The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics
Abstract
Simple Summary
Abstract
1. Introduction
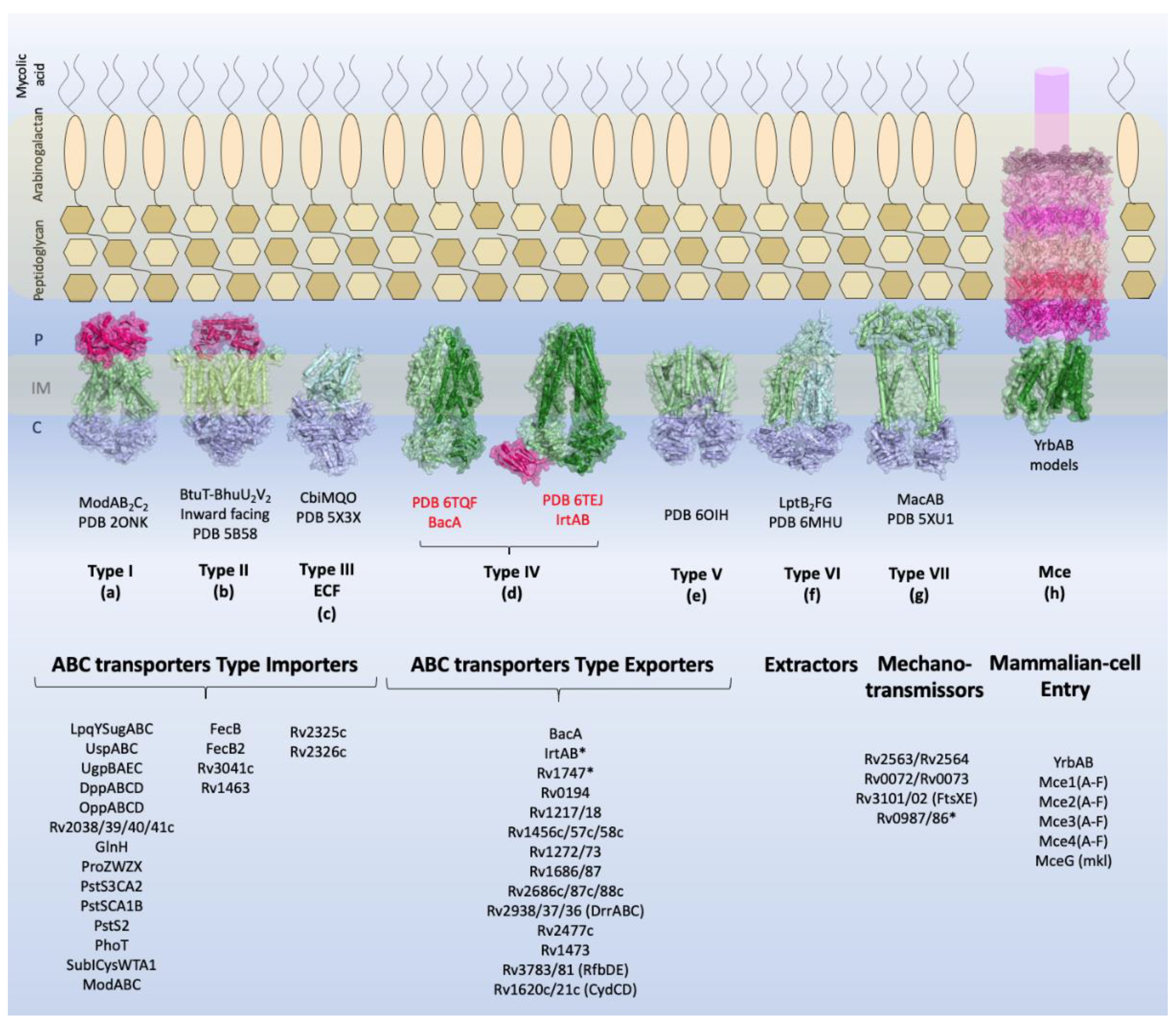
2. Identified ABC transporters in Mycobacterium tuberculosis
2.1. ABC Transporters Type Importer
2.1.1. Sugars Transporters
2.1.2. Peptides Transporters
2.1.3. Amino Acid Transporters
2.1.4. Anion Transporters
2.1.5. Metal Transporters
2.1.6. Hydrophilic Compounds
2.1.7. Energy-Coupling Factor Transporter (ECF)
2.2. ABC Transporters Type Exporters
2.2.1. Transporters Involved in the Recycling and Transport of Membrane Components and Liposaccharides
2.2.2. Electron Transport Chain (ETC)
2.2.3. Virulence and Adaptation
2.2.4. Drug Efflux and Resistance in M. tuberculosis
2.3. Distribution of ABC Transporters across Different Species of M. tuberculosis Genus
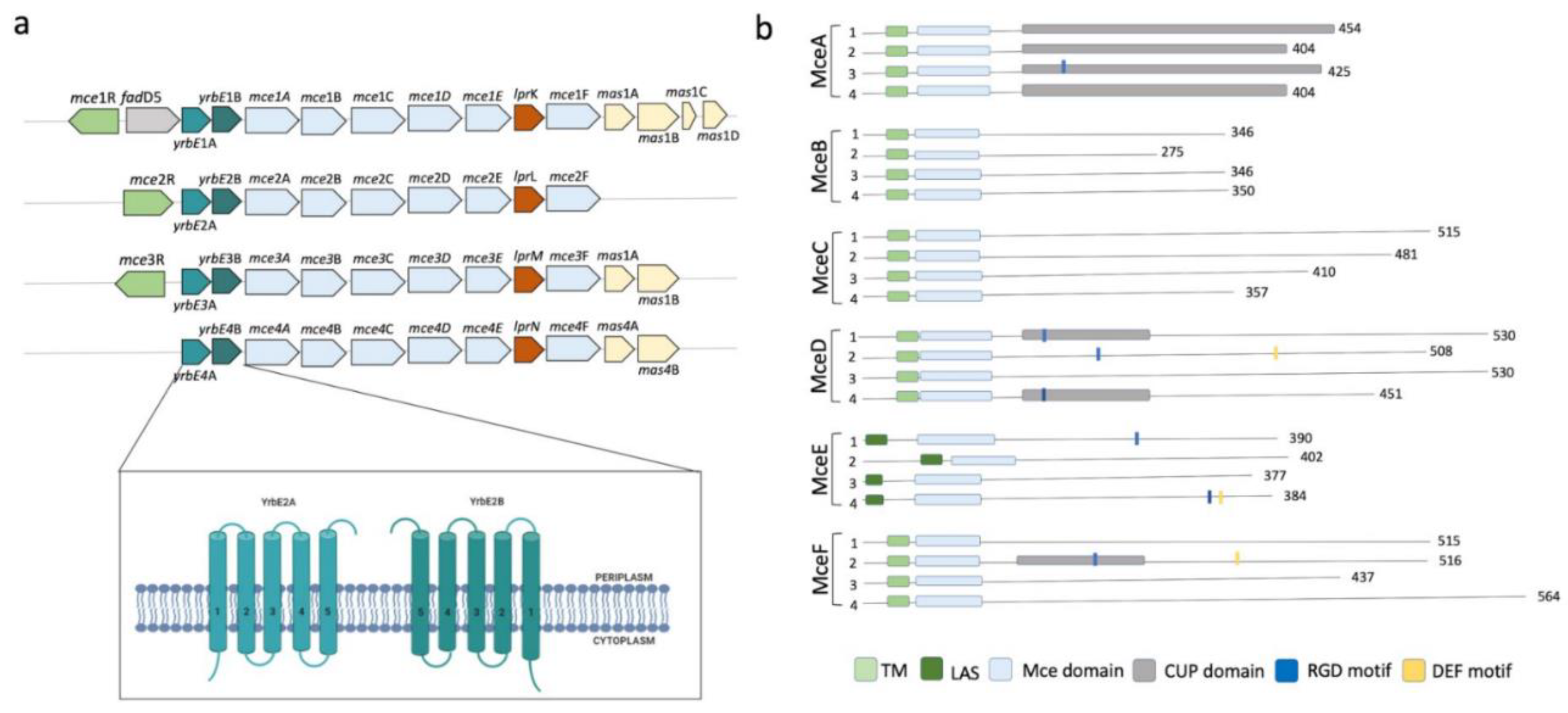
2.4. Mce Components of Mycobacterium Tuberculosis
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2019; Global Tuberculosis Report 2019; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Dulberger, C.L.; Rubin, E.J.; Boutte, C.C. The mycobacterial cell envelope—A moving target. Nat. Rev. Microbiol. 2020, 18, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, L.D.H.; Tetu, S.G.; Hassan, K.A.; Paulsen, I.T. TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017, 45, D320–D324. [Google Scholar] [CrossRef]
- Singh, R.; Dwivedi, S.P.; Gaharwar, U.S.; Meena, R.; Rajamani, P.; Prasad, T. Recent updates on drug resistance in Mycobacterium tuberculosis. J. Appl. Microbiol. 2020, 128, 1547–1567. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.; Moraes, I. Structural biology and structure–function relationships of membrane proteins. Biochem. Soc. Trans. 2018, 47, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Chen, J. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 2004, 73, 241–268. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Tampé, R. Structural and mechanistic principles of ABC Transporters. Annu. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Braibant, M.; Gilot, P.; Content, J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Bhabha, G.; Isom, G.L.; Greenan, G.; Ovchinnikov, S.; Henderson, I.R.; Cox, J.S.; Vale, R.D. Architectures of lipid transport systems for the bacterial outer membrane. Cell 2017, 169, 273–285.e17. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandolia, A.; Chaudhry, U.; Brahmachari, V.; Bose, M. Comparison of mammalian cell entry operons of mycobacteria: In silico analysis and expression profiling. FEMS Immunol. Med. Microbiol. 2005, 43, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Wolber, J.M.; Urbanek, B.L.; Meints, L.M.; Piligian, B.F.; Lopez-Casillas, I.C.; Zochowski, K.M.; Woodruff, P.J.; Swarts, B.M. The trehalose-specific transporter LpqY-SugABC is required for antimicrobial and anti-biofilm activity of trehalose analogues in Mycobacterium smegmatis. Carbohydr. Res. 2017, 450, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, R.; Weinrick, B.; Veeraraghavan, U.; Besra, G.S.; Jacobs, W.R. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010. [Google Scholar] [CrossRef]
- Fullam, E.; Prokes, I.; Fütterer, K.; Besra, G.S. Structural and functional analysis of the solute-binding protein UspC from Mycobacterium tuberculosis that is specific for amino sugars. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- Fenn, J.S.; Nepravishta, R.; Guy, C.S.; Harrison, J.; Angulo, J.; Cameron, A.D.; Fullam, E. Structural Basis of glycerophosphodiester recognition by the Mycobacterium tuberculosis substrate-binding protein UgpB. ACS Chem. Biol. 2019, 14, 1879–1887. [Google Scholar] [CrossRef]
- Jiang, D.; Zhang, Q.; Zheng, Q.; Zhou, H.; Jin, J.; Zhou, W.; Bartlam, M.; Rao, Z. Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine. FEBS J. 2014, 281, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Shin, A.R.; Kim, H.J.; Cho, S.N.; Park, J.K.; Shin, S.J. Identification of Rv2041c, a Novel Immunogenic Antigen from Mycobacterium tuberculosis with serodiagnostic potential. Scand. J. Immunol. 2009, 70, 457–464. [Google Scholar] [CrossRef]
- Mitra, A.; Ko, Y.H.; Cingolani, G.; Niederweis, M. Heme and hemoglobin utilization by Mycobacterium tuberculosis. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Dasgupta, A.; Sureka, K.; Mitra, D.; Saha, B.; Sanyal, S.; Das, A.K.; Chakrabarti, P.; Jackson, M.; Gicquel, B.; Kundu, M.; et al. An oligopeptide transporter of Mycobacterium tuberculosis regulates cytokine release and apoptosis of infected macrophages. PLoS ONE 2010, 5, e12225. [Google Scholar] [CrossRef]
