Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and RNA Preparation
2.2. Electrophoretic Mobility Shift Assay (EMSA)
2.3. Ribozyme Activity Assay
3. Results and Discussion
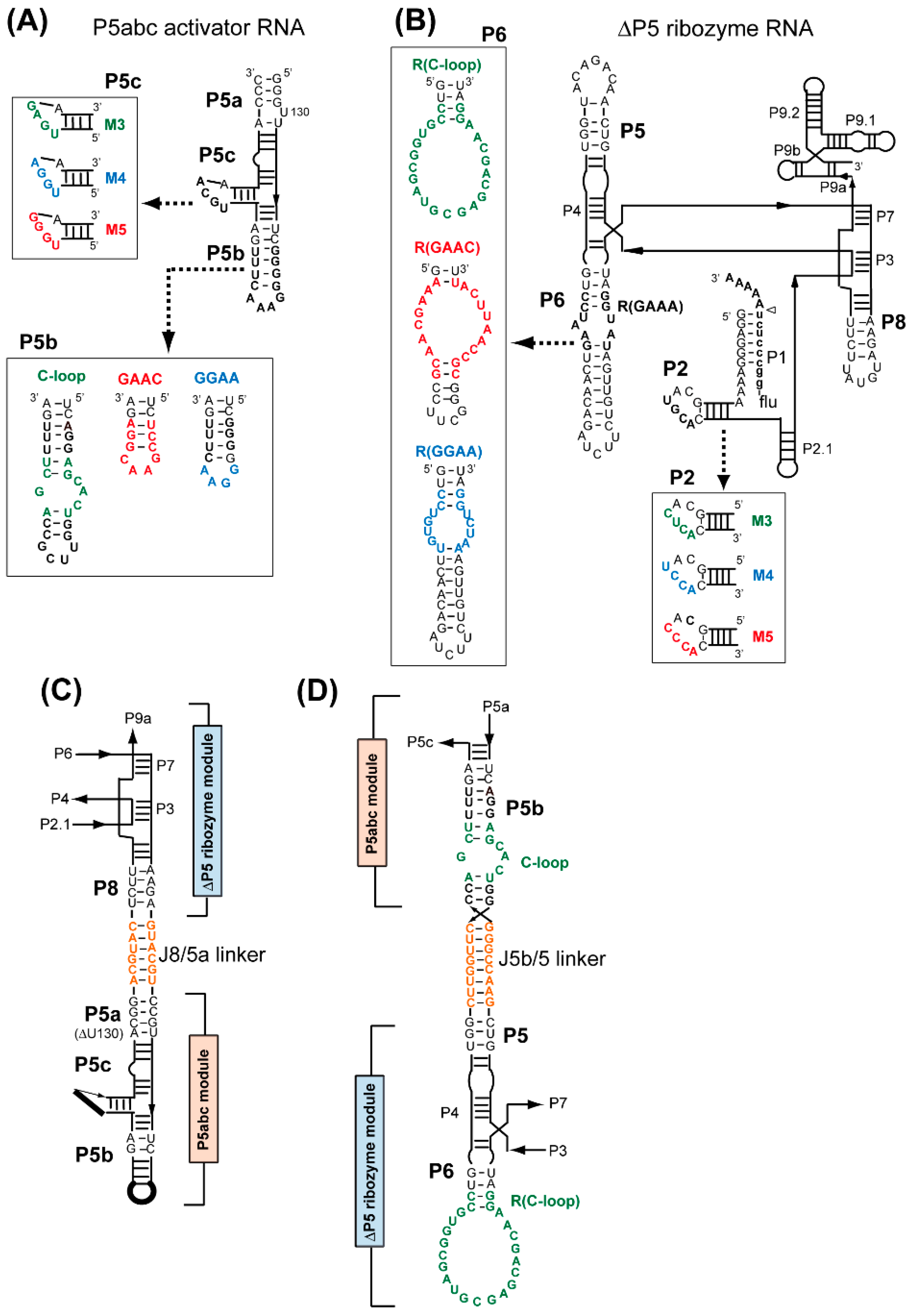
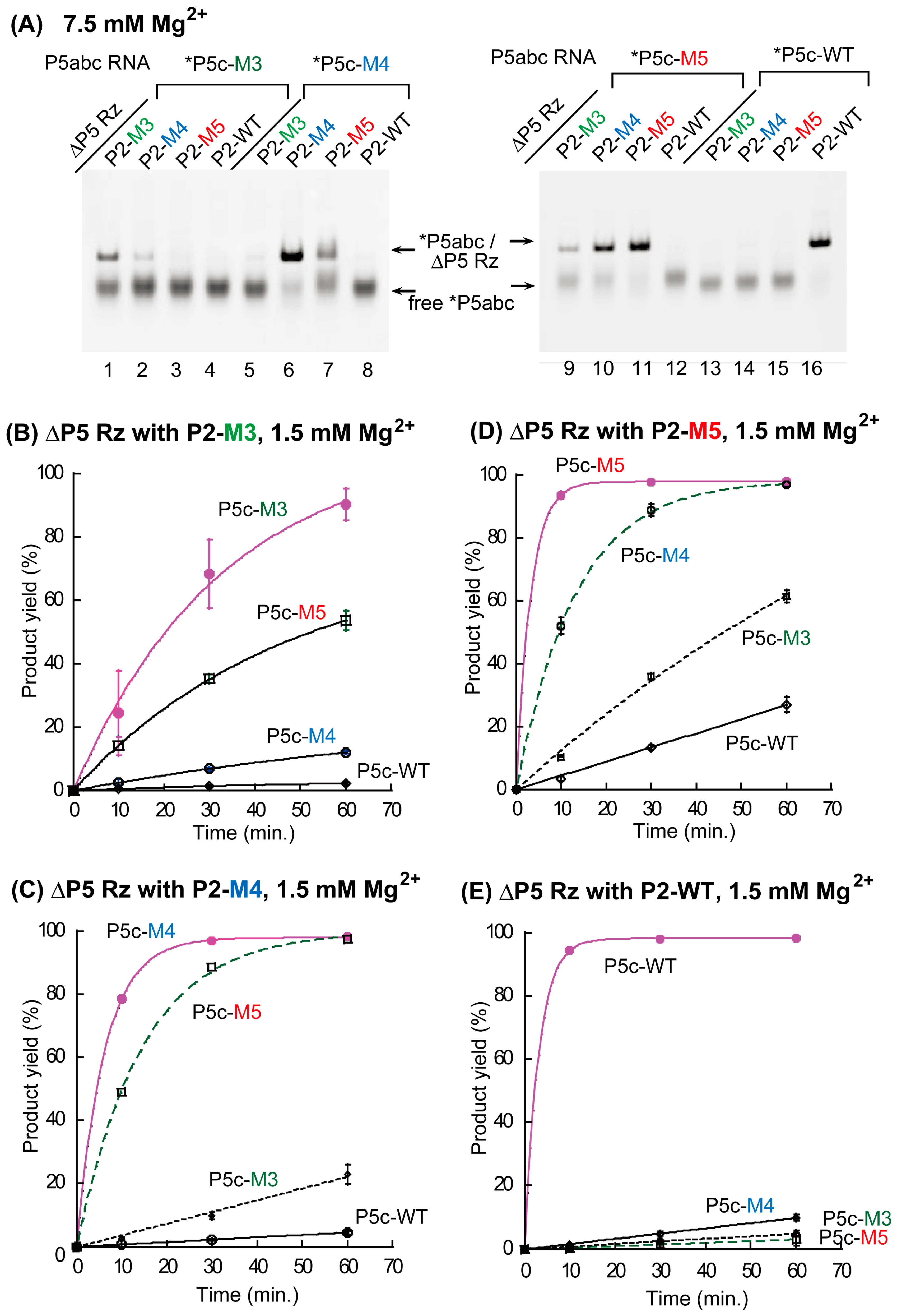
3.1. Assembly of Bimolecular Group I Ribozymes through