Glutathione Restores Hg-Induced Morpho-Physiological Retardations by Inducing Phytochelatin and Oxidative Defense in Alfalfa
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Culture and Treatment
2.2. Determination of Growth and Photosynthesis Parameters
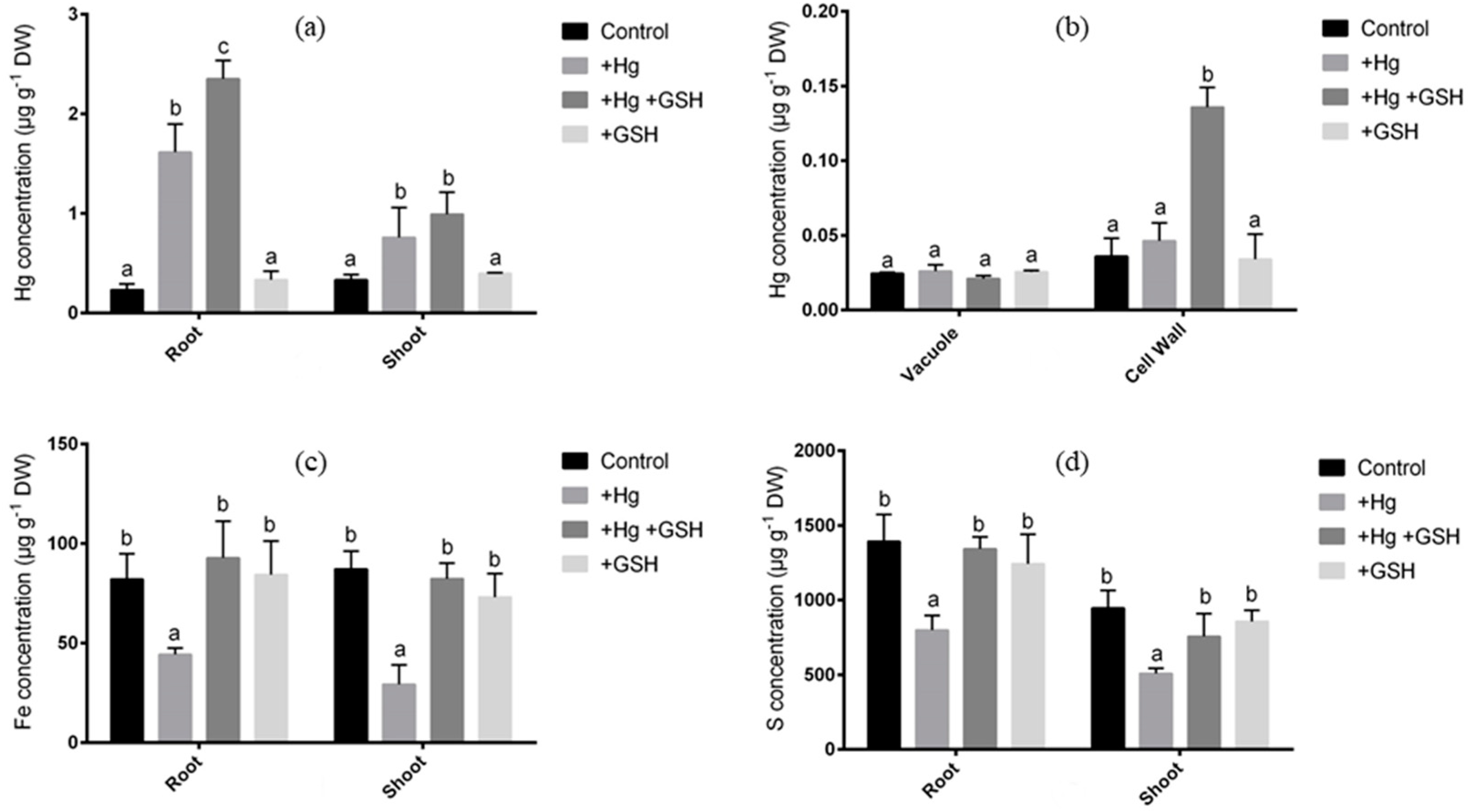
2.3. Determination of Elemental Concentration in Root, Shoot, Vacuole, and Cell Wall
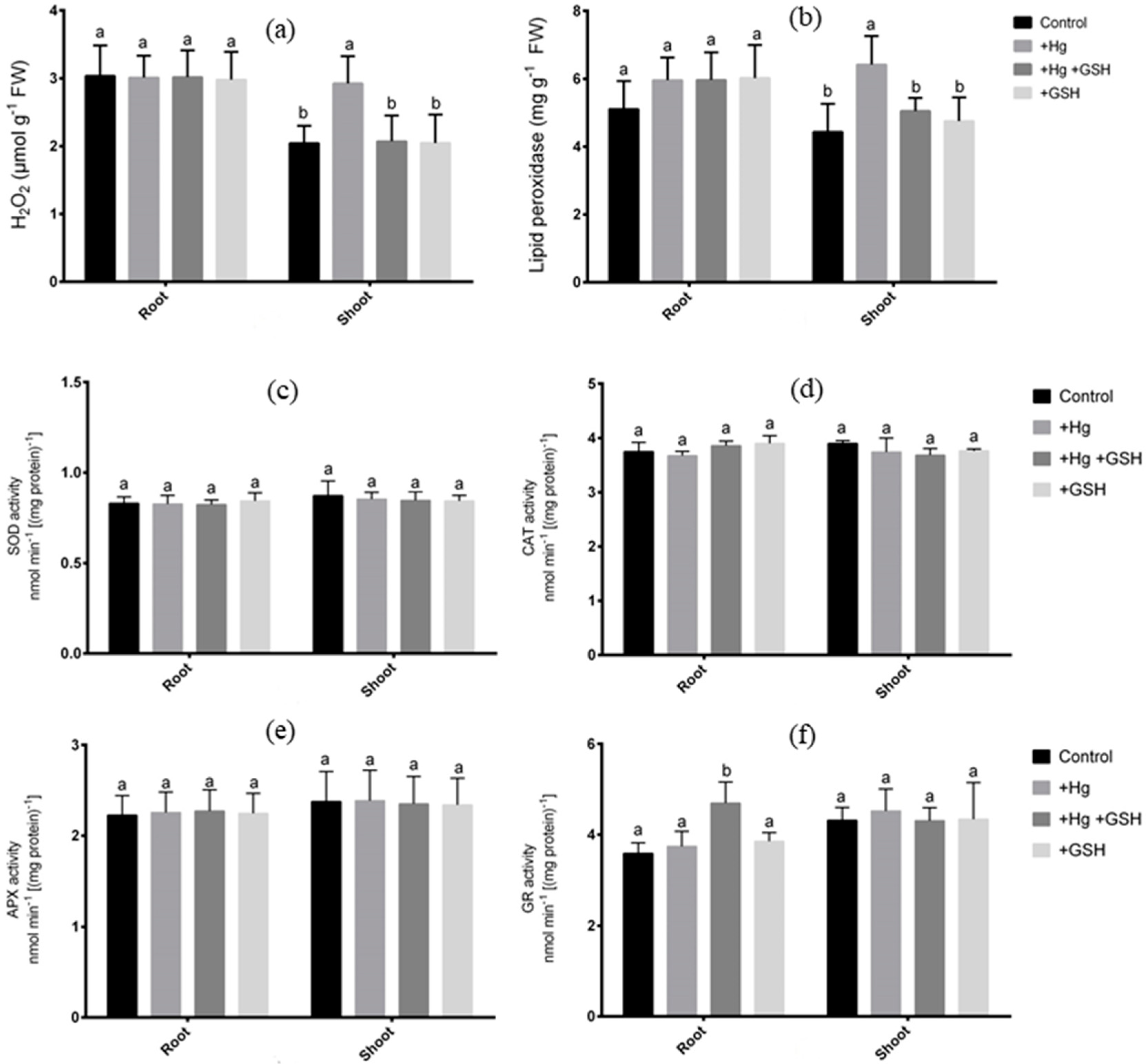
2.4. Measurement of MDA and H2O2 Levels
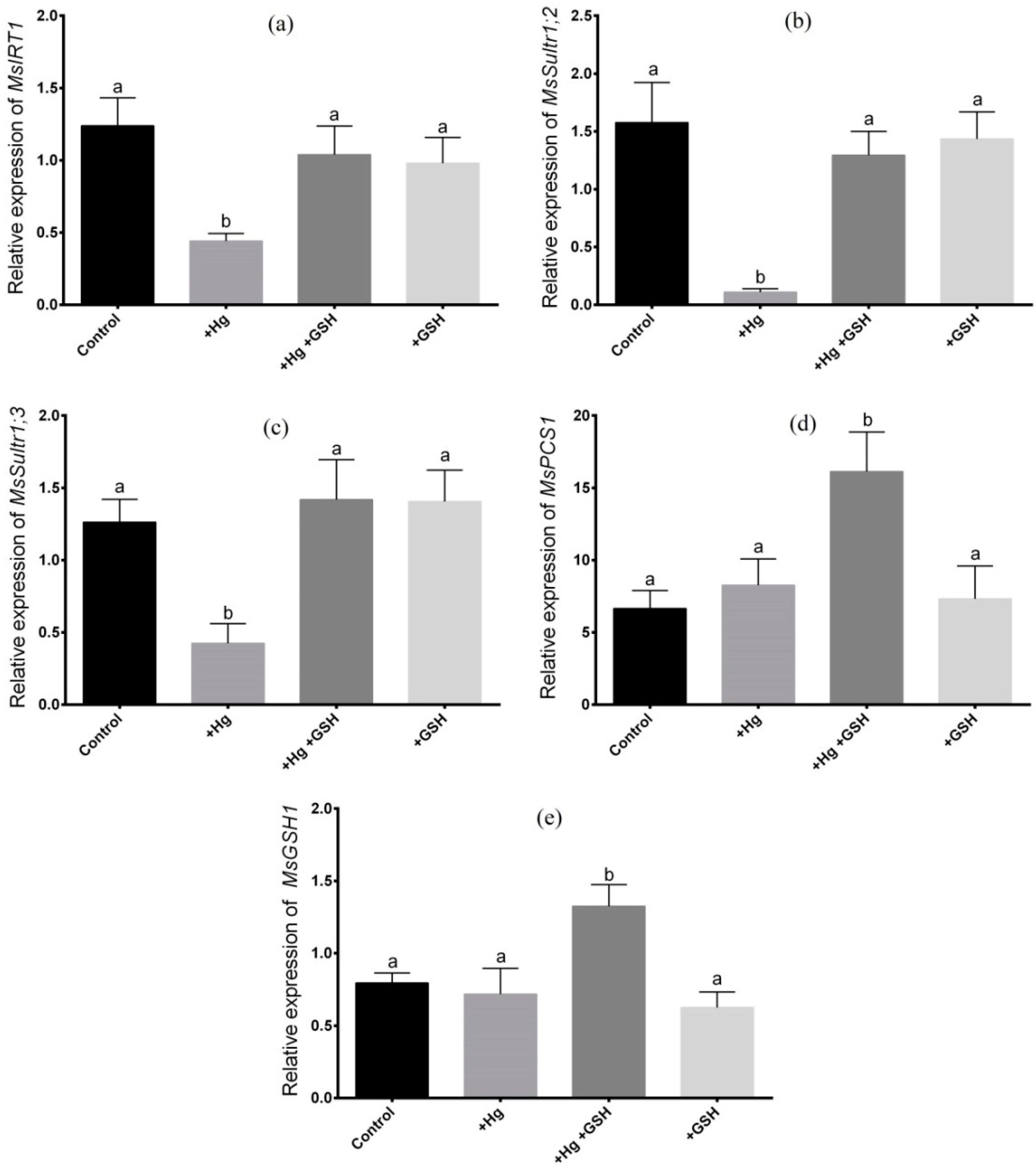
2.5. RNA Isolation, cDNA Synthesis, and Gene Expression Analysis by qRT-PCR
2.6. In Silico Characterization of the MsPCS1 Gene
2.7. Measurement of Antioxidant Enzymes
2.8. Determination of Glutathione (GSH) and Phytochelatins (PCs)
2.9. Statistical Analysis
3. Results
3.1. Phenotypic and Morphological Features
3.2. Characterization of Photosynthesis and Metal Elements
3.3. Expression of Candidate Genes in Roots
3.4. In Silico Characterization of the MsPCS1 Gene
3.5. Changes in Reactive Oxygen Species and Antioxidant Enzymes
3.6. Changes in Phytochelatin and Glutathione
4. Discussion
4.1. Improvement in Morphological and Physiological Features
4.2. Mechanisms of Hg Detoxifications
4.3. Improvement of the Redox Status of the Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hernández, L.E.; Sobrino-Plata, J.; Montero-Palmero, M.B.; Carrasco-Gil, S.; Flores-Cáceres, M.L.; Ortega-Villasante, C.; Escobar, C. Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J. Exp. Bot. 2015, 66, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Pacyna, J.M. Recent advances in mercury research. Sci. Total Environ. 2020, 738, 139955. [Google Scholar] [CrossRef] [PubMed]
- Alloway, B.J. Sources of Heavy Metals and Metalloids in Soils. In Heavy Metals in Soils: Trace Metals and Metalloids in Soils and their Bioavailability; Alloway, B.J., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 11–50. [Google Scholar]
- Fernández-Martínez, R.; Larios, R.; Gómez-Pinilla, I.; Gómez-Mancebo, B.; López-Andrés, S.; Loredo, J.; Ordóñez, A.; Rucandio, I. Mercury accumulation and speciation in plants and soils from abandoned cinnabar mines. Geoderma 2015, 253–254, 30–38. [Google Scholar] [CrossRef]
