Ascovirus P64 Homologs: A Novel Family of Large Cationic Proteins That Condense Viral Genomic DNA for Encapsidation
Abstract
1. Introduction
2. Materials and Methods
2.1. Virion Purification
2.2. Isolation of Recombinant 6x-Histidine-Tagged Proteins
2.3. SDS-PAGE, Western and Southwestern Blotting
2.4. DNA-Protein Complex Aggregation
2.5. Transmission Electron Microscopy
2.6. Database Searches
3. Results
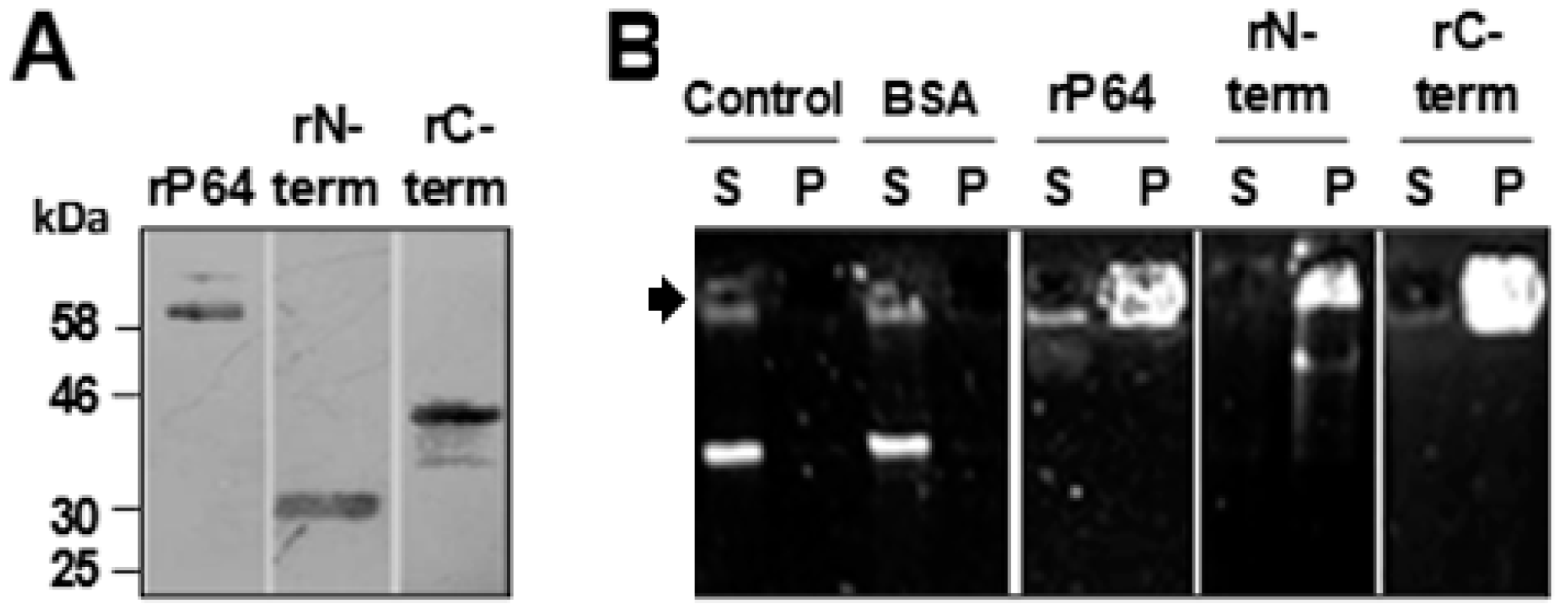
3.1. P64 and its N-Terminal and C-Terminal Domains Condense and Precipitate SfAV-1a gDNA
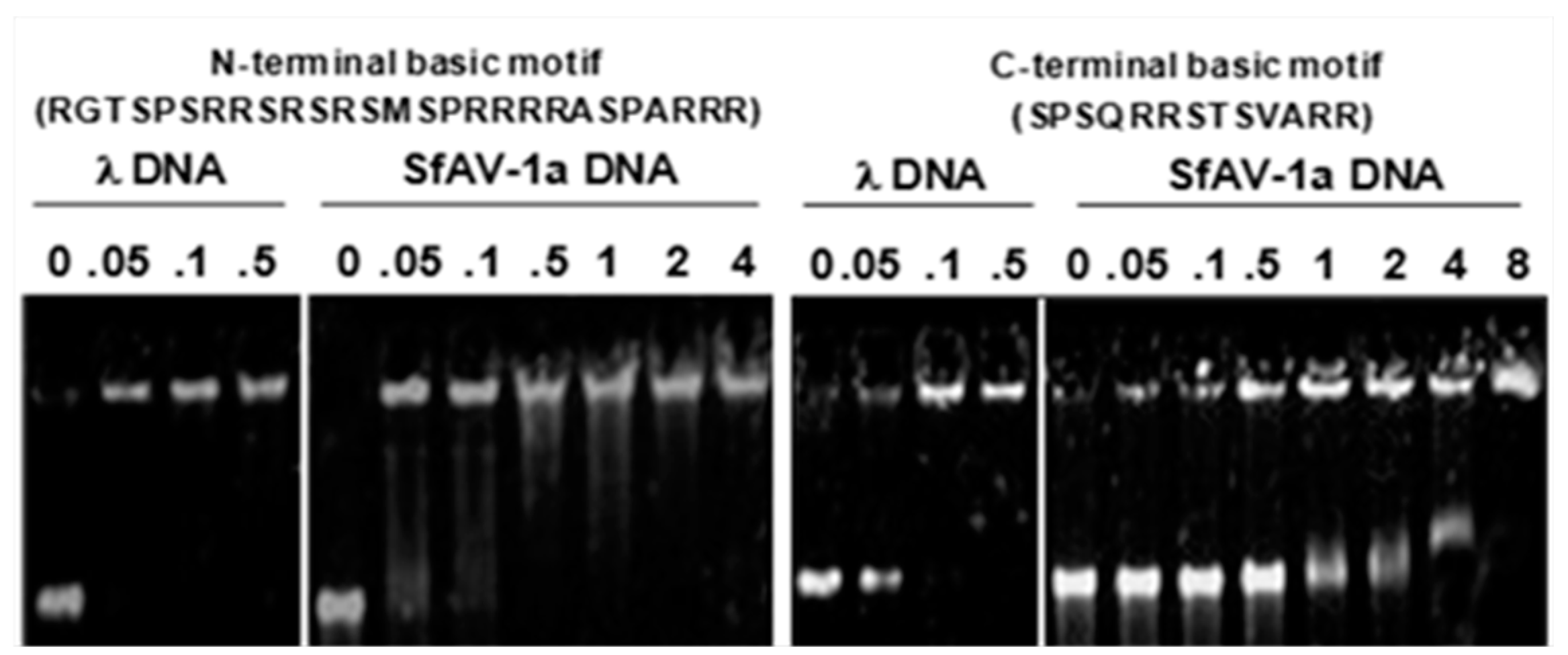
3.2. Basic Motifs in P64’s N-Terminal and C-Terminal Domains Bind DNA Non-Specifically
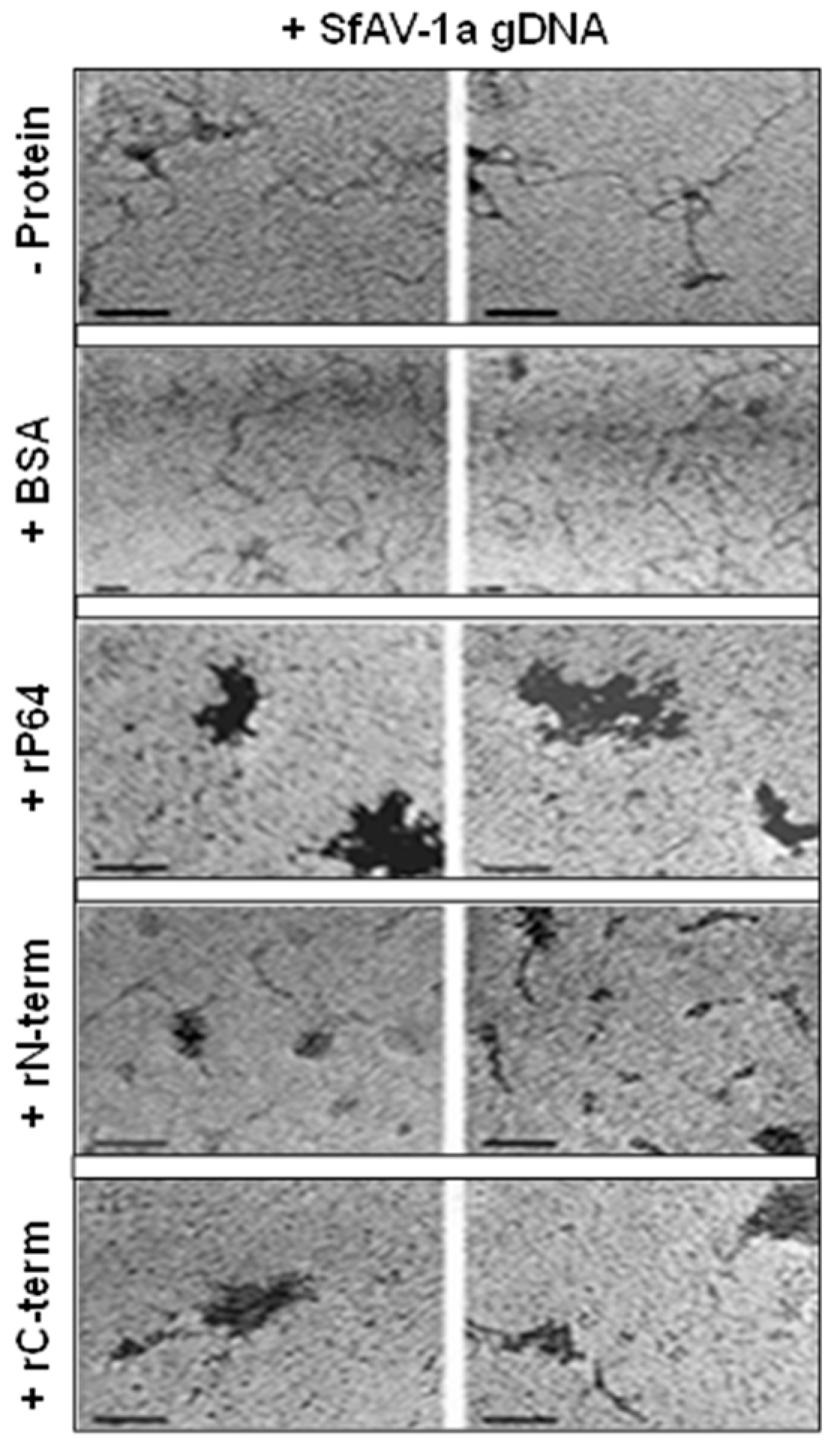
3.3. Transmission Electron Microscopy (TEM) Demonstrates P64 and its C-terminal Domain SfAV-1a Condense gDNA
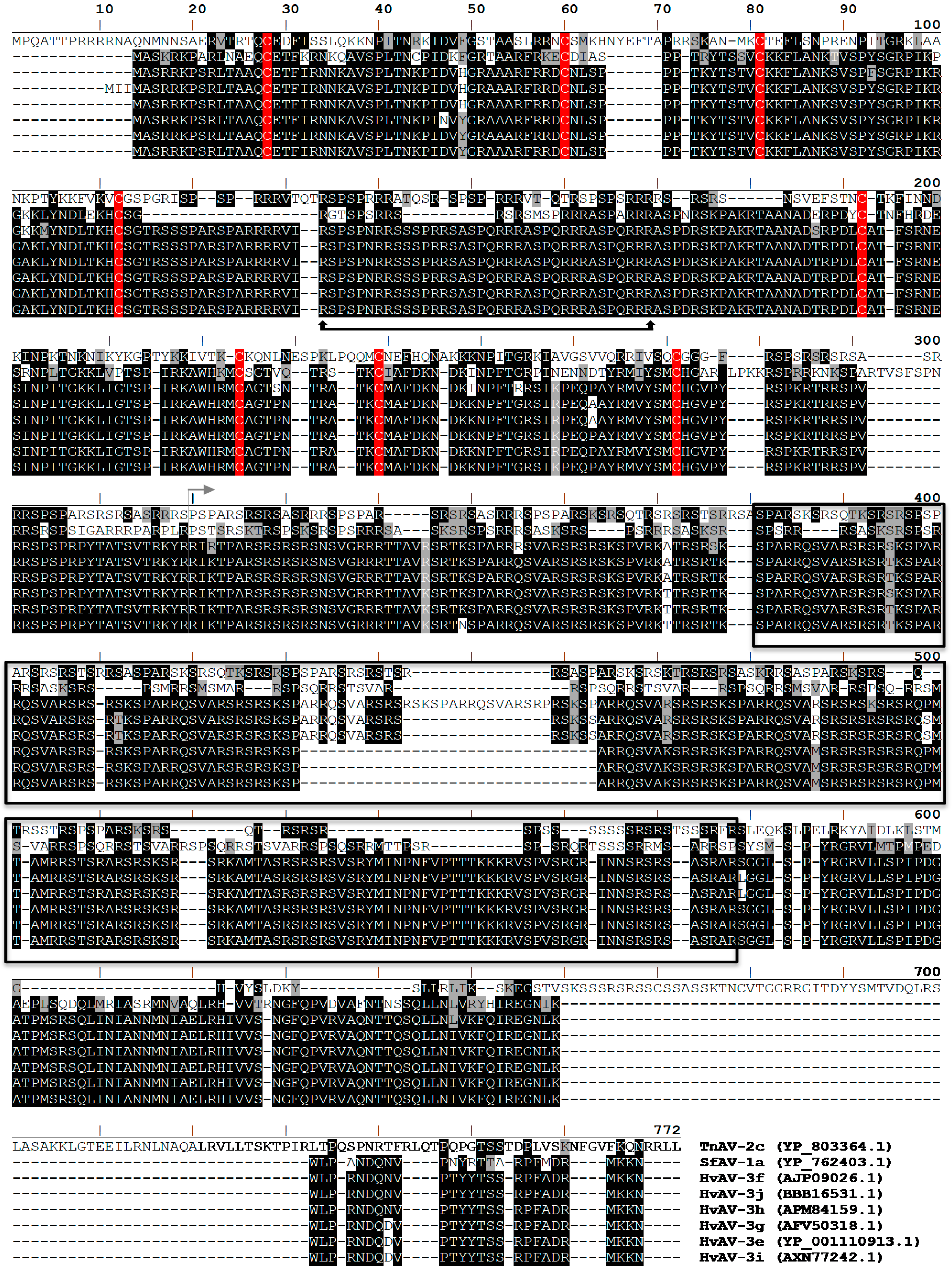
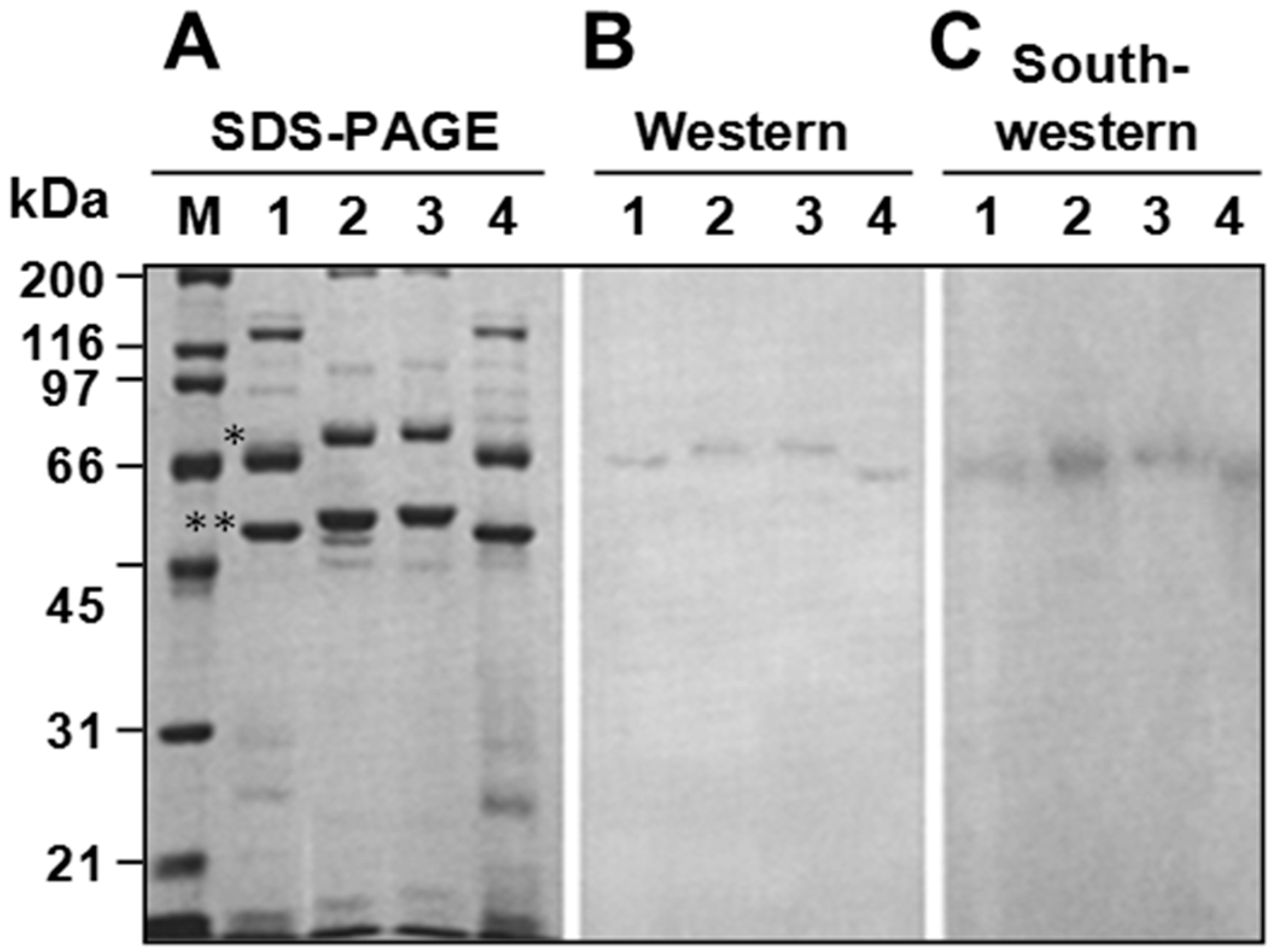
3.4. P64 Homologs Are Abundant DNA-Binding Proteins in Virions of Ascoviruses
3.5. Ascovirus P64 Homologs Comprise a Novel Family of Unusually Large Cationic Proteins That Condense Viral DNA for Encapsidation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chelikan, V.; Rajan, T.; Kondabagil, K. Revisiting the genome packaging in viruses with lessons from the “Giants”. Virology 2014, 466, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.; Dauden, M.I.; Carroscosa, J.L. Nucleic acid packaging in viruses. Subcell. Biochem. 2013, 68, 361–394. [Google Scholar] [PubMed]
