The Foreign Oligochaete Species Quistadrilus multisetosus (Smith, 1900) in Lake Geneva: Morphological and Molecular Characterization and Environmental Influences on Its Distribution
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sites and Repartition of the Analyses
2.2. Sampling
2.3. Morphological Examination of Oligochaete Communities
2.4. Examination of Specimens from Collections
2.5. Genetic Analyses
2.5.1. Acquisition of the COI Barcode of Quistadrilus Multisetosus
2.5.2. Construction of a COI Phylogenetic Tree
2.5.3. eDNA Metabarcoding
DNA Extraction, PCR Amplification, Library Preparation and Illumina Sequencing
Sequence Analysis
3. Results
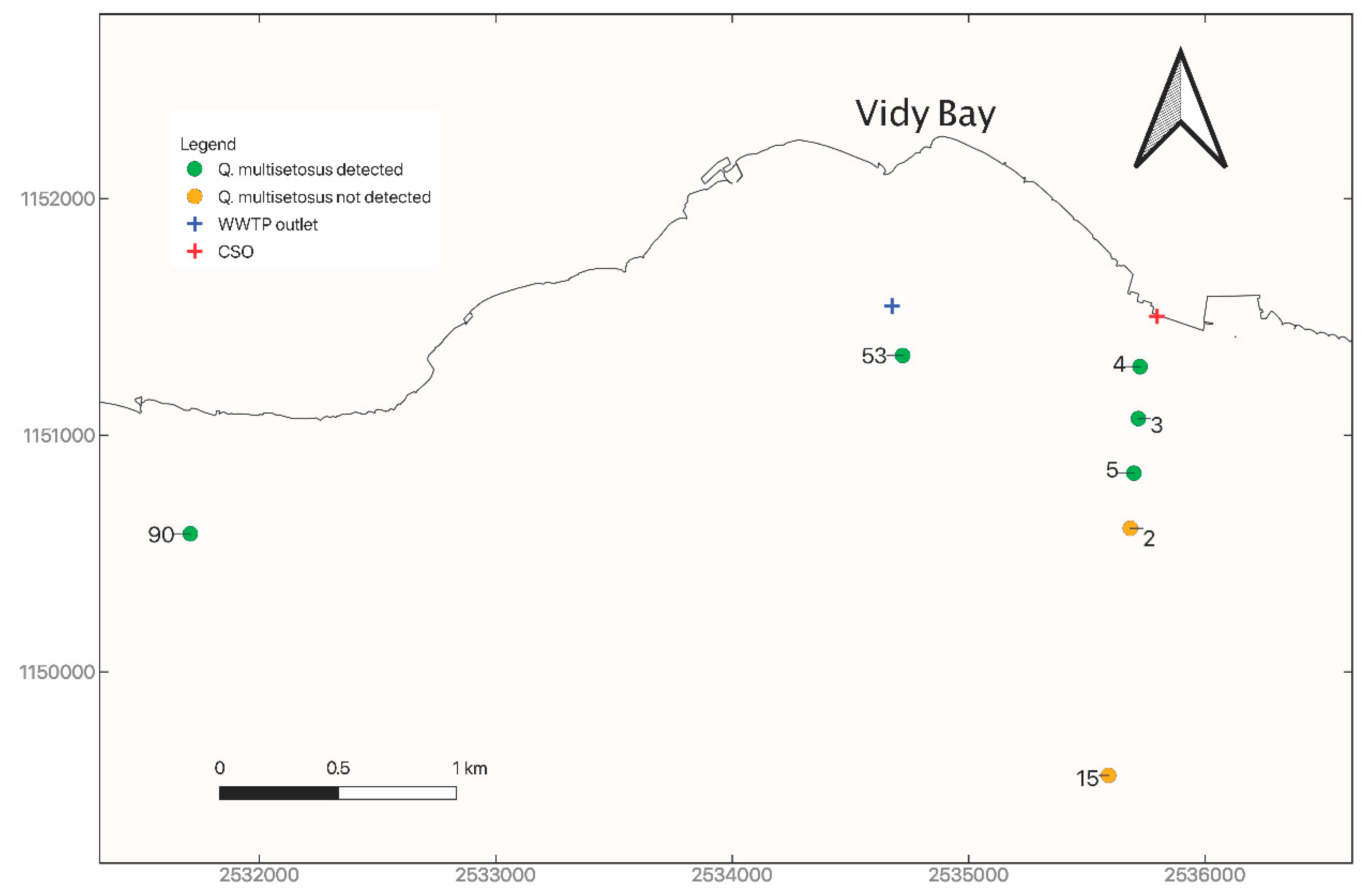
3.1. Distribution and Abundance of Quistadrilus Multisetosus
3.2. Morphological Differentiation of Quistadrilus Multisetosus from Spirosperma Ferox and Embolocephalus Velutinus
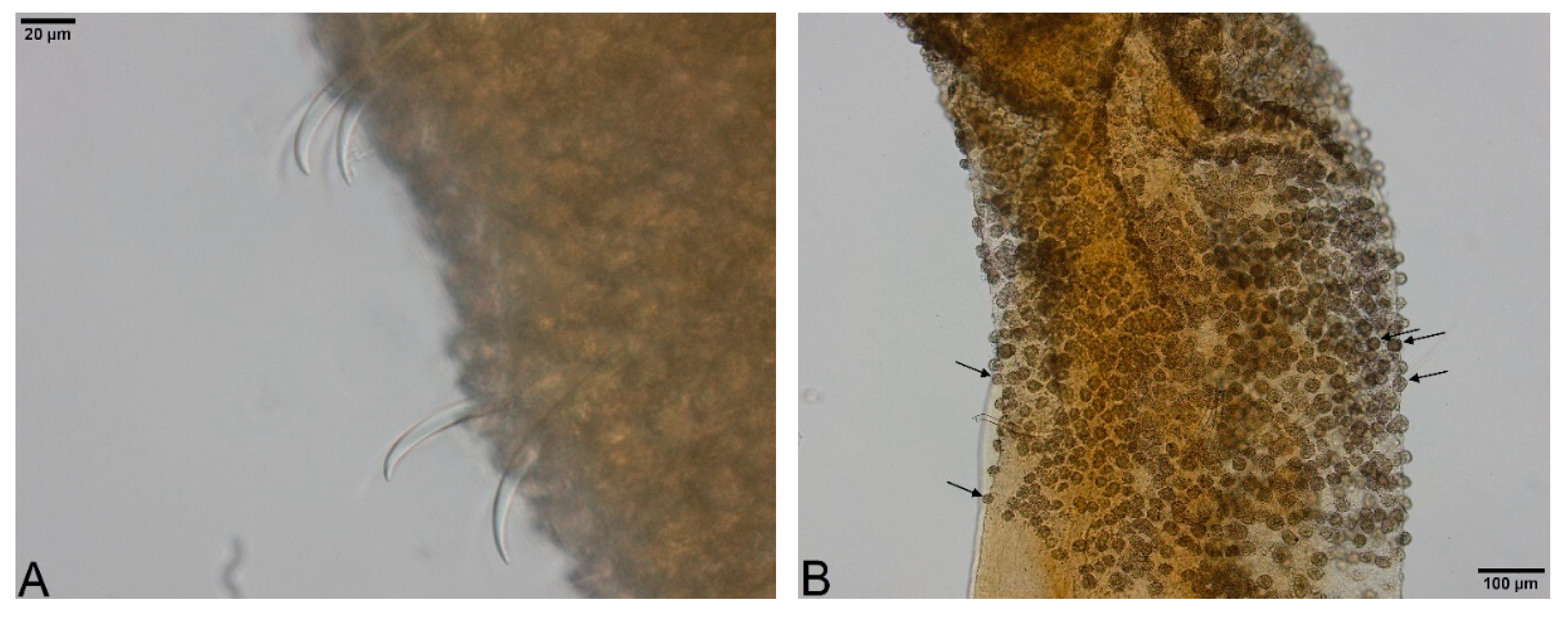
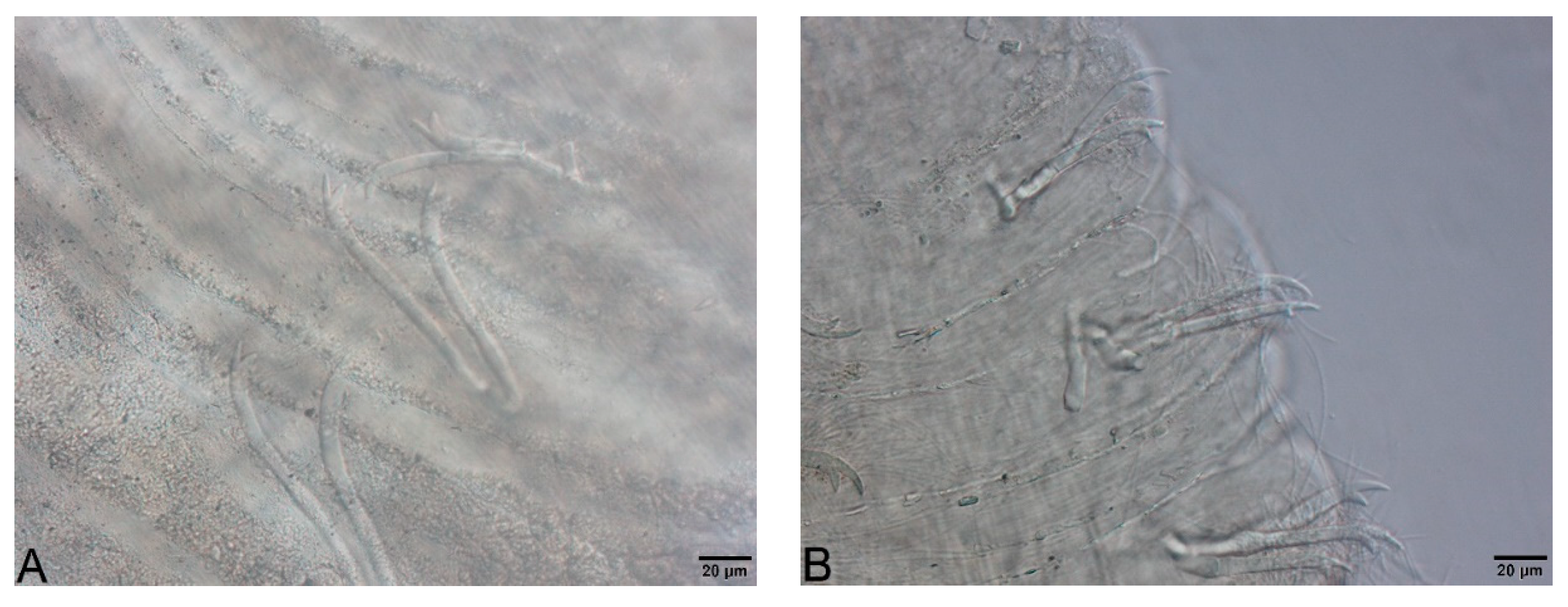
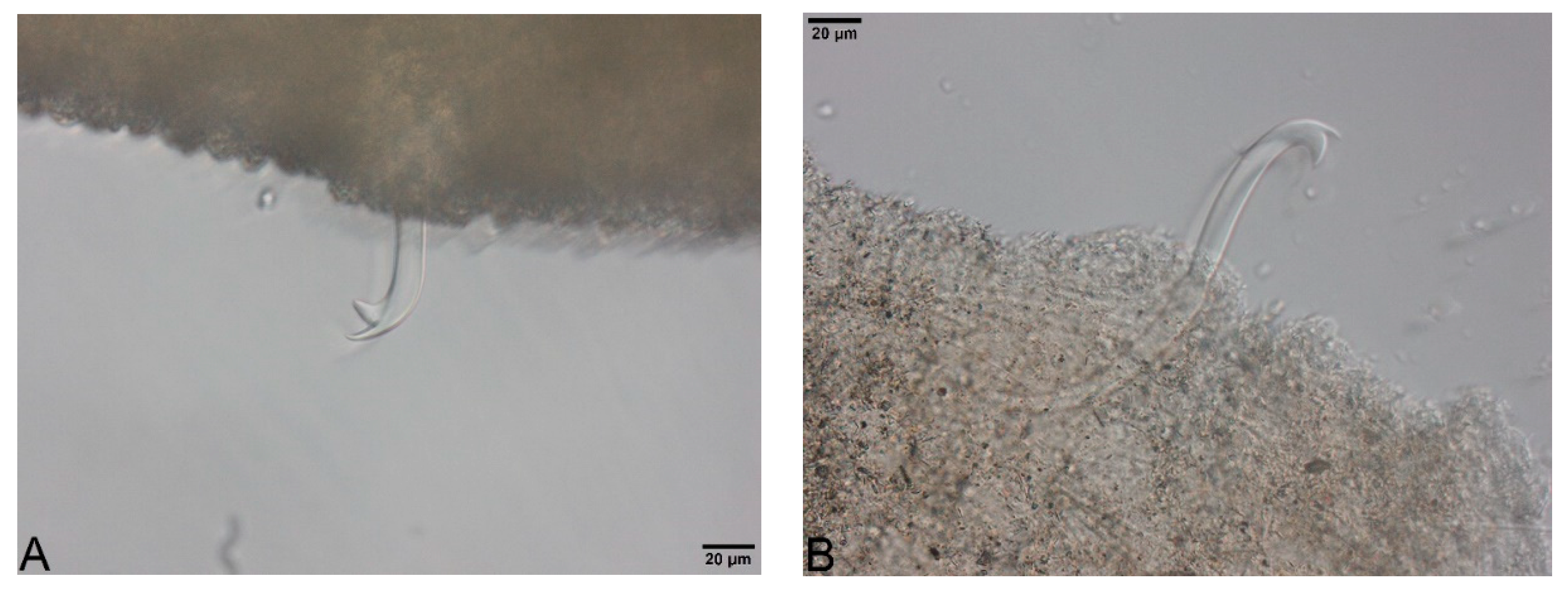
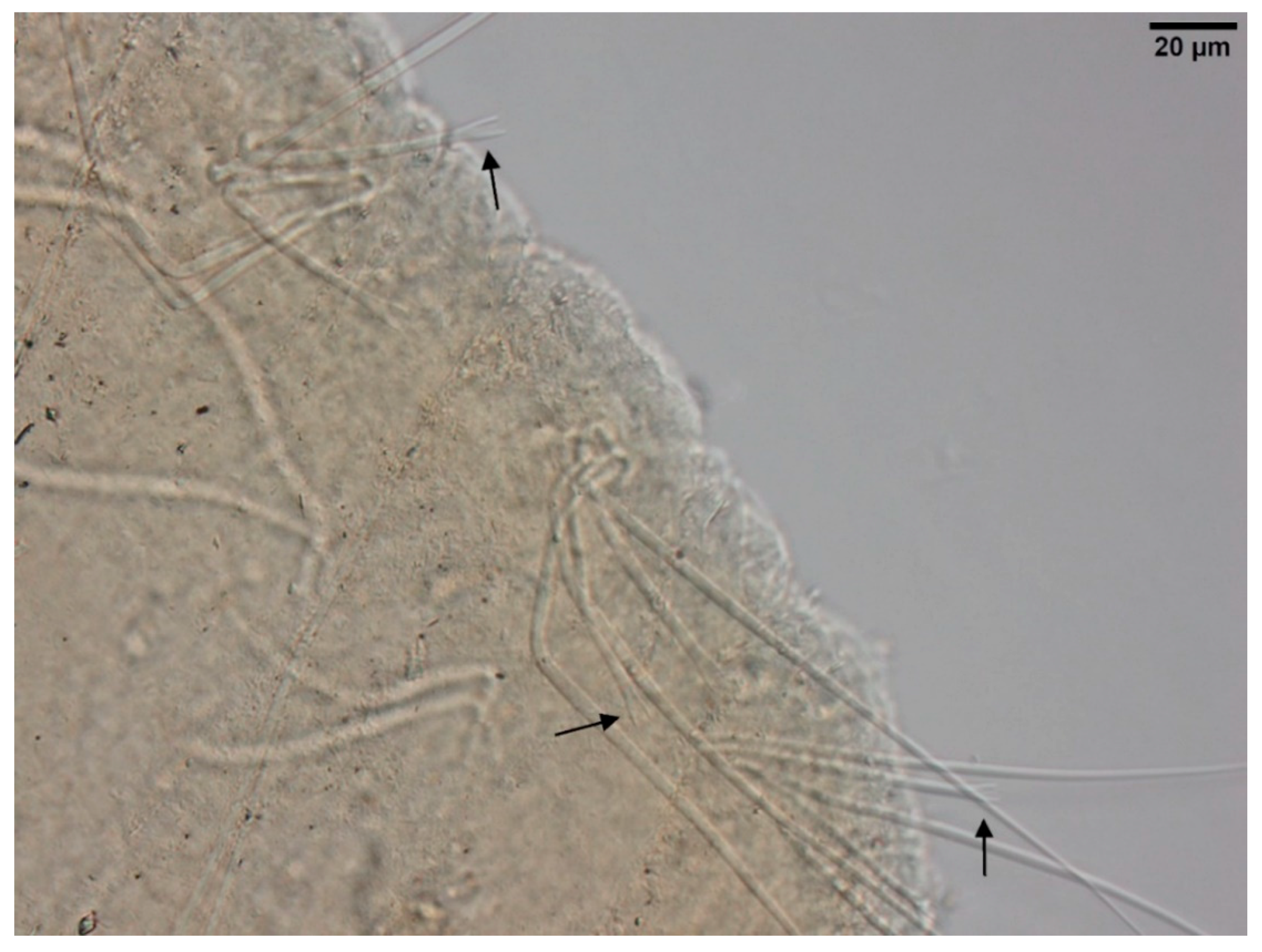