- Israr, M.; Sahi, S.; Datta, R.; Sarkar, D. Bioaccumulation and physiological effects of mercury in Sesbania drummondii. Chemosphere 2006, 65, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-O.; Bae, H.-J.; Cho, E.; Kang, H. Exogenous glutathione enhances mercury tolerance by inhibiting mercury entry into plant cells. Front. Plant Sci. 2017, 8, 683. [Google Scholar] [CrossRef]
- Marques, A.P.G.C.; Rangel, A.O.S.S.; Castro, P.M.L. Remediation of heavy metal contaminated soils: Phytoremediation as a potentially promising clean-up technology. Crit. Rev. Environ. Sci. Technol. 2009, 39, 622–654. [Google Scholar] [CrossRef]
- Zhang, W.-H.; Tyerman, S.D. Inhibition of water channels by HgCl2 in intact wheat root cells. Plant Physiol 1999, 120, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Bricker, T.J.; Pichtel, J.; Brown, H.J.; Simmons, M. Phytoextraction of Pb and Cd from a superfund soil: Effects of amendments and croppings. J. Environ. Sci. Health A 2001, 36, 1597–1610. [Google Scholar] [CrossRef] [PubMed]
- Salt, D.E.; Rauser, W.E. MGATP-dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiol. 1995, 107, 1293–1301. [Google Scholar] [CrossRef]
- Mendoza-Cózatl, D.G.; Jobe, T.O.; Hauser, F.; Schroeder, J.I. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr. Opin. Plant Biol. 2011, 14, 554–562. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, J.; Wang, Y.; Zhang, X.; Shen, Z.; Hu, Z. Ectopic expression of Vicia sativa caffeoyl-CoA O-methyltransferase (VsCCoAOMT) increases the uptake and tolerance of cadmium in Arabidopsis. Environ. Exp. Bot. 2018, 145, 47–53. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ueda, Y.; Mukai, A.; Ochiai, K.; Matoh, T. Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance. Plant Direct. 2018, 2, e00034. [Google Scholar] [CrossRef]
- Ha, S.B.; Smith, A.P.; Howden, R.; Dietrich, W.M.; Bugg, S.; O’Connell, M.J.; Goldsbrough, P.B.; Cobbett, C.S. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 1999, 11, 1153–1164. [Google Scholar] [CrossRef]