- Deng, J.; Bi, L.; Tao, S.-C.; Zhang Correspondence, X.-E.; Zhou, L.; Guo, S.-J.; Fleming, J.; Jiang, H.-W.; Zhou, Y.; Gu, J.; et al. Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 2014, 9, 2317–2329. [Google Scholar] [CrossRef]
- Bhattacharyya, N.; Nkumama, N.; Newland-smith, Z.; Lin, L.; Yin, W.; Cullen, R.E. An aspartate-specific solute-binding protein regulates protein kinase G activity to controls glutamate metabolism in Mycobacteria. mBio 2018, 9, e00931-18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Chen, J.H.; Wang, Y.; Wang, Z.P.; He, Y.X. The putative compatible solute-binding protein ProX from Mycobacterium tuberculosis H37Rv: Biochemical characterization and crystallographic data. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Campanini, B.; Pieroni, M.; Raboni, S.; Bettati, S.; Benoni, R.; Pecchini, C.; Costantino, G.; Mozzarelli, A. Inhibitors of the sulfur assimilation pathway in bacterial pathogens as enhancers of antibiotic therapy. Curr. Med. Chem. 2015, 22, 187–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, N.E.; Lee, J.S.; Shin, J.H.; Lee, J.Y.; Ko, J.H.; Chang, C.L.; Kima, Y.S. Identification of mycobacterial antigens in human urine by use of immunoglobulin g isolated from sera of patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2016, 54, 1631–1637. [Google Scholar] [CrossRef]
- Målen, H.; Søfteland, T.; Wiker, H.G. Antigen analysis of Mycobacterium tuberculosis H37Rv culture filtrate proteins. Scand. J. Immunol. 2008, 67, 245–252. [Google Scholar] [CrossRef]
- Hwang, W.H.; Lee, W.K.; Ryoo, S.W.; Yoo, K.Y.; Tae, G.S. Expression, purification and improved antigenicity of the Mycobacterium tuberculosis PstS1 antigen for serodiagnosis. Protein Expr. Purif. 2014. [Google Scholar] [CrossRef]
- Xu, W.; DeJesus, M.A.; Rücker, N.; Engelhart, C.A.; Wright, M.G.; Healy, C.; Lin, K.; Wang, R.; Park, S.W.; Ioerger, T.R.; et al. Chemical genetic interaction profiling antibiotic resistance in Mycobacterium. Antimicrob. Chemother 2017, 61, 1–15. [Google Scholar] [CrossRef]
- Willemse, D.; Weber, B.; Masino, L.; Warren, R.M.; Adinolfi, S.; Pastore, A.; Williams, M.J. Rv1460, a SufR homologue, is a repressor of the suf operon in Mycobacterium tuberculosis. PLoS ONE 2018, 13, e0200145. [Google Scholar] [CrossRef]
- Arnold, F.M.; Weber, M.S.; Gonda, I.; Gallenito, M.J.; Adenau, S.; Egloff, P.; Zimmermann, I.; Hutter, C.A.J.; Hürlimann, L.M.; Peters, E.E.; et al. The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 2020, 580, 413–417. [Google Scholar] [CrossRef]
- Slotboom, D.J.; Ettema, T.W.; Nijland, M.; Thangaratnarajah, C. Bacterial multi-solute transporters. FEBS Lett. 2020, 1–10. [Google Scholar] [CrossRef]
- Rempel, S.; Gati, C.; Nijland, M.; Thangaratnarajah, C.; Karyolaimos, A.; de Gier, J.W.; Guskov, A.; Slotboom, D.J. A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature 2020, 580, 409–412. [Google Scholar] [CrossRef]
- Warner, D.F. Mycobacterium tuberculosis metabolism. Cold Spring Harb. Perspect. Med. 2015, 5, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.K.; Dubey, S.K.; Bhatnagar, R. ATP-binding cassette (ABC) import systems of Mycobacterium tuberculosis: Target for drug and vaccine development. Emerg. Microbes Infect. 2020, 9, 207–220. [Google Scholar] [CrossRef]
- Niderweis, M. Nutrient acquisition by mycobacteria. Microbiology 2008, 154, 679–692. [Google Scholar] [CrossRef]
- Titgemeyer, F.; Amon, J.; Parche, S.; Mahfoud, M.; Bail, J.; Schlicht, M.; Rehm, N.; Hillmann, D.; Stephan, J.; Walter, B.; et al. A genomic view of sugar transport in Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Bacteriol. 2007, 189, 5903–5915. [Google Scholar] [CrossRef]
- Sabharwal, N.; Varshney, K.; Rath, P.P.; Gourinath, S.; Das, U. Biochemical and biophysical characterization of nucleotide binding domain of Trehalose transporter from Mycobacterium tuberculosis. Int. J. Biol. Macromol. 2020, 152, 109–116. [Google Scholar] [CrossRef]
- Rengarajan, J.; Bloom, B.R.; Rubin, E.J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 2005, 102, 8327–8332. [Google Scholar] [CrossRef]
- Griffin, J.E.; Gawronski, J.D.; DeJesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Hingley-Wilson, S.M.; Lougheed, K.E.; Ferguson, K.; Leiva, S.; Williams, H.D. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis 2010, 90, 236–244. [Google Scholar] [CrossRef]
- Ferraris, D.M.; Spallek, R.; Oehlmann, W.; Singh, M.; Rizzi, M. Crystal structure of the Mycobacterium tuberculosis phosphate binding protein PstS3. Proteins Struct. Funct. Bioinform. 2014, 82, 2268–2274. [Google Scholar] [CrossRef]
- Vyas, N.K.; Vyas, M.N.; Quiocho, F.A. Crystal structure of M. tuberculosis ABC phosphate transport receptor: Specificity and charge compensation dominated by ion-dipole interactions. Structure 2003. [Google Scholar] [CrossRef]