- Large, dark and roundish particle aggregates arranged randomly, covering the whole body surface, often hiding the chaetae (Figure 3A); Irregular and small dark particle aggregates arranged in transversal lines in some parts of the body surface, but almost always completely hidden by the large dark particle aggregates; In ventral bundles, simple-pointed and finely bifid chaetae (Figure 4A); In anterior dorsal bundles, chaetae are bifid with short inconspicuous teeth, these chaetae are mostly hidden by the large dark particle aggregates; Presence of prominent light sensory papillae arranged in a transversal row in each segment on the chaetal line but hidden by the large dark particle aggregates; Prostomium not elongated (Figure 3A) Embolocephalus velutinus *Shape of the chaetae different, all the ventral chaetae bifid 2
- Large, dark and roundish particle aggregates similar to those of E. velutinus, arranged randomly, covering a large part of the body surface (Figure 4B); Irregular and small dark particle aggregates arranged in transversal lines in some parts of the body surface, often also present in the anterior part (Figure 3B); Absence of prominent light sensory papillae; Prostomium not or slightly elongated (Figure 3B); In anterior dorsal bundles, pectinate lyre-shaped chaetae with short teeth (Figure 5); In anterior ventral bundles, chaetae are bifid with upper tooth as long or 1.5-fold longer than the lower one (Figure 6A); In posterior ventral bundles, chaetae are bifid with a large lower tooth and a thin upper tooth (Figure 7A); Posterior ventral chaetae sometimes absent or inconspicuous (hidden by the large dark particle aggregates) in some segments Spirosperma ferox *
- Irregular and small dark particle aggregates arranged in transversal lines in some parts of the body surface (Figure 3C); Sometimes, presence of large, dark and roundish particle aggregates on the body surface, but few and localized; Presence of prominent light sensory papillae arranged in a transversal row in each segment on the chaetal line (Figure 8A,B) but often not well visible on fixed specimens; Prostomium elongated (Figure 3C); In anterior dorsal bundles, pectinate chaetae with long and straight teeth (Figure 9); In anterior ventral bundles, chaetae are bifid with upper tooth generally 1.5 to 2.5 fold longer than the lower tooth (Figure 6B); In posterior ventral bundles, chaetae are bifid and strongly sigmoid, with a large and curved lower tooth and a thinner and shorter