- Lieberman, P.M. Chromatin organization and virus gene expression. J. Cell. Physiol. 2008, 216, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Locker, R.C.; Fuller, S.D.; Harvey, S.C. DNA organization and thermodynamics during viral packing. Biophys. J. 2007, 93, 2861–2869. [Google Scholar] [CrossRef] [PubMed]
- Brewer, L.R.; Corzett, M.R.; Balhorn, R. Protamine-induced condensation and decondensation of small DNA molecules. Science 1999, 286, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Koltover, I.; Wagner, K.; Safinya, C.R. DNA condensation in two dimensions. Proc. Natl. Acad. Sci. USA 2000, 97, 14046–14051. [Google Scholar] [CrossRef] [PubMed]
- Bartova, E.; Krejci, J.; Harnicarova, A.; Galiova, G.; Kozubek, S. Histone modifications and nuclear architecture: A review. J. Histochem. Cytochem. 2008, 56, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Bulach, D.M.; Kumar, C.A.; Zaia, A.; Liang, B.; Tribe, D.E. Group II nucleopolyhedrovirus subgroups revealed by phylogenetic analysis of polyhedron and DNA polymerase gene sequences. J. Invertebr. Pathol. 1999, 73, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.I.; Reinberg, D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009, 43, 559–599. [Google Scholar] [CrossRef] [PubMed]