- Heinkel, F.; Shen, L.; Richard-Greenblatt, M.; Okon, M.; Bui, J.M.; Gee, C.L.; Gay, L.M.; Alber, T.; Av-Gay, Y.; Gsponer, J.; et al. Biophysical characterization of the tandem fha domain regulatory module from the Mycobacterium tuberculosis ABC Transporter Rv1747. Structure 2018, 26, 972–986.e6. [Google Scholar] [CrossRef] [PubMed]
- Mavrici, D.; Marakalala, M.J.; Holton, J.M.; Prigozhin, D.M.; Gee, C.L.; Zhang, Y.J.; Rubin, E.J.; Alber, T. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc. Natl. Acad. Sci. USA 2014, 111, 8037–8042. [Google Scholar] [CrossRef]
- Dejesus, M.A.; Gerrick, E.R.; Xu, W.; Park, S.W.; Long, J.E.; Boutte, C.C.; Rubin, E.J.; Schnappinger, D.; Ehrt, S.; Fortune, S.M.; et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 2017, 8. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Kim, S.Y.; Shin, A.R.; Lee, B.S.; Kim, H.J.; Jeon, B.Y.; Cho, S.N.; Park, J.K.; Shill, S.J. Characterization of immune responses to Mycobacterium tuberculosis Rv2041c protein. J. Bacteriol. Virol. 2009, 39, 183–193. [Google Scholar] [CrossRef]
- Mitra, A.; Speer, A.; Lin, K.; Ehrt, S.; Niederweis, M. PPE Surface Proteins Are Required for Heme Utilization by Mycobacterium tuberculosis. mBio 2017, 8. [Google Scholar] [CrossRef]
- Stewart, G.R.; Patel, J.; Robertson, B.D.; Rae, A.; Young, D.B. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005, 1, e33. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.P.; Llabrés, S.; Neuberger, A.; Blaza, J.N.; Bai, X.C.; Okada, U.; Murakami, S.; Van Veen, H.W.; Zachariae, U.; Scheres, S.H.W.; et al. Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017, 2. [Google Scholar] [CrossRef]
- Crow, A.; Greene, N.P.; Kaplan, E.; Koronakis, V. Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. USA 2017, 114, 12572–12577. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.P.; Kaplan, E.; Crow, A.; Koronakis, V. Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Okada, U.; Yamashita, E.; Neuberger, A.; Morimoto, M.; van Veen, H.W.; Murakami, S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 2017, 8, 1336. [Google Scholar] [CrossRef]
- Nguyen, L.; Walburger, A.; Houben, E.; Koul, A.; Muller, S.; Morbitzer, M.; Klebl, B.; Ferrari, G.; Pieters, J. Role of protein kinase G in growth and glutamine metabolism of Mycobacterium bovis BCG. J. Bacteriol. 2005, 16, 5852–5856. [Google Scholar] [CrossRef][Green Version]
- Nohno, T.; Saito, T.; Hong, J.S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ). Mol. Gen. Genet. 1986, 205, 260–269. [Google Scholar] [CrossRef]
- Meza, A.N.; Cambui, C.C.N.; Moreno, A.C.R.; Fessel, M.R.; Balan, A. Mycobacterium tuberculosis CysA2 is a dual sulfurtransferase with activity against thiosulfate and 3-mercaptopyruvate and interacts with mammalian cells. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Choi, G.E.; Eom, S.H.; Jung, K.H.; Son, J.W.; Shin, A.R.; Shin, S.J.; Kim, K.H.; Chang, C.L.; Kim, H.J. CysA2: A candidate serodiagnostic marker for Mycobacterium tuberculosis infection. Respirology 2010, 15, 636–642. [Google Scholar] [CrossRef]
- Wooff, E.; Michell, S.L.; Gordon, S.V.; Chambers, M.A.; Bardarov, S.; Jacobs, W.R.; Hewinson, R.G.; Wheeler, P.R. Functional genomics reveals the sole sulphate transporter of the Mycobacterium tuberculosis complex and its relevance to the acquisition of sulphur in vivo. Mol. Microbiol. 2002, 43, 653–663. [Google Scholar] [CrossRef]
- Williams, M.J.; Kana, B.D.; Mizrahi, V. Functional analysis of molybdopterin biosynthesis in mycobacteria identifies a fused molybdopterin synthase in Mycobacterium tuberculosis. J. Bacteriol. 2011, 193, 98–106. [Google Scholar] [CrossRef]
- Camacho, L.R.; Ensergueix, D.; Perez, E.; Gicquel, B.; Guilhot, C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 1999, 34, 257–267. [Google Scholar] [CrossRef]
- Braibant, M.; Lefèvre, P.; De Wit, L.; Ooms, J.; Peirs, P.; Huygen, K.; Wattiez, R.; Content, J. Identification of a second Mycobacterium tuberculosis gene cluster encoding proteins of an ABC phosphate transporter. FEBS Lett. 1996. [Google Scholar] [CrossRef]
- Lefèvre, P.; Braibant, M.; De Wit, L.; Kalai, M.; Röeper, D.; Grötzinger, J.; Delville, J.P.; Peirs, P.; Ooms, J.; Huygen, K.; et al. Three different putative phosphate transport receptors are encoded by the Mycobacterium tuberculosis genome and are present at the surface of Mycobacterium bovis BCG. J. Bacteriol. 1997, 179, 2900–2906. [Google Scholar] [CrossRef]
