerability in brain hypoxia. Crit. Rev. Neurobiol. 1991, 6, 149–182. [Google Scholar]
- Semenza, G.L. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 2011, 365, 537–547. [Google Scholar] [CrossRef]
- Peng, J.H.; Feng, Y.; LeBlanc, M.H.; Rhodes, P.G.; Parker, J.C., Jr. Apoptosis and necrosis in developing cerebellum and brainstem induced after focal cerebral hypoxic-ischemic injury. Brain Res. Dev. Brain Res. 2005, 156, 87–92. [Google Scholar] [CrossRef]
- Tu, L.; Wang, Y.; Chen, D.; Xiang, P.; Shen, J.; Li, Y.; Wang, S. Protective Effects of Notoginsenoside R1 via Regulation of the PI3K-Akt-mTOR/JNK Pathway in Neonatal Cerebral Hypoxic-Ischemic Brain Injury. Neurochem. Res. 2018, 43, 1210–1226. [Google Scholar] [CrossRef]
- Fan, X.; Heijnen, C.J.; van der Kooij, M.A.; Groenendaal, F.; van Bel, F. The role and regulation of hypoxia-inducible factor-1alpha expression in brain development and neonatal hypoxic-ischemic brain injury. Brain Res. Rev. 2009, 62, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Barth, D.A.; Prinz, F.; Teppan, J.; Jonas, K.; Klec, C.; Pichler, M. Long-Noncoding RNA (lncRNA) in the Regulation of Hypoxia-Inducible Factor (HIF) in Cancer. Nc RNA 2020, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.W.; Kung, H.J. Long non-coding RNA and tumor hypoxia: New players ushered toward an old arena. J. Biomed. Sci. 2017, 24, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Han, K.; Zhang, D.; Chen, J.; Xu, Z.; Hou, L. The role of long noncoding RNA in traumatic brain injury. Neuropsychiatr. Dis. Treat. 2019, 15, 1671–1677. [Google Scholar] [CrossRef]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef]
- Li, L.; Qu, Y.; Li, J.; Xiong, Y.; Mao, M.; Mu, D. Relationship between HIF-1alpha expression and neuronal apoptosis in neonatal rats with hypoxia-ischemia brain injury. Brain Res. 2007, 1180, 133–139. [Google Scholar] [CrossRef]
- Sen, T.; Sen, N. Treatment with an activator of hypoxia-inducible factor 1, DMOG provides neuroprotection after traumatic brain injury. Neuropharmacology 2016, 107, 79–88. [Google Scholar] [CrossRef]
- Ding, J.Y.; Kreipke, C.W.; Speirs, S.L.; Schafer, P.; Schafer, S.; Rafols, J.A. Hypoxia-inducible factor-1alpha signaling in aquaporin upregulation after traumatic brain injury. Neurosci. Lett. 2009, 453, 68–72. [Google Scholar] [CrossRef]
- Umschweif, G.; Alexandrovich, A.G.; Trembovler, V.; Horowitz, M.; Shohami, E. Hypoxia-inducible factor 1 is essential for spontaneous recovery from traumatic brain injury and is a key mediator of heat acclimation induced neuroprotection. J. Cereb. Blood Flow Metab. 2013, 33, 524–531. [Google Scholar] [CrossRef]
- Dharap, A.; Nakka, V.P.; Vemuganti, R. Effect of focal ischemia on long noncoding RNAs. Stroke 2012, 43, 2800–2802. [Google Scholar] [CrossRef]
- Voellenkle, C.; Garcia-Manteiga, J.M.; Pedrotti, S.; Perfetti, A.; De Toma, I.; Da Silva, D.; Maimone, B.; Greco, S.; Fasanaro, P.; Creo, P.; et al. Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA-sequencing. Sci. Rep. 2016, 6, 24141. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, X.; Cheng, R.; Hou, X.; Chen, Y.; Feng, Y.; Qiu, J. Analysis of long non-coding RNA expression profiles in neonatal rats with hypoxic-ischemic brain damage. J. Neurochem. 2019, 149, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Y.; Qu, Y. Long non-coding RNAs and hypoxic-ischemic brain damage. Chin. J. Contemp. Pediatrics 2016, 18, 1183–1187. [Google Scholar]
- Zhao, R.B.; Zhu, L.H.; Li, H.J.; Fan, Z.M.; Xia, Z.K. High-throughput sequencing analysis of lncRNAs in hippocampus tissues with hypoxic-ischemic brain damage. Int. J. Clin. Exp. Pathol. 2018, 11, 5265–5277. [Google Scholar]
- Zhao, F.; Qu, Y.; Liu, J.; Liu, H.; Zhang, L.; Feng, Y.; Wang, H.; Gan, J.; Lu, R.; Mu, D. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Neonatal Rats Following Hypoxic-ischemic Brain Damage. Sci. Rep. 2015, 5, 13850. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Y.; Tang, J.; Zhang, L.; Li, S.P.; Feng, Y.; Liu, H.T.; Qu, Y.; Mu, D.Z. [Role of long non-coding RNA BC088414 in hypoxic-ischemic injury of neural cells]. Chin. J. Contemp. Pediatrics 2015, 17, 1348–1353. [Google Scholar]