- Ramos, J.; Clemente, M.R.; Naya, L.; Loscos, J.; Pérez-Rontomé, C.; Sato, S.; Tabata, S.; Becana, M. Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol. 2007, 143, 1110–1118. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Z.; Zhu, C. Heteroexpression of the wheat phytochelatin synthase gene (TaPCS1) in rice enhances cadmium sensitivity. Acta Biochim. Biophys. Sin. 2012, 44, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.-i.; Suzui, N.; Nagasaka, T.; Komatsu, F.; Ishioka, N.S.; Ito-Tanabata, S.; Kawachi, N.; Rai, H.; Hattori, H.; Chino, M.; et al. Application of glutathione to roots selectively inhibits cadmium transport from roots to shoots in oilseed rape. J. Exp. Bot. 2013, 64, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-Y.; Wang, X.-Y.; Dai, H.-X.; Zhang, G.-P.; Wu, F.-B. Effect of exogenous glutathione and selenium on cadmium-induced changes in cadmium and mineral concentrations and antioxidative metabolism in maize seedlings. Asian J. Chem. 2013, 25, 2970–2976. [Google Scholar] [CrossRef]
- Nakazawa, R.; Ikawa, M.; Yasuda, K.; Takenaga, H. Synergistic inhibition of the growth of suspension cultured tobacco cells by simultaneous treatment with cadmium and arsenic in relation to phytochelatin synthesis. Soil Sci. Plant Nutr. 2000, 46, 271–275. [Google Scholar] [CrossRef]
- Sahu, G.K.; Upadhyay, S.; Sahoo, B.B. Mercury induced phytotoxicity and oxidative stress in wheat (Triticum aestivum L.) plants. Physiol. Mol. Biol. Plants 2012, 18, 21–31. [Google Scholar] [CrossRef]
- Demiral, T.; Türkan, İ. Comparative lipid peroxidation, antioxidant defense systems and proline content in roots of two rice cultivars differing in salt tolerance. Environ. Exp. Bot. 2005, 53, 247–257. [Google Scholar] [CrossRef]
- Szalai, G.; Kellős, T.; Galiba, G.; Kocsy, G. Glutathione as an Antioxidant and Regulatory Molecule in Plants Under Abiotic Stress Conditions. J. Plant. Growth Regul. 2009, 28, 66–80. [Google Scholar] [CrossRef]