- La Bella, F.; Vesco, C. Late modifications of simian virus 40 chromatin during the lytic cycle occur in an immature form of virion. J. Virol. 1980, 33, 1138–1150. [Google Scholar] [PubMed]
- Milavetz, B. Hyperacetylation and differential deacetylation of histones H4 and H3 define two distinct classes of acetylated SV40 chromosome early in infection. Virology 2004, 319, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Kamio, N.; Cueno, M.E.; Saito, Y.; Inoue, H.; Saito, I.; Ochiai, K. Role of the histone H3 lysine 9 methyltransferase Suv39 h1 in maintaining Epstein-Barr virus latency in B95-8 cells. FEBS J. 2014, 281, 2148–2158. [Google Scholar] [CrossRef] [PubMed]
- Jeng, M.Y.; Ali, I.; Ott, M. Manipulation of the host protein acetylation network by human immunodeficiency virus type 1. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 314–325. [Google Scholar] [PubMed]
- Segura, M.M.; Garnier, A.; Di Falco, M.R.; Whisell, G.; Meneses-Acosta, A.; Archand, N.; Kamen, A. Identification of host proteins associated with retroviral vector particles by proteonomic analysis of highly purified vector preparations. J. Virol. 2008, 82, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Mainprizw, T.H.; Friesen, P.D.; Miller, L.K. Location, transcription, and sequence of a baculovirus gene encoding a small arginine-rich polypeptide. J. Virol. 1987, 61, 661–666. [Google Scholar] [PubMed]