- Zhao, R.B.; Zhu, L.H.; Shu, J.P.; Qiao, L.X.; Xia, Z.K. GAS5 silencing protects against hypoxia/ischemia-induced neonatal brain injury. Biochem. Biophys. Res. Commun. 2018, 497, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.W.; Lou, X.Q.; Liu, Z.L.; Zhang, N.; Pang, L. LncRNA SNHG1 protects SH-SY5Y cells from hypoxic injury through miR-140-5p/Bcl-XL axis. Int. J. Neurosci. 2020, 1–10. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, H.; Fan, Z.; Li, G.; Ma, Q.; Tao, Z.; Wang, R.; Feng, J.; Luo, Y. Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke 2017, 48, 2211–2221. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, X.; Guo, R.; Meng, J. Protective effects of lncRNA H19 silence against hypoxia-induced injury in PC-12 cells by regulating miR-28. Int. J. Biol. Macromol. 2019, 121, 546–555. [Google Scholar] [CrossRef]
- Zhong, J.; Jiang, L.; Huang, Z.; Zhang, H.; Cheng, C.; Liu, H.; He, J.; Wu, J.; Darwazeh, R.; Wu, Y.; et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav. Immun. 2017, 65, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, L.; Yin, L. lncRNA NEAT1 Binds to MiR-339-5p to Increase HOXA1 and Alleviate Ischemic Brain Damage in Neonatal Mice. Mol. Ther. Nucleic Acids 2020, 20, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, F.; Zhang, M.; Shang, X.Y.; Xie, X.; Fu, T.; Li, J.P.; Li, H.L. Role of LncRNA MALAT-1 in hypoxia-induced PC12 cell injury via regulating p38MAPK signaling pathway. Neurosci. Lett. 2018, 670, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Xu, H.; Chen, G.; Wang, H.; Bi, Y.; Gao, H.; Luo, Y. Roles of lncRNA UCA1-miR-18a-SOX6 axis in preventing hypoxia injury following cerebral ischemia. Int. J. Clin. Exp. Pathol. 2017, 10, 8187–8198. [Google Scholar] [PubMed]
- Li, H.; Xu, L.X.; Yu, J.; Tan, L.; Miao, P.; Yang, X.; Tian, Q.; Li, M.; Feng, C.X.; Yang, Y.; et al. The role of a lncRNA (TCONS_00044595) in regulating pineal CLOCK expression after neonatal hypoxia-ischemia brain injury. Biochem. Biophys. Res. Commun. 2020, 528, 1–6. [Google Scholar] [CrossRef]
- Li, E.Y.; Zhao, P.J.; Jian, J.; Yin, B.Q.; Sun, Z.Y.; Xu, C.X.; Tang, Y.C.; Wu, H. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic-ischemic injury through miR-211/GDNF. Cell Cycle 2019, 18, 156–166. [Google Scholar] [CrossRef]
- Rubenstein, N.M.; Callahan, J.A.; Lo, D.H.; Firestone, G.L. Selective glucocorticoid control of Rho kinase isoforms regulate cell-cell interactions. Biochem. Biophys. Res. Commun. 2007, 354, 603–607. [Google Scholar] [CrossRef]
- Ark, M.; Ozdemir, A.; Polat, B. Ouabain-induced apoptosis and Rho kinase: A novel caspase-2 cleavage site and fragment of Rock-2. Apoptosis 2010, 15, 1494–1506. [Google Scholar] [CrossRef]
- Chen, H.; Li, X. LncRNA ROR is involved in cerebral hypoxia/reoxygenation-induced injury in PC12 cells via regulating miR-135a-5p/ROCK1/2. Am. J. Transl. Res. 2019, 11, 6145–6158. [Google Scholar]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef]
- Luoto, K.R.; Kumareswaran, R.; Bristow, R.G. Tumor hypoxia as a driving force in genetic instability. Genome Integr. 2013, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug. Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Nair, L.; Chung, H.; Basu, U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat. Rev. Mol. Cell Biol. 2020, 21, 123–136. [Google Scholar] [CrossRef]
- Yang, H.; Jiang, Y.; Zhang, Y.; Xu, Y.; Zhang, C.; Han, J.; Su, F.; Liu, X.; Mi, K.; Liu, B.; et al. System level characterization of small molecule drugs and their affected long noncoding RNAs. Aging 2019, 11, 12428–12451. [Google Scholar] [CrossRef]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef]
- McCown, P.J.; Wang, M.C.; Jaeger, L.; Brown, J.A. Secondary Structural Model of Human MALAT1 Reveals Multiple Structure-Function Relationships. Int. J. Mol. Sci. 2019, 20, 5610. [Google Scholar] [CrossRef]
- Frank, F.; Kavousi, N.; Bountali, A.; Dammer, E.B.; Mourtada-Maarabouni, M.; Ortlund, E.A. The lncRNA Growth Arrest Specific 5 Regulates Cell Survival via Distinct Structural Modules with Independent Functions. Cell Rep. 2020, 32, 107933. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Rinaldi, C.; Wood, M.J.A. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Katsushima, K.; Natsume, A.; Ohka, F.; Shinjo, K.; Hatanaka, A.; Ichimura, N.; Sato, S.; Takahashi, S.; Kimura, H.; Totoki, Y.; et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016, 7, 13616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lee, J.H.; Ruan, H.; Ye, Y.; Krakowiak, J.; Hu, Q.; Xiang, Y.; Gong, J.; Zhou, B.; Wang, L.; et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019, 10, 4562. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]