upper tooth (Figure 7B); Posterior ventral chaetae always present and well visible in each segment Quistadrilus multisetosus* one specimen of S. ferox and one specimen of E. velutinus without any large dark and roundish particle aggregates were found in Lake Geneva; the specimen of E. velutinus presented small dark particle aggregates arranged in transversal lines in some parts of the body surface.
3.3. Examination of Specimens from Collections
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Holmquist, C. Revision of the genus Peloscolex (Oligochaeta, Tubificidae): 2. Scrutiny of the species. Zool. Scr. 1976, 8, 37–60. [Google Scholar] [CrossRef]
- Spencer, D.R.; Hudson, P.L. The Oligochaeta (Annelida, Clitellata) of the St. Lawrence Great Lakes region: An update. J. Great Lakes Res. 2003, 29, 89–104. [Google Scholar] [CrossRef]
- Lafont, M. Contribution à La Gestion Des Eaux Continentales: Utilisation Des Oligochètes Comme Descripteurs De L’état Biologique Et Du Degré De Pollution Des Eaux Et Des Sédiments. Ph.D. Thesis, UCBL, Lyon, France, 1989. [Google Scholar]
- Dumnicka, E. Alien Naididae species (Annelida: Clitellata) and their role in aquatic habitats in Poland. Biologia 2016, 71, 16–23. [Google Scholar] [CrossRef]