- Hasan, M.K.; Liu, C.; Wang, F.; Ahammed, G.J.; Zhou, J.; Xu, M.-X.; Yu, J.-Q.; Xia, X.-J. Glutathione-mediated regulation of nitric oxide, S-nitrosothiol and redox homeostasis confers cadmium tolerance by inducing transcription factors and stress response genes in tomato. Chemosphere 2016, 161, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.S.; Huang, S.Q.; Guo, K.; Mehta, S.K.; Zhang, P.C.; Yang, Z.M. Metabolic adaptations to mercury-induced oxidative stress in roots of Medicago sativa L. J. Inorg. Biochem. 2007, 101, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Teng, Y.; Wang, J.; Li, J. Vanadium Uptake by Alfalfa Grown in V–Cd-Contaminated Soil by Pot Experiment. Biol. Trace Elem. Res. 2011, 142, 787–795. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; California Agricultural Experiment Station: Los Angeles, CA, USA, 1950; Volume 347, p. 32. [Google Scholar]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2013, 11, 459–469. [Google Scholar] [CrossRef]
- Yuan, Z.; Cao, Q.; Zhang, K.; Ata-Ul-Karim, S.T.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Optimal leaf positions for SPAD meter measurement in rice. Front. Plant Sci. 2016, 7, 719. [Google Scholar] [CrossRef]
- Uddling, J.; Gelang-Alfredsson, J.; Piikki, K.; Pleijel, H. Evaluating the relationship between leaf chlorophyll concentration and SPAD-502 chlorophyll meter readings. Photosynth. Res. 2007, 91, 37–46. [Google Scholar] [CrossRef]
- Rahman, M.A.; Parvin, M.; Das, U.; Ela, E.J.; Lee, S.-H.; Lee, K.-W.; Kabir, A.H. Arbuscular mycorrhizal symbiosis mitigates iron (Fe)-deficiency retardation in alfalfa (Medicago sativa L.) through the enhancement of Fe accumulation and sulfur-assisted antioxidant defense. Int. J. Mol. Sci. 2020, 21, 2219. [Google Scholar] [CrossRef]
- Pourghasemian, N.; Landberg, T.; Ehsanzadeh, P.; Greger, M. Different response to Cd stress in domesticated and wild safflower (Carthamus spp.). Ecotoxicol. Environ. Saf. 2019, 171, 321–328. [Google Scholar] [CrossRef]
- Peever, T.L.; Higgins, V.J. Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 1989, 90, 867–875. [Google Scholar] [CrossRef]