| Brain Injury | lncRNA | Targets | Function | Ref. |
|---|---|---|---|---|
| TBI | MALAT1 | MAPK pathways | Beneficially regulates deficits in TBI models | [38] |
| MEG3 | IL-1β, IL-6, IL-8, and TNF-α | Upregulates pro-inflammatory cytokines in the plasma of TBI patients | [39] | |
| p21 | Microglial activation | [41] | ||
| Gm4419 | TNF-α | Astrocyte apoptosis | [42] | |
| HOTAIR | MYD88 | Activates the microglia and regulates the inflammatory response molecule | [43] | |
| Stroke | MALAT1 | Contributes to the pathological process of ischemic stroke | [45] | |
| TUG1 | ||||
| FosDT | ||||
| SNHG14 | ||||
| ANRIL | Roles the negative correlation with the severity of ischemic stroke | [45,47] | ||
| H19 | Contributes to the pathological process of ischemic stroke and shows differences in specific allele | [45,49] | ||
| N1LR | p53 and Nck1 | Attenuates ischemic brain injury by inhibiting the apoptotic process and associates the pathological process of ischemic stroke | [45,50] | |
| C2dat1 | CaMKIIδ | Modulates CaMKIIδ under oxygen-glucose deprivation (OGD)/reperfusion via promoting NF-κB signaling and accounting for focal ischemia in vitro and in vivo | [45,51] | |
| GAS5 | miR-137 | Inhibits the protection of neurons and decreases cell viability through the Notch1 signaling | [52] | |
| MEG3 | Participates in pathological process of hemorrhagic and ischemic stroke and enhances neuronal apoptosis via the suppression of the PI3K/AKT pathway | [13,45] | ||
| Hypoxia | ENSRNOG 00000021987 | Promote neuronal apoptosis induced hypoxic-ischemic brain damage (HIBD) via increasing apoptosis-related genes and proteins | [74] | |
| BC088414 | Casp6 and Adrb2 | Reduces PC-12 cell apoptosis and increases proliferation via decreasing apoptosis-related gene | [75,76] | |
| GAS5 | miR-23a | Protects against HIBD via modulating hippocampal neuron function | [77] | |
| miR-221/PUMA | Promotes the neuronal apoptosis under hypoxia | [14] | ||
| SNHG1 | miR-140-5p /Bcl-XL | Inhibits neuroblastoma cell viability and enhances cell apoptosis under hypoxia | [78] | |
| H19 | miR-28/SP1 | Protects from the hypoxia-induced injury in PC-12 cell via deactivating the PDK/AKT and JAK/STAT pathways | [80] | |
| miR-19/Id2 | Promotes the hypoxic-ischemic neuronal apoptosis | [15] | ||
| NEAT1 | miR-339-5p /HOXA1 | Attenuates HIBD progression via reducing neuronal cell apoptosis | [82] | |
| MALAT1 | p38 MAPK pathway | Enhances the cell apoptosis and decreases cell viability under hypoxia in PC-12 | [83] | |
| UCA1 | miR-18a/SOX6 | Prevents hypoxia injury following cerebral ischemia via promoting migration, invasion, and viability, and reducing apoptosis in PC-12 | [84] | |
| TCONSA_ 00044595 | miR-182/CLOCK | Relates to abnormal changes in the pineal gene upon HIBD | [85] | |
| MIAT | miR-211/GDNF | Suppresses hypoxic/ischemic injury in neonatal rat via inhibiting neurons apoptosis | [86] | |
| ROR | miR-135a-5p/ ROCK1/2 | Promotes cerebral hypoxia/reoxygenation-induced injury via increasing caspase-related apoptosis in PC-12 | [89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, K.-H.; Yang, S.; Kim, S.-H.; Chun, S.; Joo, J.-Y. Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury. Biology 2020, 9, 458. https://doi.org/10.3390/biology9120458
Lim K-H, Yang S, Kim S-H, Chun S, Joo J-Y. Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury. Biology. 2020; 9(12):458. https://doi.org/10.3390/biology9120458
Chicago/Turabian StyleLim, Key-Hwan, Sumin Yang, Sung-Hyun Kim, Sungkun Chun, and Jae-Yeol Joo. 2020. "Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury" Biology 9, no. 12: 458. https://doi.org/10.3390/biology9120458
APA StyleLim, K.-H., Yang, S., Kim, S.-H., Chun, S., & Joo, J.-Y. (2020). Discoveries for Long Non-Coding RNA Dynamics in Traumatic Brain Injury. Biology, 9(12), 458. https://doi.org/10.3390/biology9120458