- Vetricek, S.; Sporka, F. First record of Quistadrilus multisetosus (Tibificidae, Oligochaeta) (Smith, 1900) from the Czech Republic. Lauterbornia 2016, 81, 21–26. [Google Scholar]
- Pryanichnikova, E.G.; Perova, S.N.; Semernoy, V.P. First finding of Quistadrilus multisetosus (Smith, 1900) (Oligochaeta: Tubificidae) in the Rybinsk Reservoir. Inland Water Biol. 2017, 10, 328–330. [Google Scholar] [CrossRef]
- Perova, S.N.; Pryanichnikova, E.G.; Zhgareva, N.N. Appearance and Distribution of New Alien Macrozoobenthos Species in the Upper Volga Reservoirs. Russ. J. Biol. Invasions 2019, 10, 30–38. [Google Scholar] [CrossRef]
- Kasprzak, K. A new species of Tubificidae (Oligochaeta) found in Poland. Bull. Acad. Pol. Sci. 1971, 19, 261–267. [Google Scholar]
- Vivien, R.; Lafont, M. Note faunistique sur les oligochètes aquatiques de la région genevoise et de Suisse. Rev. Suisse Zool. 2015, 122, 207–2012. [Google Scholar]
- Aquatische Noezoen im Bodensee. Available online: http://www.neozoen-bodensee.de/aktuelles/neuer-schlammroehrenwurm-im-bodensee-quistadrilus-multisetosus (accessed on 8 October 2020).