- Rahman, M.A.; Alam, I.; Kim, Y.-G.; Ahn, N.-Y.; Heo, S.-H.; Lee, D.-G.; Liu, G.; Lee, B.-H. Screening for salt-responsive proteins in two contrasting alfalfa cultivars using a comparative proteome approach. Plant Physiol. Biochem. 2015, 89, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Timothy, B.L.; Charles, E. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Sun, M.; Zigman, S. An improved spectrophotometric assay for superoxide dismutase based on epinephrine autoxidation. Anal. Biochem. 1978, 90, 81–89. [Google Scholar] [CrossRef]
- Halliwell, B.; Foyer, C.H. Properties and physiological function of a glutathione reductase purified from spinach leaves by affinity chromatography. Planta 1978, 139, 9–17. [Google Scholar] [CrossRef]
- Kabir, A.H.; Hossain, M.M.; Khatun, M.A.; Mandal, A.; Haider, S.A. Role of silicon counteracting cadmium toxicity in alfalfa (Medicago sativa L.). Front. Plant Sci. 2016, 7, 1117. [Google Scholar] [CrossRef]
- Lindberg, S.; Landberg, T.; Greger, M. Cadmium uptake and interaction with phytochelatins in wheat protoplasts. Plant Physiol. Biochem. 2007, 45, 47–53. [Google Scholar] [CrossRef]
- Larbi, A.; Morales, F.; Abadía, A.; Gogorcena, Y.; Lucena, J.J.; Abadía, J. Effects of Cd and Pb in sugar beet plants grown in nutrient solution: Induced Fe deficiency and growth inhibition. Funct. Plant Biol. 2002, 29, 1453–1464. [Google Scholar] [CrossRef]
- Ortega-Villasante, C.; Hernández, L.E.; Rellán-Álvarez, R.; Del Campo, F.F.; Carpena-Ruiz, R.O. Rapid alteration of cellular redox homeostasis upon exposure to cadmium and mercury in alfalfa seedlings. New Phytol. 2007, 176, 96–107. [Google Scholar] [CrossRef]
- Patra, M.; Bhowmik, N.; Bandopadhyay, B.; Sharma, A. Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance. Environ. Exp. Bot. 2004, 52, 199–223. [Google Scholar] [CrossRef]
- Chen, J.; Shiyab, S.; Han, F.X.; Monts, D.L.; Waggoner, C.A.; Yang, Z.; Su, Y. Bioaccumulation and physiological effects of mercury in Pteris vittata and Nephrolepis exaltata. Ecotoxicology 2008, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Rucińska-Sobkowiak, R. Water relations in plants subjected to heavy metal stresses. Acta Physiol. Plant. 2016, 38, 257. [Google Scholar] [CrossRef]
- Popova, L.P.; Maslenkova, L.T.; Yordanova, R.Y.; Ivanova, A.P.; Krantev, A.P.; Szalai, G.; Janda, T. Exogenous treatment with salicylic acid attenuates cadmium toxicity in pea seedlings. Plant Physiol. Biochem. 2009, 47, 224–231. [Google Scholar] [CrossRef] [PubMed]