- Collins, D.M.; Kawakami, R.P.; Buddle, B.M.; Wards, B.J.; de Lisle, G.W. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis identifies phoT as having roles in tuberculosis virulence and phosphate transport. Microbiology 2003, 149, 3203–3212. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vanzembergh, F.; Peirs, P.; Lefevre, P.; Celio, N.; Mathys, V.; Content, J.; Kalai, M. Effect of PstS sub-units or PknD deficiency on the survival of Mycobacterium tuberculosis. Tuberculosis 2010, 90, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Rifat, D.; Bishai, W.R.; Karakousis, P.C. Phosphate depletion: A novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 2009, 200, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.; Wolff, K.; Nguyen, L. Molecular Biology of Drug Resistance in Mycobacterium tuberculosis. In Pathogenesis of Mycobacterium Tuberculosis and its Interaction with the Host Organism; Springer: Berlin/Heidelberg, Germany, 2012; ISBN 978-3-642-40231-9. [Google Scholar]
- Neyrolles, O.; Wolschendorf, F.; Mitra, A.; Niederweis, M. Mycobacteria, metals, and the macrophage. Immunol. Rev. 2015, 264, 249–263. [Google Scholar] [CrossRef]
- Pandey, M.; Talwar, S.; Bose, S.; Pandey, A.K. Iron homeostasis in Mycobacterium tuberculosis is essential for persistence. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Shitharan, M. Iron Homeostasis in Mycobacterium. J. Bacteriol. 2016, 198, 2399–2409. [Google Scholar] [CrossRef]
- Chao, A.; Sieminski, P.J.; Owens, C.P.; Goulding, C.W. Iron Acquisition in Mycobacterium tuberculosis. Chem. Rev. 2019, 119, 1193–1220. [Google Scholar] [CrossRef]
- Braun, V.; Herrmann, C. Docking of the periplasmic FecB binding protein to the FecCD transmembrane proteins in the ferric citrate transport system of Escherichia coli. J. Bacteriol. 2007. [Google Scholar] [CrossRef]
- Ryndak, M.B.; Wang, S.; Smith, I.; Rodriguez, G.M. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. J. Bacteriol. 2010, 192, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Sangari, F.J.; Parker, A.; Bermudez, L.E. fecB, a gene potentially involved in iron transport in Mycobacterium avium, is not induced within macrophages. FEMS Microbiol. Lett. 2005, 247, 185–191. [Google Scholar] [CrossRef]
- Scheepers, G.H.; Lycklama a Nijeholt, J.A.; Poolman, B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016, 590, 4393–4401. [Google Scholar] [CrossRef] [PubMed]
- Kavvas, E.S.; Catoiu, E.; Mih, N.; Yurkovich, J.T.; Seif, Y.; Dillon, N.; Heckmann, D.; Anand, A.; Yang, L.; Nizet, V.; et al. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gao, F. Iron–Sulfur cluster biogenesis and iron homeostasis in Cyanobacteria. Front. Microbiol. 2020, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Farhana, A.; Kumar, S.; Rathore, S.S.; Ghosh, P.C.; Ehtesham, N.Z.; Tyagi, A.K.; Hasnain, S.E. Mechanistic insights into a novel exporter-importer system of Mycobacterium tuberculosis unravel its role in trafficking of iron. PLoS ONE 2008, 3, e2087. [Google Scholar] [CrossRef] [PubMed]
- De Voss, J.J.; Rutter, K.; Schroeder, B.G.; Barry, C.E. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 1999, 181, 4443–4451. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Smith, I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 2006, 188, 424–430. [Google Scholar] [CrossRef]
- Domenech, P.; Kobayashi, H.; Levier, K.; Walker, G.C.; Barry, C.E. BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 2009, 191, 477–485. [Google Scholar] [CrossRef]
- Rempel, S.; Stanek, W.K.; Slotboom, D.J. Energy-Coupling Factor—Type ATP-Binding Cassette Transporters. Annu. Rev. Biochem. 2019, 88, 551–576. [Google Scholar] [CrossRef]
- Heinkel, F.; Abraham, L.; Ko, M.; Chao, J.; Bach, H.; Hui, L.T.; Li, H.; Zhu, M.; Ling, Y.M.; Rogalski, J.C.; et al. Phase separation and clustering of an ABC transporter in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2019, 116, 16326–16331. [Google Scholar] [CrossRef] [PubMed]
- Kana, B.D.; Weinstein, E.A.; Avarbock, D.; Dawes, S.S.; Rubin, H.; Mizrahi, V. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 2001, 183, 7076–7086. [Google Scholar] [CrossRef] [PubMed]
- Be, N.A.; Lamichhane, G.; Grosset, J.; Tyagi, S.; Cheng, Q.J.; Kim, K.S.; Bishai, W.R.; Jain, S.K. Murine model to study the invasion and survival of Mycobacterium tuberculosis in the central nervous system. J. Infect. Dis. 2008, 198, 1520–1528. [Google Scholar] [CrossRef]
- Mir, M.A.; Arumugam, M.; Mondal, S.; Rajeswari, H.S.; Ramakumar, S.; Ajitkumar, P. Mycobacterium tuberculosis Cell division protein, FtsE, is an ATPase in dimeric form. Protein J. 2015, 34, 35–47. [Google Scholar] [CrossRef]