- Brinkhurst, R.O.; Jamieson, B.G.M. Aquatic Oligochaeta of the World; Oliver and Boyd: Edinburgh, UK, 1971; p. 860. [Google Scholar]
- Timm, T. A guide to the freshwater oligochaeta and Polychaeta of Northern and Central Europe. Lauterbornia 2009, 66, 1–235. [Google Scholar]
- Timm, T.; Martin, P. Phylum Annelida. Class Clitellata: Subclass Oligochaeta. In Thorp and Covich’s Freshwater Invertebrates, 4th ed.; Keys to Palaearctic Fauna; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Lods-Crozet, B. Evolution du zoobenthos du Léman. Campagne 2009. Rapp. Comm. Int. Prot. Eaux Léman Contre Pollut. 2011, 143–151. [Google Scholar]
- Lods-Crozet, B.; Chevalley, P.A. Evolution du zoobenthos profond du Léman. Campagne 2015. Rapp. Comm. Int. Prot. Eaux Léman Contre Pollut. 2016, 132–141. [Google Scholar]
- Vivien, R.; Apothéloz-Perret-Gentil, L.; Pawlowski, P.; Werner, I.; Ferrari, B.J.D. High-throughput DNA barcoding of oligochaetes for abundance-based indices to assess the biological quality of sediments in streams and lakes. Sci. Rep. 2020, 10, 2041. [Google Scholar] [CrossRef] [PubMed]
- Loizeau, J.-L.; Makri, S.; Arpagaus, P.; Ferrari, B.; Casado-Martinez, C.; Benejam, T.; Marchand, P. Micropolluants métalliques et organiques dans les sédiments superficiels du Léman. Campagne 2016. Rapp. Comm. Int. Prot. Eaux Léman Contre Pollut. 2017, 153–207. [Google Scholar]
- Reymond, O. Préparations microscopiques permanentes d’oligochètes: Une meéthode simple. Bull. Soc. Vaud. Sci. Nat. 1994, 83, 1–3. [Google Scholar]
- Lang, C. Influence des rejets de la station d’épuration de Vidy sur la faune benthique du Léman. Int. Ver. Theor. Angew. Limnol. Verh. 1975, 19, 1182–1192. [Google Scholar] [CrossRef]
- Lods-Crozet, B.; Reymond, O. Ten years trends in the oligochaete and chironomid fauna of Lake Neuchâtel (Switzerland). Rev. Suisse Zool. 2005, 112, 543–558. [Google Scholar] [CrossRef]
- Erséus, C.; Gustafsson, D.R. Cryptic speciation in Clitellate model organism. In Annelids in Modern Biology; Shain, D.H., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 31–46. [Google Scholar]
- Zhou, H.; Fend, S.V.; Gustafson, D.L.; De Wit, P.; Erséus, C. Molecular phylogeny of Nearctic species of Rhynchelmis (Annelida). Zool. Scr. 2010, 39, 378–393. [Google Scholar] [CrossRef]
- Vivien, R.; Holzmann, M.; Werner, I.; Pawlowski, J.; Lafont, M.; Ferrari, B.J.D. Cytochrome c oxidase barcodes for aquatic oligochaete identification: Development of a Swiss reference database. PeerJ 2017, 5, e4122. [Google Scholar] [CrossRef]
- Prantoni, A.L.; Belmonte-Lopes, R.; Lana, P.C.; Erséus, C. Genetic diversity of marine oligochaetous clitellates in selected areas of the South Atlantic as revealed by DNA barcoding. Invertebr. Syst. 2018, 32, 524–532. [Google Scholar] [CrossRef]
- Elbrecht, V.; Vamos, E.E.; Meissner, K.; Aroviita, J.; Leese, F. Assessing strengths and weaknesses of DNA metabarcoding-based macroinvertebrate identification for routine stream monitoring. Meth. Ecol. Evol. 2017, 8, 1265–1275. [Google Scholar] [CrossRef]
- Xiong, W.; Li, H.; Zhan, A. Early detection of invasive species in marine ecosystems using high-throughput sequencing: Technical challenges and possible solutions. Mar. Biol. 2016, 163, 139. [Google Scholar] [CrossRef]
- Blackman, R.C.; Constable, D.; Hahn, C.; Sheard, A.M.; Durkota, J.; Hänfling, B.; Handley, L.L. Detection of a new non-native freshwater species by DNA metabarcoding of environmental samples—First record of Gammarus fossarum in the UK. Aquat. Invasions 2017, 12, 177–189. [Google Scholar] [CrossRef]
- Deiner, K.; Walser, J.C.; Mächler, E.; Altermatt, F. Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol. Conserv. 2015, 183, 53–63. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef]
