Digital PCR: What Relevance to Plant Studies?
Abstract
Simple Summary
Abstract
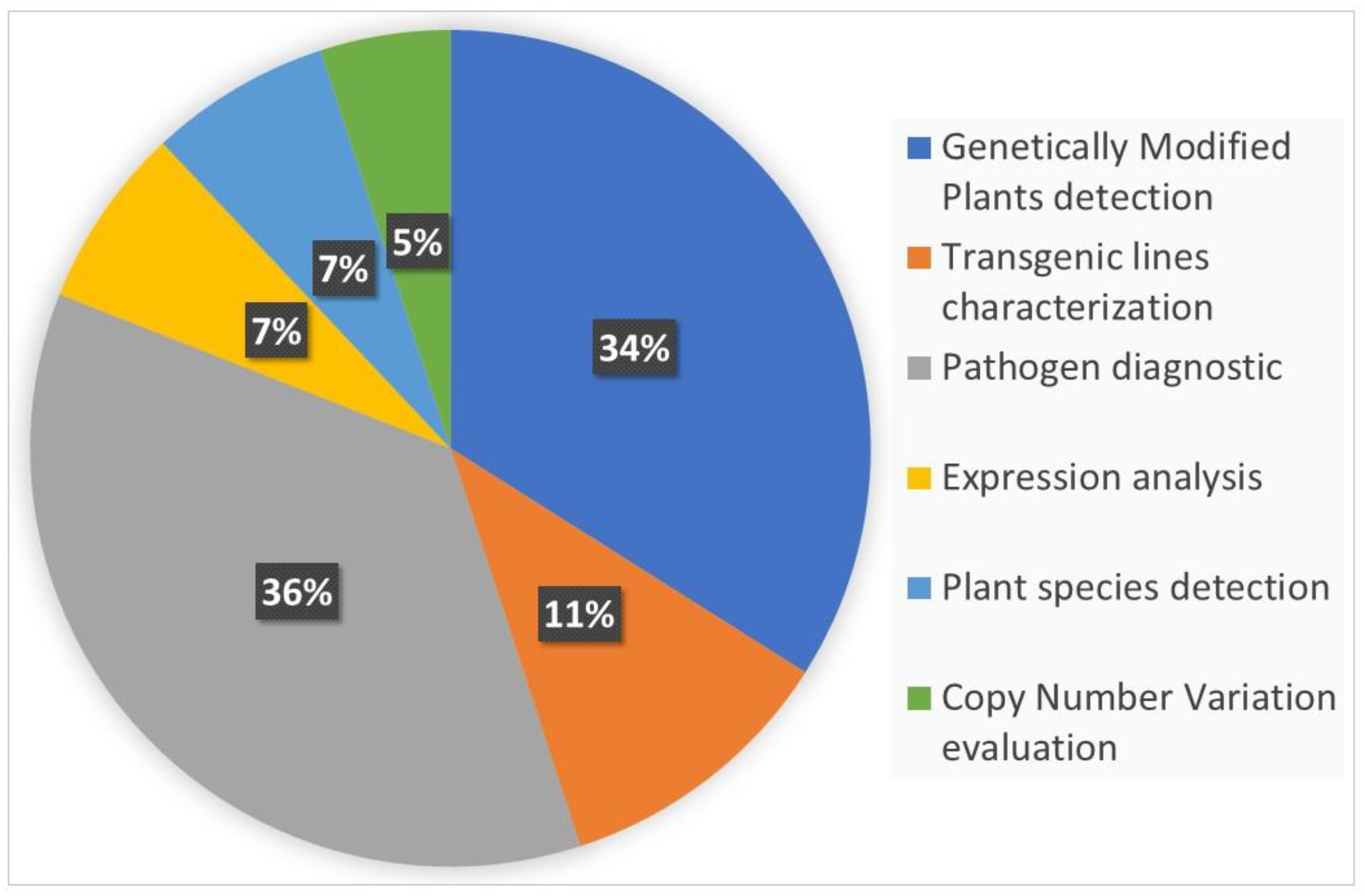
1. Introduction
2. Genetically Modified Plants Detection
- Sample preparation and DNA extraction.
- Digital PCR analysis for the amplification of the target transgenic sequence.
- Digital PCR analysis for amplification of a native, reference sequence.
- Data evaluation.