- Kanji, A.; Hasan, R.; Ali, A.; Zaver, A.; Zhang, Y.; Imtiaz, K.; Shi, W.; Clark, T.G.; McNerney, R.; Phelan, J.; et al. Single nucleotide polymorphisms in efflux pumps genes in extensively drug resistant Mycobacterium tuberculosis isolates from Pakistan. Tuberculosis 2017, 107, 20–30. [Google Scholar] [CrossRef]
- Pasca, M.R.; Guglierame, P.; Arcesi, F.; Bellinzoni, M.; Rossi, E.; De Riccardi, G. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux. Antimicrob. Agents Chemother. 2004, 48, 3175–3178. [Google Scholar] [CrossRef]
- Hao, P.; Shi-Liang, Z.; Ju, L.; Ya-Xin, D.; Biao, H.; Xu, W.; Min-Tao, H.; Shou-Gang, K.; Ke, W. The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Narang, A.; Garima, K.; Porwal, S.; Bhandekar, A.; Shrivastava, K.; Giri, A.; Sharma, N.K.; Bose, M.; Varma-Basil, M. Potential impact of efflux pump genes in mediating rifampicin resistance in clinical isolates of Mycobacterium tuberculosis from India. PLoS ONE 2019, 14, e0223163. [Google Scholar] [CrossRef]
- Kumar, N.; Radhakrishnan, A.; Wright, C.C.; Chou, T.H.; Lei, H.T.; Bolla, J.R.; Tringides, M.L.; Rajashankar, K.R.; Su, C.C.; Purdy, G.E.; et al. Crystal structure of the transcriptional regulator Rv1219c of Mycobacterium tuberculosis. Protein Sci. 2014, 23, 423–432. [Google Scholar] [CrossRef]
- Murugasu-Oei, B.; Tay, A.; Dick, T. Upregulation of stress response genes and ABC transporters in anaerobic stationary-phase Mycobacterium smegmatis. Mol. Gen. Genet. 1999, 262, 677–682. [Google Scholar] [CrossRef]
- Faksri, K.; Tan, J.H.; Disratthakit, A.; Xia, E.; Prammananan, T.; Suriyaphol, P.; Khor, C.C.; Teo, Y.Y.; Ong, R.T.H.; Chaiprasert, A. Whole-genome sequencing analysis of serially isolated multi-drug and extensively drug resistant Mycobacterium tuberculosis from Thai patients. PLoS ONE 2016, 11, e0160992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Al-Hendy, A.; Toivanen, P.; Skumik, M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: Similarities to the dTDP-L-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol. Microbiol. 1993, 9, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Manning, P.A.; Stroeher, U.H.; Karageorgos, L.E.; Morona, R. Putative O-antigen transport genes within the rfb region of Vibrio cholerae O1 are homologous to those for capsule transport. Gene 1995, 158, 1–7. [Google Scholar] [CrossRef]
- Vissa, V.D.; Brennan, P.J. The genome of Mycobacterium leprae: A minimal mycobacterial gene set. Genome Biol. 2001, 2, 1–8. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Kos, V.; Whitfield, C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 2010, 74, 341–362. [Google Scholar] [CrossRef]
- Trutneva, K.A.; Shleeva, M.O.; Demina, G.R.; Vostroknutova, G.N.; Kaprelyans, A.S. One-Year Old Dormant, “non-culturable” Mycobacterium tuberculosis preserves significantly diverse protein profile. Front. Cell. Infect. Microbiol. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Phong, T.Q.; Ha, D.T.T.; Volker, U.; Hammer, E. Using a label free quantitative proteomics approach to identify changes in protein abundance in multidrug-resistant Mycobacterium tuberculosis. Indian J. Microbiol. 2015, 55, 219–230. [Google Scholar] [CrossRef][Green Version]
- Spivey, V.L.; Molle, V.; Whalan, R.H.; Rodgers, A.; Leiba, J.; Stach, L.; Walker, K.B.; Smerdon, S.J.; Buxton, R.S. Forkhead-associated (FHA) domain containing ABC transporter Rv1747 is positively regulated by Ser/Thr phosphorylation in Mycobacterium tuberculosis. J. Biol. Chem. 2011, 286, 26198–26209. [Google Scholar] [CrossRef]
- Spivey, V.L.; Whalan, R.H.; Hirst, E.M.A.; Smerdon, S.J.; Buxton, R.S. An attenuated mutant of the Rv1747 ATP-binding cassette transporter of Mycobacterium tuberculosis and a mutant of its cognate kinase, PknF, show increased expression of the efflux pump-related iniBAC operon. Cambridge., UK. FEMS Microbiol. Lett. 2013, 347, 107–115. [Google Scholar] [CrossRef]
- Curry, J.M.; Whalan, R.; Hunt, D.M.; Gohil, K.; Strom, M.; Rickman, L.; Colston, M.J.; Smerdon, S.J.; Buxton, R.S. An ABC transporter containing a forkhead-associated domain interacts with a serine-threonine protein kinase and is required for growth of Mycobacterium tuberculosis in mice. Infect. Immun. 2005, 73, 4471–4477. [Google Scholar] [CrossRef]
- Molle, V.; Soulat, D.; Jault, J.M.; Grangeasse, C.; Cozzone, A.J.; Prost, J.F. Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2004. [Google Scholar] [CrossRef]
- Gee, C.L.; Papavinasasundaram, K.G.; Blair, S.R.; Baer, C.E.; Falick, A.M.; King, D.S.; Griffin, J.E.; Venghatakrishnan, H.; Zukauskas, A.; Wei, J.-R.; et al. A phosphorylated pseudokinase complex controls cell wall synthesis in Mycobacteria. Sci. Signal. 2012, 5, ra7. [Google Scholar] [CrossRef] [PubMed]