- He, J.Y.; Ren, Y.F.; Zhu, C.; Yan, Y.P.; Jiang, D.A. Effect of Cd on growth, photosynthetic gas exchange, and chlorophyll fluorescence of wild and Cd-sensitive mutant rice. Photosynthetica 2008, 46, 466. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Ye, H.; Garifullina, G.F.; Zhang, L.; Pilon-Smits, E.A.H.; Pilon, M. Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 2005, 138, 161–172. [Google Scholar] [CrossRef]
- Singh, S.; Parihar, P.; Singh, R.; Singh, V.P.; Prasad, S.M. Heavy metal tolerance in plants:role of transcriptomics, proteomics, metabolomics, and ionomics. Front. Plant Sci. 2016, 6, 1143. [Google Scholar] [CrossRef] [PubMed]
- DalCorso, G.; Manara, A.; Furini, A. An overview of heavy metal challenge in plants: From roots to shoots. Metallomics 2013, 5, 1117–1132. [Google Scholar] [CrossRef]
- Leitenmaier, B.; Küpper, H. Compartmentation and complexation of metals in hyperaccumulator plants. Front. Plant Sci. 2013, 4, 374. [Google Scholar] [CrossRef]
- Greger, M.; Kabir, A.H.; Landberg, T.; Maity, P.J.; Lindberg, S. Silicate reduces cadmium uptake into cells of wheat. Environ. Pollut. 2016, 211, 90–97. [Google Scholar] [CrossRef]
- Carrier, P.; Baryla, A.; Havaux, M. Cadmium distribution and microlocalization in oilseed rape (Brassica napus) after long-term growth on cadmium-contaminated soil. Planta 2003, 216, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K. Heavy metals toxicity in plants: An overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. S. Afr. J. Bot. 2010, 76, 167–179. [Google Scholar] [CrossRef]
- Picault, N.; Cazalé, A.C.; Beyly, A.; Cuiné, S.; Carrier, P.; Luu, D.T.; Forestier, C.; Peltier, G. Chloroplast targeting of phytochelatin synthase in Arabidopsis: Effects on heavy metal tolerance and accumulation. Biochimie 2006, 88, 1743–1750. [Google Scholar] [CrossRef] [PubMed]