3. Transgenic Lines Characterization
4. Expression Analysis and Regulation
5. Plant Species Traceability
6. Phytopathogens Diagnostics
7. Other dPCR Applications
8. Minimum Information for Publication of Quantitative Digital PCR Experiments
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hansen, C.; Quake, S.R. Solving the “world-to-chip” interface problem with a microfluidic matrix. Anal. Chem. 2003, 75, 4718–4723. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.; Huang, Y. Digital PCR: Endless Frontier of ‘Divide and Conquer’. Micromachines 2017, 8, 231. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.; Mingam, A.; Charrier, B. Real-time PCR: What relevance to plant studies? J. Exp. Bot. 2004, 55, 1445–1454. [Google Scholar] [CrossRef]
- ISAAA. Global Status of Commercialized Biotech/GM Crops 2018, ISAABrief n. 54. Available online: http://www.isaaa.org/resources/publications/briefs/54/executivesummary/default.asp (accessed on 28 October 2020).
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef]
- Corbisier, P.; Barbante, A.; Berben, G.; Broothaerts, W.; De Loose, M.; Emons, H.; Georgieva, T.; Lievens, A.; Mazzara, M.; Papazova, N.; et al. Recommendation for the unit of measurement and the measuring system to report traceable and comparable results expressing GM content in accordance with EU legislation. EU Publ. 2017. [Google Scholar] [CrossRef]
- Zhu, P.; Fu, W.; Wang, C.; Du, Z.; Huang, K.; Zhu, S.; Xu, W. Development and application of absolute quantitative detection by duplex chamber-based digital PCR of genetically modified maize events without pretreatment steps. Anal. Chim. Acta 2016, 916, 60–66. [Google Scholar] [CrossRef]
- Deng, T.; Huang, W.; Ren, J.; Ma, X.; Ge, Y.; Chen, Y. Verification and applicability of endogenous reference genes for quantifying GM rice by digital PCR. Anal. Biochem. 2019, 587, 113442. [Google Scholar] [CrossRef]
- Iwobi, A.; Gerdes, L.; Busch, U.; Pecoraro, S. Droplet digital PCR for routine analysis of genetically modified foods (GMO)–A comparison with real-time quantitative PCR. Food Control 2016, 69, 205–213. [Google Scholar] [CrossRef]
- Wang, X.; Tang, T.; Miao, Q.; Xie, S.; Chen, X.; Tang, J.; Xu, J. Detection of transgenic rice line TT51-1 in processed foods using conventional PCR, real-time PCR, and droplet digital PCR. Food Control 2019, 98, 380–388. [Google Scholar] [CrossRef]
- Gerdes, L.; Iwobi, A.; Busch, U.; Pecoraro, S. Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol. Det. Quant. 2016, 7, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Demeke, T.; Gräfenhan, T.; Holigroski, M.; Fernando, U.; Bamforth, J.; Lee, S.J. Assessment of droplet digital PCR for absolute quantification of genetically engineered OXY235 canola and DP305423 soybean samples. Food Control 2014, 46, 470–474. [Google Scholar] [CrossRef]
- Demeke, T.; Eng, M. Effect of endogenous reference genes on digital PCR assessment of genetically engineered canola events. Biomol. Det. Quant. 2018, 15, 24–29. [Google Scholar] [CrossRef]
- Köppel, R.; Bucher, T.; Frei, A.; Waiblinger, H.U. Droplet digital PCR versus multiplex real-time PCR method for the detection and quantification of DNA from the four transgenic soy traits MON87769, MON87708, MON87705 and FG72, and lectin. Eur. Food Res. Technol. 2015, 241, 521–527. [Google Scholar] [CrossRef]
- Köppel, R.; Bucher, T.; Bär, D.; van Velsen, F.; Ganeshan, A. Validation of 13 duplex droplet digital PCR systems for quantitative GMO analysis of most prevalent GMO traits. Eur. Food Res. Technol. 2018, 244, 313–321. [Google Scholar] [CrossRef]
- Xu, X.; Peng, C.; Wang, X.; Chen, X.; Wang, Q.; Xu, J. Comparison of droplet digital PCR with quantitative real-time PCR for determination of zygosity in transgenic maize. Transgenic Res. 2016, 25, 855–864. [Google Scholar] [CrossRef]
- Sun, Y.; Joyce, P.A. Application of droplet digital PCR to determine copy number of endogenous genes and transgenes in sugarcane. Plant Cell Rep. 2017, 36, 1775–1783. [Google Scholar] [CrossRef]