- Glass, L.N.; Swapna, G.; Chavadi, S.S.; Tufariello, J.A.M.; Mi, K.; Drumm, J.E.; Lam, T.K.T.; Zhu, G.; Zhan, C.; Vilchéze, C.; et al. Mycobacterium tuberculosis universal stress protein Rv2623 interacts with the putative ATP binding cassette (APC) transporter Rv1747 to regulate mycobacterial growth. PLoS Pathog. 2017, 13, e1006515. [Google Scholar] [CrossRef]
- Drumm, J.E.; Mi, K.; Bilder, P.; Sun, M.; Lim, J.; Bielefeldt-Ohmann, H.; Basaraba, R.; So, M.; Zhu, G.; Tufariello, J.M.; et al. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding: Requirement for establishing chronic persistent infection. PLoS Pathog. 2009, 5, e1000460. [Google Scholar] [CrossRef]
- Berube, B.J.; Parish, T. Combinations of respiratory chain inhibitors have enhanced bactericidal activity against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2018, 62, 1–10. [Google Scholar] [CrossRef]
- Cruz-Ramos, H.; Cook, G.M.; Wu, G.; Cleeter, M.W.; Poole, R.K. Membrane topology and mutational analysis of Escherichia coli CydDC, an ABC-type cysteine exporter required for cytochrome assembly. Microbiology 2004, 150, 3415–3427. [Google Scholar] [CrossRef]
- Saini, V.; Chinta, K.C.; Reddy, V.P.; Glasgow, J.N.; Stein, A.; Lamprecht, D.A.; Rahman, M.A.; Mackenzie, J.S.; Truebody, B.E.; Adamson, J.H.; et al. Hydrogen sulfide stimulates Mycobacterium tuberculosis respiration, growth and pathogenesis. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Dhar, N.; McKinney, J.D. Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc. Natl. Acad. Sci. USA 2010, 107, 12275–12280. [Google Scholar] [CrossRef]
- Cook, G.M.; Greening, C.; Hards, K.; Berney, M. Energetics of Pathogenic Bacteria and Opportunities for Drug Development, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; Volume 65. [Google Scholar]
- Matthysse, A.G.; Yarnall, H.A.; Young, N. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 1996, 178, 5302–5308. [Google Scholar] [CrossRef]
- Wang, J.; Behr, M.A. Building a better bacillus: The emergence of Mycobacterium tuberculosis. Front. Microbiol. 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Pethe, K.; Swenson, D.L.; Alonso, S.; Anderson, J.; Wang, C.; Russell, D.G. Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. USA 2004, 101, 13642–13647. [Google Scholar] [CrossRef] [PubMed]
- Balganesh, M.; Dinesh, N.; Sharma, S.; Kuruppath, S.; Nair, A.V.; Sharma, U. Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 2012. [Google Scholar] [CrossRef] [PubMed]
- Day, T.A.; Mittler, J.E.; Nixon, M.R.; Thompson, C.; Miner, M.D.; Hickey, M.J.; Liao, R.P.; Pang, J.M.; Shayakhmetov, D.M.; Sherman, D.R. Mycobacterium tuberculosis strains lacking surface lipid phthiocerol dimycocerosate are susceptible to killing by an early innate host response. Infect. Immun. 2014, 82, 5214–5222. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, B.S.; Bhakta, S.; Barik, R.; Basu, J.; Kundu, M.; Chakrabarti, P. Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 2002. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Sirous, M.; Absalan, Z.; Tabandeh, M.R.; Savari, M. Comparison of drrA and drrB Efflux pump genes expression in drug-susceptible and -resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran. Infect. Drug Resist. 2019, 12, 3437–3444. [Google Scholar] [CrossRef]
- Liu, J.; Tariff, H.E.; Nikaido, H. Active efflux of fluoroquinolones in Mycobacterium smegmatis mediated by LfrA, a multidrug efflux pump. J. Bacteriol. 1996, 178, 3791–3795. [Google Scholar] [CrossRef] [PubMed]
- Raheem, T.Y.; Iwalokun, B.; Fowora, M.; Adesesan, A.; Oluwadun, A. Are drug efflux genes present among Mycobacterium tuberculosis isolates from patients in Lagos, Nigeria? J. Biosci. Med. 2020, 8, 86–98. [Google Scholar] [CrossRef]
- Chatterjee, A.; Saranath, D.; Bhatter, P.; Mistry, N. Global Transcriptional Profiling of Longitudinal Clinical isolates of Mycobacterium tuberculosis exhibiting rapid accumulation of drug resistance. PLoS ONE 2013, 8, e54717. [Google Scholar] [CrossRef][Green Version]
- Dutta, N.K.; Mehra, S.; Didier, P.J.; Roy, C.J.; Doyle, L.A.; Alvarez, X.; Ratterree, M.; Be, N.A.; Lamichhane, G.; Jain, S.K.; et al. Genetic requirements for the survival of tubercle bacilli in primates. J. Infect. Dis. 2010, 201, 1743–1752. [Google Scholar] [CrossRef]
- Garima, K.; Pathak, R.; Tandon, R.; Rathor, N.; Sinha, R.; Bose, M.; Varma-Basil, M. Differential expression of efflux pump genes of Mycobacterium tuberculosis in response to varied subinhibitory concentrations of antituberculosis agents. Tuberculosis 2015, 95, 155–161. [Google Scholar] [CrossRef]