- Wilson, M.E.; Price, K.H. Association of Autographa californica nuclear polyhedrosis virus with nuclear matrix. Virology 1988, 167, 233–341. [Google Scholar] [CrossRef]
- Wang, M.; Tuladhar, E.; Shen, S.; Wang, H.; van Oers, M.M.; Vlak, J.M.; Westenberg, M. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 2010, 84, 8821–8828. [Google Scholar] [CrossRef] [PubMed]
- Van Oers, M.M.; Vlak, J.M. Baculovirus genomics. Curr. Drug Targets 2007, 8, 1051–1068. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S.; Bideshi, D.K.; Bigot, Y.; Cheng, X.W. ICTV Virus Taxonomy Profile: Ascoviridae. ICTV Report Consortium. J. Gen. Virol. 2017, 98, 4–5. [Google Scholar] [PubMed]
- Bideshi, D.K.; Bigot, Y.; Federici, B.A.; Spears, T. Ascoviruses. In Insect Virology; Asgari, S., Johnson, K.N., Eds.; Caister Academic Press: Great Britian, UK, 2010; pp. 2–34. ISBN 978-1-904455-71-4. [Google Scholar]
- Tan, Y.; Bideshi, D.K.; Johnson, J.J.; Bigot, Y.; Federici, B.A. Proteomic analysis of the Spodoptera frigiperda ascovirus 1a virion reveals 21 proteins. J. Gen. Virol. 2008, 90, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Spears, T.; Bideshi, D.K.; Johnson, J.J.; Hice, R.; Bigot, Y.; Federici, B.A. P64, a novel major virion DNA-binding protein potentially involved in condensing the Spodoptera frugiperda ascovirus 1a genome. J. Virol. 2009, 83, 2708–2714. [Google Scholar] [CrossRef] [PubMed]