- Wan, J.; Song, L.; Wu, Y.; Brzoska, P.; Keys, D.; Chen, C.; Nguyen, H.T. Application of digital PCR in the analysis of transgenic soybean plants. Adv. Biosci. Biotech. 2016, 7, 403–417. [Google Scholar] [CrossRef]
- Dobnik, D.; Spilsberg, B.; Bogožalec Košir, A.; Holst-Jensen, A.; Žel, J. Multiplex quantification of 12 European Union authorized genetically modified maize lines with droplet digital polymerase chain reaction. Anal. Chem. 2015, 87, 8218–8226. [Google Scholar] [CrossRef]
- Collier, R.; Dasgupta, K.; Xing, Y.P.; Hernandez, B.T.; Shao, M.; Rohozinski, D.; McCue, K.F. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 2017, 90, 1014–1025. [Google Scholar] [CrossRef]
- Giraldo, P.A.; Cogan, N.O.; Spangenberg, G.C.; Smith, K.F.; Shinozuka, H. Development and application of droplet digital PCR tools for the detection of transgenes in pastures and pasture-based products. Front. Plant Sci. 2019, 9, 1923. [Google Scholar] [CrossRef]
- Gao, R.; Feyissa, B.A.; Croft, M.; Hannoufa, A. Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta 2018, 247, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Wang, C.; Huang, K.; Luo, Y.; Xu, W. A novel pretreatment-free duplex chamber digital PCR detection system for the absolute quantitation of GMO samples. Int. J. Mol. Sci. 2016, 17, 402. [Google Scholar] [CrossRef] [PubMed]
- Morisset, D.; Štebih, D.; Milavec, M.; Gruden, K.; Žel, J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS ONE 2013, 8, e62583. [Google Scholar] [CrossRef] [PubMed]
- Głowacka, K.; Kromdijk, J.; Leonelli, L.; Niyogi, K.K.; Clemente, T.E.; Long, S.P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 2016, 39, 908–917. [Google Scholar] [CrossRef]
- Gao, H.; Yu, X.; Deng, T.; Sun, M.; Xiao, X.; Huang, X.; Li, R. Event-specific detection of transgenic potato AV43-6-G7 using real-time and digital PCR methods. BMC Biotechnol. 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Košir, A.B.; Spilsberg, B.; Holst-Jensen, A.; Žel, J.; Dobnik, D. Development and inter-laboratory assessment of droplet digital PCR assays for multiplex quantification of 15 genetically modified soybean lines. Sci. Rep. 2017, 7, 8601. [Google Scholar] [CrossRef]
- Fu, W.; Zhu, P.; Wang, C.; Huang, K.; Du, Z.; Tian, W.; Zhu, S. A highly sensitive and specific method for the screening detection of genetically modified organisms based on digital PCR without pretreatment. Sci. Rep. 2015, 5, 12715. [Google Scholar] [CrossRef]
- Dobnik, D.; Štebih, D.; Blejec, A.; Morisset, D.; Žel, J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci. Rep. 2016, 6, 35451. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Thomson, J.G.; Thilmony, R. A versatile and robust Agrobacterium-based gene stacking system generates high-quality transgenic Arabidopsis plants. Plant J. 2018, 95, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Paternò, A.; Verginelli, D.; Bonini, P.; Misto, M.; Quarchioni, C.; Dainese, E.; Marchesi, U. In-house validation and comparison of two wheat (Triticum aestivum) taxon-specific real-time PCR methods for GMO quantification supported by droplet digital PCR. Food Anal. Methods 2018, 11, 1281–1290. [Google Scholar] [CrossRef]
- Félix-Urquídez, D.; Pérez-Urquiza, M.; Valdez Torres, J.B.; León-Félix, J.; García-Estrada, R.; Acatzi-Silva, A. Development, Optimization, and Evaluation of a Duplex Droplet Digital PCR Assay To Quantify the T-nos/hmg Copy Number Ratio in Genetically Modified Maize. Anal Chem. 2016, 88, 812–819. [Google Scholar] [PubMed]
- Köppel, R.; Peterseil, V.; Dagand, E.; Schütz, E.; Kolberg, N.; Milavec, M.; Moor, D. Collaborative trial to assess the performance of digital PCR in the field of GMO analysis using an artificial sample material. Eur. Food Res. Technol. 2017, 243, 1091–1096. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Li, X.; Liang, J.; Li, Y.; Zeng, X.; Wu, G. Determining copy number of exogenous DNA and zygosity in transgenic rapeseed by droplet digital PCR. Oil Crop Sci. 2017, 1, 84–94. [Google Scholar]