- Tkach, V.; Pawlowski, J. A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol. 1999, 44, 147–148. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrigenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastMe 2.0: A comprehensive, accurate and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity–application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Esling, P.; Lejzerowicz, F.; Pawlowski, J. Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res. 2015, 243, 2513–2524. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, Y.; Lejzerowicz, F.; Perret-Gentil, L.A.; Pawlowski, J.; Cordier, T. SLIM: A flexible web application for the reproducible processing of environmental DNA metabarcoding data. BMC Bioinform. 2019, 20, 88. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for Illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Howmiller, R.P.; Scott, M.A. An environmental index based on relative abundance of oligochaete species. J. Water Pollut. Control Fed. 1977, 49, 809–815. [Google Scholar]
- Lafont, M.; Tixier, G.; Marsalek, J.; Jézéquel, C.; Breil, P.; Schmitt, L. From research to operational biomonitoring of freshwaters: A suggested conceptual framework and practical solutions. Ecohydrol. Hydrobiol. 2012, 12, 9–20. [Google Scholar] [CrossRef][Green Version]
- Juget, J. La faune benthique du Léman: Modalités et déterminismes écologiques du peuplement. Ph.D. Thesis, University of Lyon, Lyon, France, 1967. [Google Scholar]
- Rapin, F.; Gerdeaux, D. La protection du Léman: Priorité à la lutte contre l’eutrophisation. Arch. Sci. 2013, 66, 103–116. [Google Scholar]
- Biol’Eau. Macroinvertébrés Benthiques des Rives Genevoises du Léman–Investigations 2017; 2018; 45p. Available online: https://www.ge.ch/document/12828/telecharger (accessed on 15 November 2020).
- Timm, T.; Martin, P.J. Clitellata: Oligochaeta. In Ecology and General Biology: Thorp and Covich’s Freshwater Invertebrates; Thorp, J., Rogers, D.C., Eds.; Academic Press: Cambridge, UK, 2015; pp. 529–549. [Google Scholar]
- Lang, C.; Lang-Dobler, B. Structure of tubificid and lumbriculid worm communities, and three indices of trophy based upon these communities, as descriptors of eutrophication level of Lake Geneva (Switzerland). In Aquatic Oligochaete Biology; Brinkhurst, R.O., Cook, D.G., Eds.; Plenum Publishing Corporation: New York, NY, USA, 1980; pp. 457–470. [Google Scholar]
- Lang, C. Phosphorus decreases in Lake Geneva but climate warming hampers the recovery of pristine oligochaete communities whereas chironomids are less affected. J. Limnol. 2016, 75, 377–391. [Google Scholar] [CrossRef][Green Version]





| Site | Location | Date | Coordinates | Depth (m) | No Subsamples | No Specimens Identified | |
|---|---|---|---|---|---|---|---|
| x | y | ||||||
| 38 | St Prex | 20 April 2015 | 2,526,000 | 1,149,000 | 21 | 5 | 467 |
| 32 | Buchillon | 20 April 2015 | 2,522,000 | 1,146,600 | 22 | 5 | 220 |
| 30 | 20 April 2015 | 2,519,295 | 1,139,643 | 151 | 5 | 446 | |
| 35 | 20 April 2015 | 2,523,230 | 1,144,720 | 149 | 5 | 295 | |
| 49 | 20 April 2015 | 2,534,000 | 1,144,000 | 309 | 5 | 107 | |
| 58 | 20 April 2015 | 2,539,000 | 1,145,000 | 309 | 5 | 162 | |
| 32 | Buchillon | 26 October 2017 | 2,521,999 | 1,146,600 | 20–25 | 3 | 100 |
| 53 | baie de Vidy | 26 October 2017 | 2,534,721 | 1,151,336 | 42–44 | 3 | 100 |
| 78 | Grangettes | 26 October 2017 | 2,558,140 | 1,139,994 | 70 | 3 | 100 |
| 1 | Vengeron | 04 June 17 | 2,501,201 | 1,122,347 | 10 | 3 | 100 |
| 6 | Mies | 22 May 2018 | 2,503,999 | 1,127,985 | 54 | 3 | 100 |
| 21 | Yvoire | 22 May 2018 | 2,514,799 | 1,137,100 | 52 | 3 | 100 |
| 36 | Thonon | 22 May 2018 | 2,524,002 | 1,135,995 | 32 | 3 | 100 |