- Zaghloul, H.A.H.; Hice, R.; Arensburger, P.; Federici, B.A. Transcriptome analysis of the Spodoptera frugiperda ascoviruis in vivo provides insights into how its apoptosis inhibitors and caspase promote increased synthesis of viral vesicles and virion progeny. J. Virol. 2017. [CrossRef]
- Bideshi, D.K.; Demattei, M.V.; Rouleux-Bonnin, F.; Stasiak, K.; Tan, Y.; Bigot, S.; Bigot, Y.; Federici, B.A. Genomic sequence of the Spodoptera frugiperda ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 2006, 80, 11791–11805. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vollenweider, J.; Sogo, J.M.; Koller, T.A. Routine method for protein-free spreading of double and single-stranded nucleic acid molecules. Proc. Natl. Acad. Sci. USA 1975, 72, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Black, B.C.; Center, M.S. DNA-binding properties of the major core protein of adenovirus 2. Nucleic Acids Res. 1979, 6, 2339–2353. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ren, J.; Qu, X. Time-dependent DNA condensation induced by amyloid β-peptide. Biophys. J. 2007, 92, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Bigot, Y.; Renault, S.; Nicolas, J.; Moundras, C.; Demattei, M.V.; Samain, S.; Bideshi, D.K.; Federici, B.A. Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS ONE 2009, 4, e6397. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Balaji, S.; Koonin, E.V.; Aravind, L. Evolutionary genomics of nucleo-cytoplamsmic viruses. Virus Res. 2006, 111, 156–184. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.; Abrahão, J.S.; Drumond, B.P.; Kroon, E.G. Giants among larges: How gigantism impacts giant virus entry into amoebae. Curr. Opin. Microbiol. 2016, 31, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, K.; Renault, S.; Demattei, M.V.; Bigot, Y.; Federici, B.A. Evidence for the evolution of ascoviruses from iridoviruses. J. Gen. Virol. 2003, 84, 2999–3009. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; De Lamballerie, X.; Yutin, N.; Asgari, S.; Bigot, Y.; Bideshi, D.K.; Cheng, X.W.; Federici, B.A.; Van Etten, J.L.; Koonin, E.V.; et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013, 158, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chen, M.M.; Wan, X.Y.; Li, C.; Zhang, Q.L.; Wang, R.Y.; Cheng, D.Y.; Dong, X.; Yang, B.; Wang, X.H.; et al. Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei). Sci. Rep. 2107, 19, 11834. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xu, L.; Yang, F. Genomic characterization of a novel iridovirus from redclaw crayfish Cherax quadricarinatus: Evidence for a new genus within the family Iridoviridae. J. Gen. Virol. 2017, 98, 2589–2595. [Google Scholar] [CrossRef] [PubMed]
- Delhon, G.; Tulman, E.R.; Afonso, C.L.; Lu, Z.; Becnel, J.J.; Moser, B.A.; Kutish, G.F.; Rock, D.L. Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). J. Virol. 2006, 80, 8439–8449. [Google Scholar] [CrossRef] [PubMed]