- Cargnelutti, D.; Tabaldi, L.A.; Spanevello, R.M.; de Oliveira Jucoski, G.; Battisti, V.; Redin, M.; Linares, C.E.B.; Dressler, V.L.; de Moraes Flores, E.M.; Nicoloso, F.T. Mercury toxicity induces oxidative stress in growing cucumber seedlings. Chemosphere 2006, 65, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Bari, M.A.; Akther, M.S.; Reza, M.A.; Kabir, A.H. Cadmium tolerance is associated with the root-driven coordination of cadmium sequestration, iron regulation, and ROS scavenging in rice. Plant Physiol. Biochem. 2019, 136, 22–33. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Kao, C.H. Cadmium toxicity is reduced by nitric oxide in rice leaves. Plant Growth Regul. 2004, 42, 227–238. [Google Scholar] [CrossRef]
- Jozefczak, M.; Remans, T.; Vangronsveld, J.; Cuypers, A. Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 2012, 13, 3145–3175. [Google Scholar] [CrossRef]
- Freeman, J.L.; Persans, M.W.; Nieman, K.; Albrecht, C.; Peer, W.; Pickering, I.J.; Salt, D.E. Increased glutathione biosynthesis plays a role in nickel tolerance in thlaspi nickel hyperaccumulators. Plant Cell 2004, 16, 2176–2191. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.A.; Kabir, A.H.; Mandal, A.; Roy, S.K.; Song, Y.; Ji, H.C.; Lee, K.-W. Glutathione Restores Hg-Induced Morpho-Physiological Retardations by Inducing Phytochelatin and Oxidative Defense in Alfalfa. Biology 2020, 9, 364. https://doi.org/10.3390/biology9110364
Rahman MA, Kabir AH, Mandal A, Roy SK, Song Y, Ji HC, Lee K-W. Glutathione Restores Hg-Induced Morpho-Physiological Retardations by Inducing Phytochelatin and Oxidative Defense in Alfalfa. Biology. 2020; 9(11):364. https://doi.org/10.3390/biology9110364
Chicago/Turabian StyleRahman, Md Atikur, Ahmad Humayan Kabir, Abul Mandal, Swapan Kumar Roy, Yowook Song, Hee Chung Ji, and Ki-Won Lee. 2020. "Glutathione Restores Hg-Induced Morpho-Physiological Retardations by Inducing Phytochelatin and Oxidative Defense in Alfalfa" Biology 9, no. 11: 364. https://doi.org/10.3390/biology9110364
APA StyleRahman, M. A., Kabir, A. H., Mandal, A., Roy, S. K., Song, Y., Ji, H. C., & Lee, K.-W. (2020). Glutathione Restores Hg-Induced Morpho-Physiological Retardations by Inducing Phytochelatin and Oxidative Defense in Alfalfa. Biology, 9(11), 364. https://doi.org/10.3390/biology9110364