- Liu, J.; Li, Z.Y.; Dong, J.; Gao, D.W. A universal quantification of transgenic soybean event DAS-68416-4 using duplex digital PCR. J. Sci. Food Agric. 2020. [Google Scholar] [CrossRef]
- Grelewska-Nowotko, K.; Żurawska-Zajfert, M.; Żmijewska, E.; Sowa, S. Optimization and verification of droplet digital PCR even-specific methods for the quantification of GM maize DAS1507 and NK603. Appl. Biochem. Biotech. 2018, 185, 207–220. [Google Scholar] [CrossRef]
- Francia, E.; Morcia, C.; Pasquariello, M.; Mazzamurro, V.; Milc, J.A.; Rizza, F.; Terzi, V.; Pecchioni, N. Copy number variation at the HvCBF4-HvCBF2 genomic segment is a major component of frost resistance in barley. Plant Mol. Biol. 2016, 92, 161–175. [Google Scholar] [CrossRef]
- Lye, Z.N.; Purugganan, M.D. Copy number variation in domestication. Trends in Plant Science 2019, 24, 4. [Google Scholar] [CrossRef]
- Zmienko, A.; Samelak-Czajka, A.; Kozlowski, P.; Szymanska, M.; Figlerowicz, M. Arabidopsis thaliana population analysis reveals high plasticity of the genomic region spanning MSH2, AT3G18530 and AT3G18535 genes and provides evidence for NAHR-driven recurrent CNV events occurring in this location. BMC Genom. 2016, 17, 1–16. [Google Scholar] [CrossRef]
- McCord, P.H. Using droplet digital PCR (ddPCR) to detect copy number variation in sugarcane, a high-level polyploid. Euphytica 2016, 209, 439–448. [Google Scholar] [CrossRef]
- Jouanin, A.; Tenorio-Berrio, R.; Schaart, J.G.; Leigh, F.; Visser, R.G.; Smulders, M.J. Optimisation of droplet digital PCR for determining copy number variation of α-gliadin genes in mutant and gene-edited polyploid bread wheat. J. Cereal Sci. 2020, 92, 102903. [Google Scholar] [CrossRef]
- Lancíková, V.; Hricová, A. Digital absolute gene expression analysis of essential starch-related genes in a radiation developed Amaranthus cruentus L. variety in comparison with real-time PCR. Plants 2020, 9, 966. [Google Scholar] [CrossRef] [PubMed]
- Barone, V.; Bertoldo, G.; Magro, F.; Broccanello, C.; Puglisi, I.; Baglieri, A.; Nardi, S. Molecular and morphological changes induced by Leonardite-based biostimulant in Beta vulgaris L. Plants 2019, 8, 181. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Duarte, S.; Tedesco, S.; Fevereiro, P.; Costa, R.L. Expression profiling of Castanea genes during resistant and susceptible interactions with the oomycete pathogen Phytophthora cinnamomi reveal possible mechanisms of immunity. Front. Plant Sci. 2017, 8, 515. [Google Scholar] [CrossRef]
- Zmienko, A.; Samelak-Czajka, A.; Goralski, M.; Sobieszczuk-Nowicka, E.; Kozlowski, P.; Figlerowicz, M. Selection of reference genes for qPCR-and ddPCR-based analyses of gene expression in senescing barley leaves. PLoS ONE 2015, 10, e0118226. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, X.; Chen, W.; Bai, J.; Vasseur, L.; He, W.; You, M. Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS ONE 2019, 14, e0220475. [Google Scholar] [CrossRef]
- Morcia, C.; Bergami, R.; Scaramagli, S.; Ghizzoni, R.; Carnevali, P.; Terzi, V. A chip digital PCR assay for quantification of common wheat contamination in pasta production chain. Foods 2020, 9, 911. [Google Scholar] [CrossRef]
- Scollo, F.; Egea, L.A.; Gentile, A.; La Malfa, S.; Dorado, G.; Hernandez, P. Absolute quantification of olive oil DNA by droplet digital-PCR (ddPCR): Comparison of isolation and amplification methodologies. Food Chem. 2016, 213, 388–394. [Google Scholar] [CrossRef]
- Köppel, R.; Ledermann, R.; van Velsen, F.; Ganeshan, A.; Guertler, P. Duplex digital droplet PCR for the determination of apricot kernels in marzipan. Eur. Food Res. Technol. 2020, 246, 965–970. [Google Scholar] [CrossRef]