- Lamichhane, G.; Zignol, M.; Blades, N.J.; Geiman, D.E.; Dougherty, A.; Grosset, J.; Broman, K.W.; Bishai, W.R. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: Application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2003, 100, 7213–7218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, Y.; Yang, H.; Bao, H.; Qin, L.; Zhu, C.; Chen, Y.; Hu, Z. Impact of hypoxia on drug resistance and growth characteristics of Mycobacterium tuberculosis clinical isolates. PLoS ONE 2016, 11, e0166052. [Google Scholar] [CrossRef] [PubMed]
- Danilchanka, O.; Mailaender, C.; Niederweis, M. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2008. [Google Scholar] [CrossRef]
- Malinga, L.A.; Abeel, T.; Desjardins, C.A.; Dlamini, T.C.; Cassell, G.; Chapman, S.B.; Birren, B.W.; Earl, A.M.; van der Walt, M. Draft genome sequences of two extensively drug-resistant strains of Mycobacterium tuberculosis belonging to the Euro-American S lineage. Genome Announc. 2016, 4, 2–3. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Rodríguez, D.C.; Ocampo, M.; Varela, Y.; Curtidor, H.; Patarroyo, M.A.; Patarroyo, M.E. Mce4F Mycobacterium tuberculosis protein peptides can inhibit invasion of human cell lines. Pathog. Dis. 2015, 73, 1–12. [Google Scholar] [CrossRef]
- Nazarova, E.V.; Montague, C.R.; Huang, L.; La, T.; Russell, D.; Vanderven, B.C. The genetic requirements of fatty acid import by Mycobacterium tuberculosis within macrophages. Elife 2019, 8, 1–12. [Google Scholar] [CrossRef]
- Perkowski, E.F.; Miller, B.K.; Mccann, J.R.; Sullivan, J.T.; Malik, S.; Allen, I.C.; Godfrey, V.; Hayden, J.D.; Braunstein, M. An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol. Microbiol. 2016, 100, 90–107. [Google Scholar] [CrossRef]
- Khan, S.; Islam, A.; Hassan, M.I.; Ahmad, F. Purification and structural characterization of Mce4A from Mycobacterium tuberculosis. Int. J. Biol. Macromol. 2016, 93, 235–241. [Google Scholar] [CrossRef]
- Mikheecheva, N.E.; Zaychikova, M.V.; Melerzanov, A.V.; Danilenko, V.N. A nonsynonymous SNP catalog of Mycobacterium tuberculosis virulence genes and its use for detecting new potentially virulent sublineages. Genome Biol. Evol. 2017, 9, 887–899. [Google Scholar] [CrossRef] [PubMed]

| IMPORTERS | ||
|---|---|---|
| ABC Systems/Genomic Organization | Genomic Organization | Structural Organization |
| Sugars | ||
| LpqY/SugA/SugB/SugC (Rv1235/36/37/38) |  |  |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Trehalose recycling involved in virulence and biofilm formation | ||
| [13,14] | ||
| UspC/UspB/UspA (Rv2318/17/16) |  | 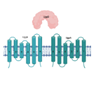 |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Amino-sugars | ||
| [9,15] | ||
| UgpB/UgpA/UgpE/UgpC (Rv2833c/32c/34c/35c) |  | 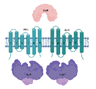 |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Glycerophosphocholine | ||
| [16,17] | ||
| Rv2041c/Rv2040c/39c/38c |  |  |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Amino-sugar | ||
| Rv2041c—potential use for serodiagnostic and | ||
| Vaccine development | ||
| [18] | ||
| Peptides | ||
| DppA/DppB/DppC/DppD (Rv3666c/65c/64c/63c) |  |  |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Dipeptides | ||
| (heme and hemoglobin?) | ||
| [19] | ||
| OppA/OppB/OppC/OppD (Rv1280c/83c/81c/82c) |  |  |
| SBP-TM1-TM2-[NBDr](2x) | ||
| Oligopeptides (glutatione and bradykinin) | ||
| [20] | ||
| Amino acids | ||
| Rv2563/Rv2564 |  | 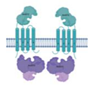 |
| Lipopolysaccharide export | ||
| [ECD/TM/NBDr](2x) | ||
| Type VII | ||
| Rv2564 is a potential biomarker for diagnosis development | ||
| [21] | ||
| Rv0072/Rv0073 | 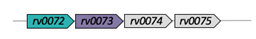 | 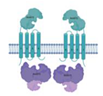 |
| Lipopolysaccharide export | ||
| [ECD/TM/NBDr](2x) | ||
| Type VII | ||
| [9] | ||
| GlnH (Rv0411c) |  |  |
| SBP | ||
| glutamine/glutamate/aspartate | ||
| [22] | ||
| ProX/ProW/ProZ/ProV (Rv3759/5756/58) | 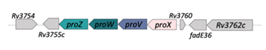 |  |
| SBP-[TM/NBDr](2x) | ||
| Glycine/betaine/L-proline/carnitine/ | ||
| choline | ||
| [23] | ||
| Anions | ||
| SubI/CysT/CysW/CysA1 (Rv2400/99/98/97) |  | 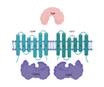 |
| SBP-TM1-TM2-[NBD](2x) | ||
| Sulfate | ||
| Members of sulfate transporter and sulfate assimilation pathway are essential and targets for drug development [24] | ||
| ModA/ModB/ModC (Rv1857/Rv1858/Rv1859) |  | 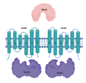 |
| SBP-[TM/NBDr](2x) | ||
| Molybdate | ||
| ModC is a good target as biomarker and potential for vaccine development [25] | ||
| PstS3/PstC2/PstA2 (Rv0928/29/30) | 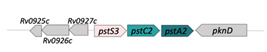 | 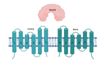 |
| SBP-TM1-TM2 | ||
| Phosphate | ||
| SBPs are potential targets for inhibition and vaccine development [26] | ||