| 4 | baie de Vidy | 18 October 2016 | 2,535,725 | 1,151,289 | 24 | 3 | 100 |
| 3 | baie de Vidy | 17 October 2016 | 2,535,718 | 1,151,070 | 46 | 3 | 100 |
| 5 | baie de Vidy | 18 October 2016 | 2,535,699 | 1,150,839 | 60 | 3 | 100 |
| 2 | baie de Vidy | 18 October 2016 | 2,535,684 | 1,150,606 | 76 | 3 | 100 |
| 15 | baie de Vidy | 20 October 2016 | 2,535,592 | 1,149,562 | 188 | 3 | 100 |
| 90 | St Sulpice | 15 August 2019 | 2,531,708 | 1,150,583 | 14 | 3 | 100 |
| 91 | Cully | 28 August 2019 | 2,545,282 | 1,148,504 | 15 | 3 | 100 |
| 92 | Chevrens | 2009 | 2,506,000 | 1,128,000 | 70 | 3 | 235 |
| 93 | Coppet | 2009 | 2,504,340 | 1,129,700 | 20–22 | 3 | 418 |
| 94 | 2009 | 2,504,885 | 1,129,170 | 40 | 3 | 358 | |
| 95 | Tougues | 2009 | 2,507,500 | 1,131,500 | 70 | 3 | 228 |
| 96 | Founex | 2009 | 2,505,360 | 1,132,000 | 20 | 3 | 281 |
| 97 | 2009 | 2,505,685 | 1,131,940 | 40 | 3 | 232 | |
| 98 | Nernier | 2009 | 2,510,800 | 1,136,350 | 70 | 5 | 209 |
| 99 | Nyon | 2009 | 2,508,200 | 1,137,000 | 20 | 5 | 341 |
| 100 | 2009 | 2,508,605 | 1,136,880 | 40 | 5 | 299 | |
| Morphological Characters | Spirosperma ferox | Quistadrilus multisetosus |
|---|---|---|
| Prominent light sensory papillae | Absent | Present but often not well visible on fixed specimens |
| Large dark and roundish particle aggregates on the body surface | Present and abundant on all or a large part of the body * | Absent or few and localized |
| Small dark particle aggregates arranged in transversal lines on the body surface | Present, often hidden by the large dark particle aggregates | Present and generally conspicuous |
| Prostomium | Flattened, rarely slightly elongated | Always elongated |
| Anterior dorsal chaetae | Lyre-shaped and short teeth | Long and straight teeth |
| Anterior ventral chaetae | Upper tooth as long or 1.5-fold longer than the lower one | Upper tooth generally 1.5 to 2.5-fold longer than the lower one |
| Posterior ventral chaetae | Not strongly sigmoid; Lower tooth not or slightly curved and upper tooth as long or slightly shorter; | Strongly sigmoid; Curved lower tooth and shorter upper tooth; |
| Sometimes absent or hidden by the large dark particle aggregates in some segments | Always well visible in each segment |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vivien, R.; Lafont, M.; Lods-Crozet, B.; Holzmann, M.; Apothéloz-Perret-Gentil, L.; Guigoz, Y.; Ferrari, B.J.D. The Foreign Oligochaete Species Quistadrilus multisetosus (Smith, 1900) in Lake Geneva: Morphological and Molecular Characterization and Environmental Influences on Its Distribution. Biology 2020, 9, 436. https://doi.org/10.3390/biology9120436
Vivien R, Lafont M, Lods-Crozet B, Holzmann M, Apothéloz-Perret-Gentil L, Guigoz Y, Ferrari BJD. The Foreign Oligochaete Species Quistadrilus multisetosus (Smith, 1900) in Lake Geneva: Morphological and Molecular Characterization and Environmental Influences on Its Distribution. Biology. 2020; 9(12):436. https://doi.org/10.3390/biology9120436
Chicago/Turabian StyleVivien, Régis, Michel Lafont, Brigitte Lods-Crozet, Maria Holzmann, Laure Apothéloz-Perret-Gentil, Yaniss Guigoz, and Benoit J. D. Ferrari. 2020. "The Foreign Oligochaete Species Quistadrilus multisetosus (Smith, 1900) in Lake Geneva: Morphological and Molecular Characterization and Environmental Influences on Its Distribution" Biology 9, no. 12: 436. https://doi.org/10.3390/biology9120436
APA StyleVivien, R., Lafont, M., Lods-Crozet, B., Holzmann, M., Apothéloz-Perret-Gentil, L., Guigoz, Y., & Ferrari, B. J. D. (2020). The Foreign Oligochaete Species Quistadrilus multisetosus (Smith, 1900) in Lake Geneva: Morphological and Molecular Characterization and Environmental Influences on Its Distribution. Biology, 9(12), 436. https://doi.org/10.3390/biology9120436