- Dong, X.; Gao, D.; Dong, J.; Chen, W.; Li, Z.; Wang, J.; Liu, J. Mass ratio quantitative detection for kidney bean in lotus seed paste using duplex droplet digital PCR and chip digital PCR. Anal. Bioanal. Chem. 2020, 412, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, Y.; Chen, C.; Zhang, Y.; Zhou, W.; Sang, Y. Identification and quantification of cassava starch adulteration in different food starches by droplet digital PCR. PLoS ONE 2020, 15, e0228624. [Google Scholar] [CrossRef] [PubMed]
- Mayer, W.; Schuller, M.; Viehauser, M.C.; Hochegger, R. Quantification of the allergen soy (Glycine max) in food using digital droplet PCR (ddPCR). Eur. Food Res. Technol. 2019, 245, 499–509. [Google Scholar] [CrossRef]
- Zhong, X.; Liu, X.L.; Lou, B.H.; Zhou, C.Y.; Wang, X.F. Development of a sensitive and reliable droplet digital PCR assay for the detection of ‘Candidatus Liberibacter asiaticus’. J. Integr. Agric. 2018, 17, 483–487. [Google Scholar] [CrossRef]
- Selvaraj, V.; Maheshwari, Y.; Hajeri, S.; Chen, J.; McCollum, T.G.; Yokomi, R. Development of a duplex droplet digital PCR assay for absolute quantitative detection of “Candidatus Liberibacter asiaticus”. PLoS ONE 2018, 13, e0197184. [Google Scholar] [CrossRef]
- Yu, L.U.; Zhang, H.J.; Zhao, Z.J.; Wen, C.L.; Ping, W.U.; Song, S.H.; Xu, X.L. Application of droplet digital PCR in detection of seed-transmitted pathogen Acidovorax citrulli. J. Integr. Agric. 2020, 19, 561–569. [Google Scholar]
- Bahder, B.W.; Helmick, E.E.; Mou, D.F.; Harrison, N.A.; Davis, R. Digital PCR technology for detection of palm-infecting phytoplasmas belonging to group 16SrIV that occur in Florida. Plant Dis. 2018, 102, 1008–1014. [Google Scholar] [CrossRef]
- Leichtfried, T.; Reisenzein, H.; Steinkellner, S.; Gottsberger, R.A. Transmission studies of the newly described apple chlorotic fruit spot viroid using a combined RT-qPCR and droplet digital PCR approach. Arch. Virol. 2020, 165, 2665–2671. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, Q.; Yin, Y.; Wang, Z. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri subsp. citri. PLoS ONE 2016, 11, e0159004. [Google Scholar] [CrossRef]
- Pandey, B.; Mallik, I.; Gudmestad, N.C. Development and application of a real-time reverse-transcription PCR and droplet digital PCR assays for the direct detection of Potato mop top virus in soil. Phytopathology 2020, 110, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Holland, T.; Bowen, P.; Kokkoris, V.; Urbez-Torres, J.R.; Hart, M. Does inoculation with arbuscular mycorrhizal fungi reduce trunk disease in grapevine rootstocks? Horticulturae 2019, 5, 61. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Wang, Q.; Zhang, Y.; Shen, W.; Li, R.; Zhou, Y. Development of a sensitive and reliable reverse transcription droplet digital PCR assay for the detection of citrus yellow vein clearing virus. Arch. Virol. 2019, 164, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Gonzalez, M.M.; Martínez Diz, M.D.P.; Andrés-Sodupe, M.; Bujanda, R.; Diaz-Losada, E.; Gramaje, D. Quantification of Cadophora luteo-olivacea from grapevine nursery stock and vineyard soil using droplet digital PCR. Plant Dis. 2020, 104, 2269–2274. [Google Scholar] [CrossRef]
- Morcia, C.; Tumino, G.; Gasparo, G.; Ceresoli, C.; Fattorini, C.; Ghizzoni, R.; Carnevali, P.; Terzi, V. Moving from qPCR to chip digital PCR assays for tracking of some Fusarium species causing Fusarium Head Blight in cereals. Microorganisms 2020, 8, 1307. [Google Scholar] [CrossRef]
- Voegel, T.M.; Nelson, L.M. Quantification of Agrobacterium vitis from grapevine nursery stock and vineyard soil using droplet digital PCR. Plant Dis. 2018, 102, 2136–2141. [Google Scholar] [CrossRef]
- Mehle, N.; Gregur, L.; Bogožalec Košir, A.; Dobnik, D. One-step reverse-transcription digital PCR for reliable quantification of different Pepino mosaic virus genotypes. Plants 2020, 9, 326. [Google Scholar] [CrossRef]
- Palumbo, J.D.; O’Keeffe, T.L.; Fidelibus, M.W. Characterization of Aspergillus section nigri species populations in vineyard soil using droplet digital PCR. Lett. Appl. Microbiol. 2016, 63, 458–465. [Google Scholar] [CrossRef]