| PstS1/PstC1/PstA1/PstB* (Rv0934/35/36/33) | 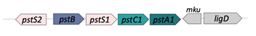 | 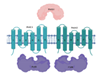 |
| SBP-TM1-TM2-[NBD](2x) | ||
| Phosphate | ||
| Potential drug targets; PstS1 is a good biomarker for diagnosis [27] | ||
| PstS2 (Rv0932) |  |  |
| SBP | ||
| Phosphate [26] | ||
| PhoT (Rv0820) |  |  |
| NBD | ||
| Phosphate [9] | ||
| Metals | ||
| FecB (Rv3040c) | 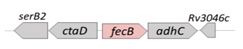 |  |
| SBP | ||
| FeIII-dicitrate | ||
| Potential drug target and vaccine development | ||
| [28] | ||
| FecB2 | 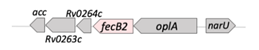 |  |
| (Rv0265c) | ||
| SBP | ||
| Iron/heme [9] | ||
| Rv3041c | 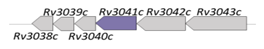 |  |
| NBD | ||
| iron-hydroxamate [9] | ||
| Rv1463 | 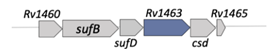 |  |
| NBD | ||
| Fe-S cluster assembly [29] | ||
| SBP/TM1/NBD1-TM2/NBD2 |  | 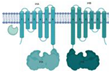 |
| Siderophore [30] | ||
| Hydrophilic compounds | ||
| BacA* (Rv1819c) |  | 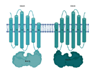 |
| [TM/NBD](2x) | ||
| Vitamin B12 [31,32] | ||
| Energy-Coupling Factors | ||
| Rv2325c/Rv2326c | 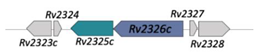 |  |
| Transporter Component | 3D Structure | PDB | Ligand | Reference |
|---|---|---|---|---|
| Rv2318 (UspC) SBP type II, sugar |  | 5K2Y; 5K2X | - | [15] |
| Rv2833c (UgpB) SBP type II, sugar |  | 6R1B; 4MF1 | GPC - | [17] |
| Rv3666c (DppA) SBP type II, peptides |  | 6E4D | SSVT | [19] |
| Rv0411c (GlnH) SBP type II, amino acids |  | 6H20 6HIU 6H2T | Asn Asp Glu | [22] |
| Rv2400c (SubI) SBP type II, anion |  | 6DDN | SO4 | - |
| Rv0928 (PstS3) SBP type II, anion |  | 4LVQ | PO4 | [42] |
| Rv0934 (PstS1) SBP type II, anion |  | 1PC3 | PO4 | [43] |
| Rv0263c (FecB2) SBP type III, iron |  | 4PM4 | - | - |
| Rv1348/Rv1349 (IrtAB), full transporter, iron |  | 6TEJ; 6TEK | - - | [30] |
| Rv1819c (BacA) Full transporter |  | 6TQF; 6TQE | AMP-PNP | [32] |
| Rv1747_FHA-1 Lipooligosaccharides, drugs |  | 6CCD | - | [44] |
| Rv1747_FHA-2 Lipooligosaccharides, drugs |  | 6CAH | - | [44] |
| Rv3101 (FtsX) ECD, division |  | 4N8N 4N8O | - | [45] |
| EXPORTERS | ||
|---|---|---|
| ABC Systems/Genomic Organization | Genomic Organization |
Structural Organization |
| Recycling of Membrane compounds/Liposaccharides | ||
| RfbD/RfbE (Rv3783/Rv3781) |  |  |
| [NBDr/TM/ECD](2x) | ||
| Cell wall biosynthesis, | ||
| ABC-2 subfamily of integral membrane proteins [9] | ||
| * Rv1747 |  |  |
| [FHA(2x)/NBD/TM](2x) | ||
| Lipo-oligosaccharides/drug efflux [83] | ||
| Electron transport chain (ETC) | ||
| CydC/CydD (Rv1620c/Rv1621c) |  | 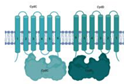 |
| TM1/NBD1-TM2/NBD2 | ||
| Cytochrome biosynthesis [84] | ||
| Virulence, adaptation | ||
| * Rv0987/Rv0986 |  | 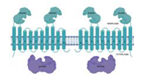 |
| Adhesion component | ||
| [ECD2x/TM/NBD](2x) | ||
| [85] | ||
| FtsX/FtsE (Rv3101/Rv3102) |  |  |
| Cell division | ||
| [ECD/TM/NBD](2x) | ||
| [86] | ||
| Drug efflux | ||
| DrrC/DrrB/DrrA (Rv2938/37/36) |  | 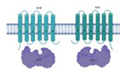 |
| TM1/TM2-[NBD](2x) | ||
| Daunorubicin/doxorubicin [87] | ||
| Rv2686c/Rv2687c/Rv2688c |  | 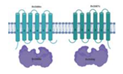 |
| TM1/TM2-[NBD](2x) | ||
| Fluoroquinolones | ||
| [88] | ||
| Rv1686c/Rv1687c |  | 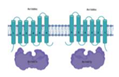 |
| [TM/NBD](2x) | ||
| Multidrug efflux [9] | ||
| Rv1273/Rv1272 |  |  |
| [TM/NBD](2x) | ||
| MSBA subfamily/Drug efflux [87] | ||
| Rv1456c/Rv1457c/Rv1458c | 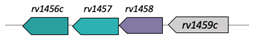 |  |
| Antibiotic transport | ||
| TM1/TM2-[NBD](2x) [89] | ||
| Rv0194 |  |  |
| Drug efflux transport | ||
| [TM/NBD](2x) [90] | ||
| Rv1217c/Rv1218c |  | 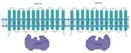 |
| [TM/NBD](2x) [91] | ||
| Rv1473 | 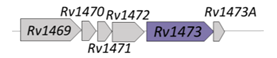 |  |
| NBD | ||
| Macrolides efflux [92,93] | ||
| Rv2477c |  |  |
| NBD | ||
| Macrolides efflux [93] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cassio Barreto de Oliveira, M.; Balan, A. The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics. Biology 2020, 9, 443. https://doi.org/10.3390/biology9120443
Cassio Barreto de Oliveira M, Balan A. The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics. Biology. 2020; 9(12):443. https://doi.org/10.3390/biology9120443
Chicago/Turabian StyleCassio Barreto de Oliveira, Marcelo, and Andrea Balan. 2020. "The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics" Biology 9, no. 12: 443. https://doi.org/10.3390/biology9120443
APA StyleCassio Barreto de Oliveira, M., & Balan, A. (2020). The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics. Biology, 9(12), 443. https://doi.org/10.3390/biology9120443