- Piégu, B.; Guizard, S.; Spears, T.; Cruaud, C.; Couloux, A.; Bideshi, D.K.; Federici, B.A.; Bigot, Y. Complete genome sequence of invertebrate iridescent virus 22 isolated from a blackfly larva. J. Gen. Virol. 2013, 94, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Piégu, B.; Guizard, S.; Spears, T.; Cruaud, C.; Couloux, A.; Bideshi, D.K.; Federici, B.A.; Bigot, Y. Complete genome sequence of an invertebrate iridovirus IIV-25 isolated from a blackfly larva. Arch. Virol. 2014, 159, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Piégu, B.; Guizard, S.; Spears, T.; Cruaud, C.; Couloux, A.; Bideshi, D.K.; Federici, B.A.; Bigot, Y. Complete genome sequence of invertebrate iridovirus IIV30 isolated from the earworm, Helicoverpa zea. J. Invertebr. Pathol. 2014, 116, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Piégu, B.; Guizard, S.; Yeping, T.; Cruaud, C.; Asgari, S.; Bideshi, D.K.; Federici, B.A.; Bigot, Y. Complete sequence of a crustacean iridoviris, IIV31, isolated from the pill bug, Armadillidium vulgare. J. Gen. Virol. 2014, 95, 1585–1590. [Google Scholar] [CrossRef] [PubMed]
- Federici, B.A. Enveloped double-stranded DNA insect virus with novel structure and cytopathology. Proc. Natl. Acad. Sci. USA 1983, 80, 7664–7668. [Google Scholar] [CrossRef] [PubMed]