- Blaya, J.; Lloret, E.; Santísima-Trinidad, A.B.; Ros, M.; Pascual, J.A. Molecular methods (digital PCR and real-time PCR) for the quantification of low copy DNA of Phytophthora nicotianae in environmental samples. Pest Manag. Sci. 2016, 72, 747–753. [Google Scholar] [CrossRef]
- Bahar, M.H.; Wist, T.J.; Bekkaoui, D.R.; Hegedus, D.D.; Olivier, C.Y. Aster leafhopper survival and reproduction, and Aster yellows transmission under static and fluctuating temperatures, using ddPCR for phytoplasma quantification. Sci. Rep. 2018, 8, 227. [Google Scholar] [CrossRef]
- Santander, R.D.; Meredith, C.L.; Aćimović, S.G. Development of a viability digital PCR protocol for the selective detection and quantification of live Erwinia amylovora cells in cankers. Sci. Rep. 2019, 9, 11530. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Yao, Z.; Liu, J.; Zhang, H.; Din, G.M.U.; Zhao, S.; Gao, L. Development of droplet digital PCR for the detection of Tilletia laevis, which causes common bunt of wheat, based on the SCAR marker derived from ISSR and real-time PCR. Sci. Rep. 2020, 10, 16106. [Google Scholar] [CrossRef] [PubMed]
- Ristaino, J.B.; Saville, A.C.; Paul, R.; Cooper, D.; Wei, Q. Detection of Phytophthora infestans by LAMP, real-time LAMP and droplet digital PCR. Plant Dis. 2019, 104, 708–716. [Google Scholar] [CrossRef]
- Gossen, B.D.; Al-Daoud, F.; Dumonceaux, T.; Dalton, J.A.; Peng, G.; Pageau, D.; McDonald, M.R. Comparison of techniques for estimation of resting spores of Plasmodiophora brassicae in soil. Plant Pathol. 2019, 68, 954–961. [Google Scholar] [CrossRef]
- Mehle, N.; Dobnik, D.; Ravnikar, M.; Novak, M.P. Validated reverse transcription droplet digital PCR serves as a higher order method for absolute quantification of Potato virus Y strains. Anal. Bioanal. Chem. 2018, 410, 3815–3825. [Google Scholar] [CrossRef]
- Maheshwari, Y.; Selvaraj, V.; Hajeri, S.; Yokomi, R. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLoS ONE 2017, 12, e0184751. [Google Scholar] [CrossRef] [PubMed]
- Dreo, T.; Pirc, M.; Ramšak, Ž.; Pavšič, J.; Milavec, M.; Zel, J.; Gruden, K. Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. J. Anal. Bioanal. Chem. 2014, 406, 6513–6528. [Google Scholar] [CrossRef]
- Hua, S.S.T.; Palumbo, J.D.; Parfitt, D.E.; Sarreal, S.B.L.; O’Keeffe, T.L. Development of a droplet digital PCR assay for population analysis of aflatoxigenic and atoxigenic Aspergillus flavus mixtures in soil. Mycotoxin Res. 2018, 34, 187–194. [Google Scholar] [CrossRef]
- Zulak, K.G.; Cox, B.A.; Tucker, M.A.; Oliver, R.P.; Lopez-Ruiz, F.J. Improved detection and monitoring of fungicide resistance in Blumeria graminis f. sp. hordei with high-throughput genotype quantification by digital PCR. Front. Microbiol. 2018, 9, 706. [Google Scholar]
- Xie, S.; Yu, H.; Wang, Q.; Cheng, Y.; Ding, T. Two rapid and sensitive methods based on TaqMan qPCR and droplet digital PCR assay for quantitative detection of Bacillus subtilis in rhizosphere. J. Appl. Microbiol. 2020, 128, 518–527. [Google Scholar] [CrossRef]
- Stevanato, P.; Biscarini, F. Digital PCR as New Approach to SNP Genotyping in Sugar Beet. Sugar Tech 2016, 18, 429–432. [Google Scholar] [CrossRef]
- Huggett, J.F. and dMIQE Group. The Digital MIQE Guidelines Update:Minimum Information for Publication of QuantitativeDigital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef] [PubMed]
- Pecoraro, S.; Berben, G.; Burns, M.; Corbisier, P.; De Giacomo, M.; De Loose, M.; Dagand, E.; Dobnik, D.; Eriksson, R.; Holst-Jensen, A.; et al. Overview and Recommendations for the Application of Digital PCR. EUR 29673 EN; Publications Office of the European Union: Luxembourg, 2019; ISBN 978-92-76-00180-5. JRC115736. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- White, R.A.; Blainey, P.C.; Fan, H.C.; Quake, S.R. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genom. 2009, 10, 116. [Google Scholar]
- GMO Methods Database. Available online: https://gmo-crl.jrc.ec.europa.eu/gmomethods/ (accessed on 25 November 2020).

| Instrumental Platform | Plant Species | Genetically Modified Line | Endogenous Reference Gene(s) | Bibliography |
|---|---|---|---|---|
| Chamber-based digital PCR | Zea mays | NK603, MON810, MON863, Bt176, 3272, MIR162, MIR604 | Adh and hmg | [10] |
| Droplet digital PCR | Oryza sativa | Kefeng-6 | Sps2, RBE4, and ppi-PPF | [11] |
| Droplet digital PCR | Zea mays Glycine max | MON88017, MON87460, MON89034, MIR162 CV127, MON87701, and MON87705 | - | [12] |
| Droplet digital PCR | Oryza sativa | TT51-1 | PLD | [13] |
| Droplet digital PCR | Glycine max | A2704-12, 356043, 305423, and 40-3-2 | Lec-1 | [14] |
| Droplet digital PCR | Brassica napus Glycine max | OXY235 DP305423 | hmg Lec-1 | [15] |
| Droplet digital PCR | Brassica napus | HCN92 | Cruciferin, CruA, FatA, and hmg-I/Y | [16] |
| Droplet digital PCR | Glycine max | MON87769, MON87708, MON87705, FG72 | Lec-1 | [17] |
| Droplet digital PCR | Glycine max, Zea mays | RR, MON89788, 2704, Bt176, Bt11, MON810, GA21, NK603, MON863, 59122, MIR604, TC1507, and T25 | Lec-1 hmg | [18] |
| Droplet digital PCR | Zea mays | IE034 | Adh | [19] |
| Droplet digital PCR | Saccharum officinarum | Q208 and Q240 | ACT | [20] |
| Digital PCR | Glycine max | 40-3-2, MON89788 | Lec | [21] |
| Droplet digital PCR | Zea mays | DAS1507, DAS59122, GA21, MIR162, MIR604, MON810, MON863, MON89034, NK603, T25, Bt11, and MON88017 | hmgA | [22] |
| Droplet digital PCR | Oryza sativa, Citrus, Solanum tuberosum, Zea mays, Lycopersicon esculentum, Triticum | Non-commercial plants | Rice-OsUBC, citrus-CsDHN, potato-StAAP2, maize-ZmADH1, tomato-SISYS, and wheat-PINb-D1b | [23] |
| Droplet digital PCR | Lolium | Non-commercial plant | LpCul4 | [24] |
| Droplet digital PCR | Medicago sativa | Non-commercial plants | - | [25] |
| Chamber-based digital PCR Droplet digital PCR | Zea mays | GA21 | Adh1 | [26] |
| Droplet digital PCR | Zea mays | MON810 | hmg | [27] |
| Droplet digital PCR | Nicotiana tabacum | Non-commercial plants | Ntactin and NtTubulin_1 | [28] |
| Droplet digital PCR | Solanum tuberosum | AV43-6-G7 | fru | [29] |
| Droplet digital PCR | Glycine max | 15 lines (authorized or with valid EFSA application) | Lec-1 | [30] |
| Chamber-based digital PCR Droplet digital PCR | Zea mays | MON810, MON863, TC1507, MIR604, MIR162, GA21, T25, NK603, and BT176 | - | [31] |
| Droplet digital PCR | Zea mays | MON863, MON810, DP98140, MIR604, GA21, MON89034, and MIR162 | hmgA | [32] |
| Droplet digital PCR | Arabidopsis thaliana | Non-commercial plant | AAP1 | [33] |
| Droplet digital PCR | Triticum | Non-commercial plant | ssII-D and waxy-D1 | [34] |
| Droplet digital PCR | Zea mays | Certified reference materials | hmg | [35] |
| Droplet digital PCR | Glycine max | multitarget DNA molecule encoding for eight transgene soy traits | Lec-1 | [36] |
| Droplet digital PCR | Brassica napus | Non-commercial transgenic lines | CruA | [37] |
| Droplet digital PCR | Glycine max | DAS-68416-4 | - | [38] |
| Droplet digital PCR | Zea mays | DAS1507 and NK603 | hmg and Adh1 | [39] |
| Target Microorganism | Disease | Affected Crop | Reference |
|---|---|---|---|
| Candidatus Liberibacter asiaticus | Huanglongbing (HLB; yellow shoot disease) | Citrus | [56] |
| Candidatus Liberibacter asiaticus | Huanglongbing (HLB; yellow shoot disease) | Citrus | [57] |
| Acidovorax citrulli | Bacterial fruit blotch | Cucurbitaceous | [58] |