- Federici, B.A.; Bideshi, D.K.; Tan, Y.; Spears, T.; Bigot, Y. Ascoviruses: Superb manipulators of apoptosis for viral replication and transmission. Curr. Top. Microbiol. Immunol. 2009, 328, 171–196. [Google Scholar] [PubMed]
- Bideshi, D.K.; Tan, Y.; Bigot, Y.; Federici, B.A. A viral caspase contributes to modified apoptosis for virus transmission. Genes Dev. 2005, 19, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bideshi, D.K.; Spears, T.; Zaghloul, H.A.H.; Tan, Y.; Bigot, Y.; Federici, B.A. Ascovirus P64 Homologs: A Novel Family of Large Cationic Proteins That Condense Viral Genomic DNA for Encapsidation. Biology 2018, 7, 44. https://doi.org/10.3390/biology7030044
Bideshi DK, Spears T, Zaghloul HAH, Tan Y, Bigot Y, Federici BA. Ascovirus P64 Homologs: A Novel Family of Large Cationic Proteins That Condense Viral Genomic DNA for Encapsidation. Biology. 2018; 7(3):44. https://doi.org/10.3390/biology7030044
Chicago/Turabian StyleBideshi, Dennis K., Tatsinda Spears, Heba A. H. Zaghloul, Yeping Tan, Yves Bigot, and Brian A. Federici. 2018. "Ascovirus P64 Homologs: A Novel Family of Large Cationic Proteins That Condense Viral Genomic DNA for Encapsidation" Biology 7, no. 3: 44. https://doi.org/10.3390/biology7030044
APA StyleBideshi, D. K., Spears, T., Zaghloul, H. A. H., Tan, Y., Bigot, Y., & Federici, B. A. (2018). Ascovirus P64 Homologs: A Novel Family of Large Cationic Proteins That Condense Viral Genomic DNA for Encapsidation. Biology, 7(3), 44. https://doi.org/10.3390/biology7030044