| Group 16SrIV phytoplasmas | Lethal yellowing (LY) | Phoenix dactylifera | [59] |
| Apscaviroid (apple chlorotic fruit spot viroid—ACFSVd) | Chlorotic fruit spots and bump-like symptoms on the skin of apples | Malus | [60] |
| Xanthomonas citri subsp. citri | Citrus bacterial canker | Citrus | [61] |
| Potato mop top virus | Potato mop top disease (tuber necrosis, internode reduction, foliar yellow spots, and plant chlorosis) | Solanum tuberosum | [62] |
| Ilyonectria | Black foot disease | Vitis vinifera | [63] |
| Citrus yellow vein clearing virus (CYVCV) | Yellow vein disease | Citrus | [64] |
| Cadophora luteo-olivacea | Petri disease and esca of grapevine | Vitis vinifera | [65] |
| Fusarium graminearum, Fusarium culmorum, Fusarium sporotrichioides, Fusarium poae, Fusarium avenaceum | Fusarium head blight | Small grain cereals | [66] |
| Agrobacterium vitis | Crown gall | Vitis vinifera | [67] |
| Pepino mosaic virus (PepMV) | Fruit marbling, leaf, and stem necrosis | Lycopersicon esculentum | [68] |
| Aspergillus niger, Aspergillus welwitschiae, Aspergillus tubingensis, Aspergillus carbonarius | Bunch rots and mycotoxin production | Vitis vinifera | [69] |
| Phytophthora nicotianae | Root rot, crown rot, fruit rot, leaf infection, and stem infection | Nicotiana tabacum | [70] |
| Candidatus Phytoplasma asteris | Aster yellows (AY) | Brassica | [71] |
| Erwinia amylovora | Fire blight | Malus | [72] |
| Tilletia laevis | Common bunt | Triticum | [73] |
| Phytophtora infestans | Late blight | Solanum | [74] |
| Plasmodiophora brassicae | Clubroot | Brassica | [75] |
| Potato virus Y strains | Mosaic symptoms | Solanum | [76] |
| Spiroplasma citri | Citrus stubborn disease | Citrus | [77] |
| Erwinia amylovora and Ralstonia solanacearum | Fire blight of rosaceous plants, potato brown rot | Solanaceae, Rosaceae | [78] |
| Advantages |
|---|
| Absolute quantification, no need to rely on reference or standard for several applications |
| Sensitivity and accuracy, useful to detect rare and low copy number targets |
| Suitability for allelic variant detection |
| Applicable to complex mixtures and complex background |
| Resistance to PCR inhibitors |
| Linear response to number of copies |
| Disadvantages |
| More expensive compared to qPCR, although questionable |
| Limited dynamic range of detection |
| Problems with very large amplicons |
| More complex work-flow compared to qPCR |
| More expensive instrumentation compared to qPCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morcia, C.; Ghizzoni, R.; Delogu, C.; Andreani, L.; Carnevali, P.; Terzi, V. Digital PCR: What Relevance to Plant Studies? Biology 2020, 9, 433. https://doi.org/10.3390/biology9120433
Morcia C, Ghizzoni R, Delogu C, Andreani L, Carnevali P, Terzi V. Digital PCR: What Relevance to Plant Studies? Biology. 2020; 9(12):433. https://doi.org/10.3390/biology9120433
Chicago/Turabian StyleMorcia, Caterina, Roberta Ghizzoni, Chiara Delogu, Lorella Andreani, Paola Carnevali, and Valeria Terzi. 2020. "Digital PCR: What Relevance to Plant Studies?" Biology 9, no. 12: 433. https://doi.org/10.3390/biology9120433
APA StyleMorcia, C., Ghizzoni, R., Delogu, C., Andreani, L., Carnevali, P., & Terzi, V. (2020). Digital PCR: What Relevance to Plant Studies? Biology, 9(12), 433. https://doi.org/10.3390/biology9120433