Artificial RNA–RNA Interacting Motifs
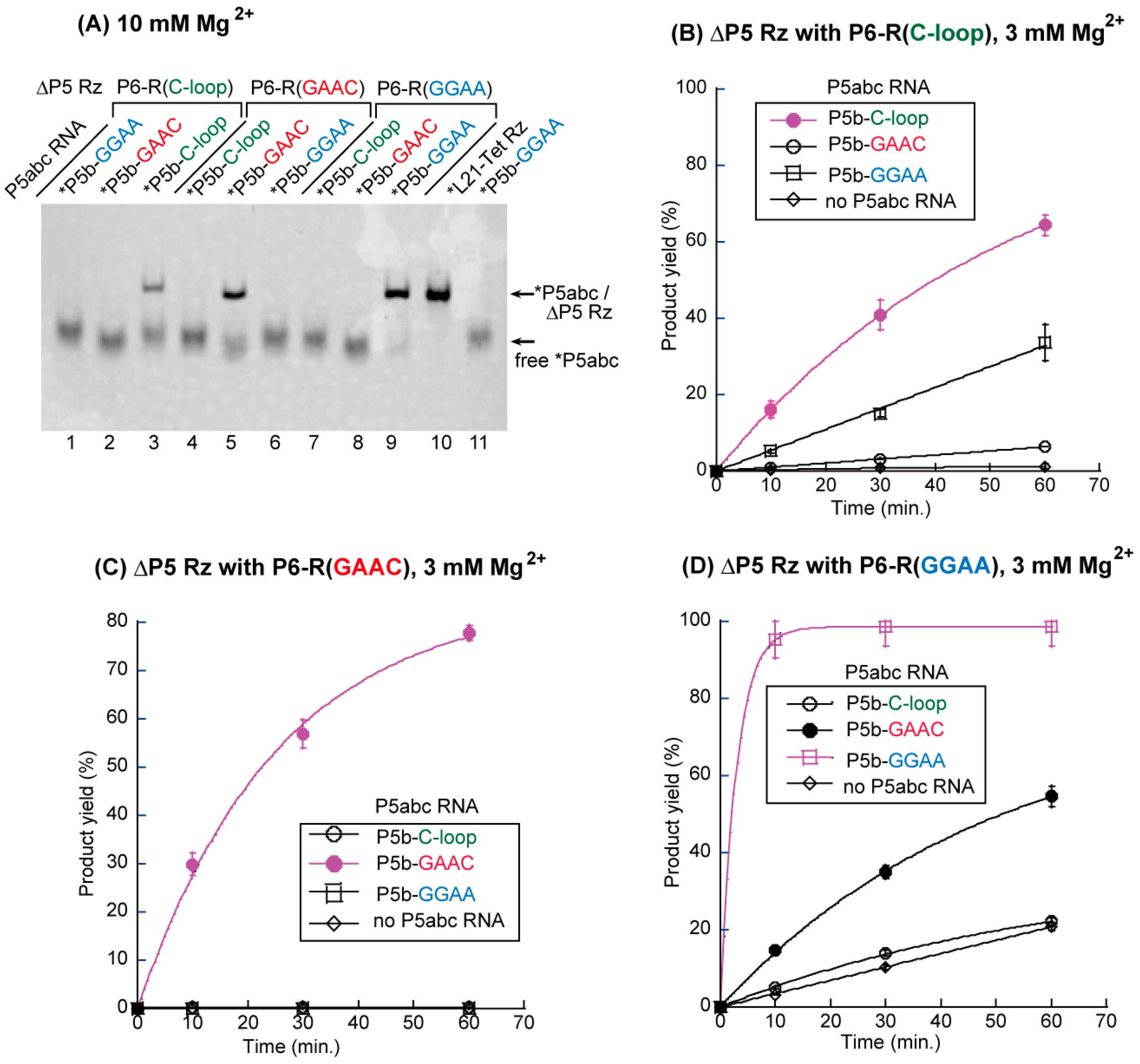
3.2. Artificial Kissing Loop Interactions to Assemble Bimolecular Group I Ribozymes
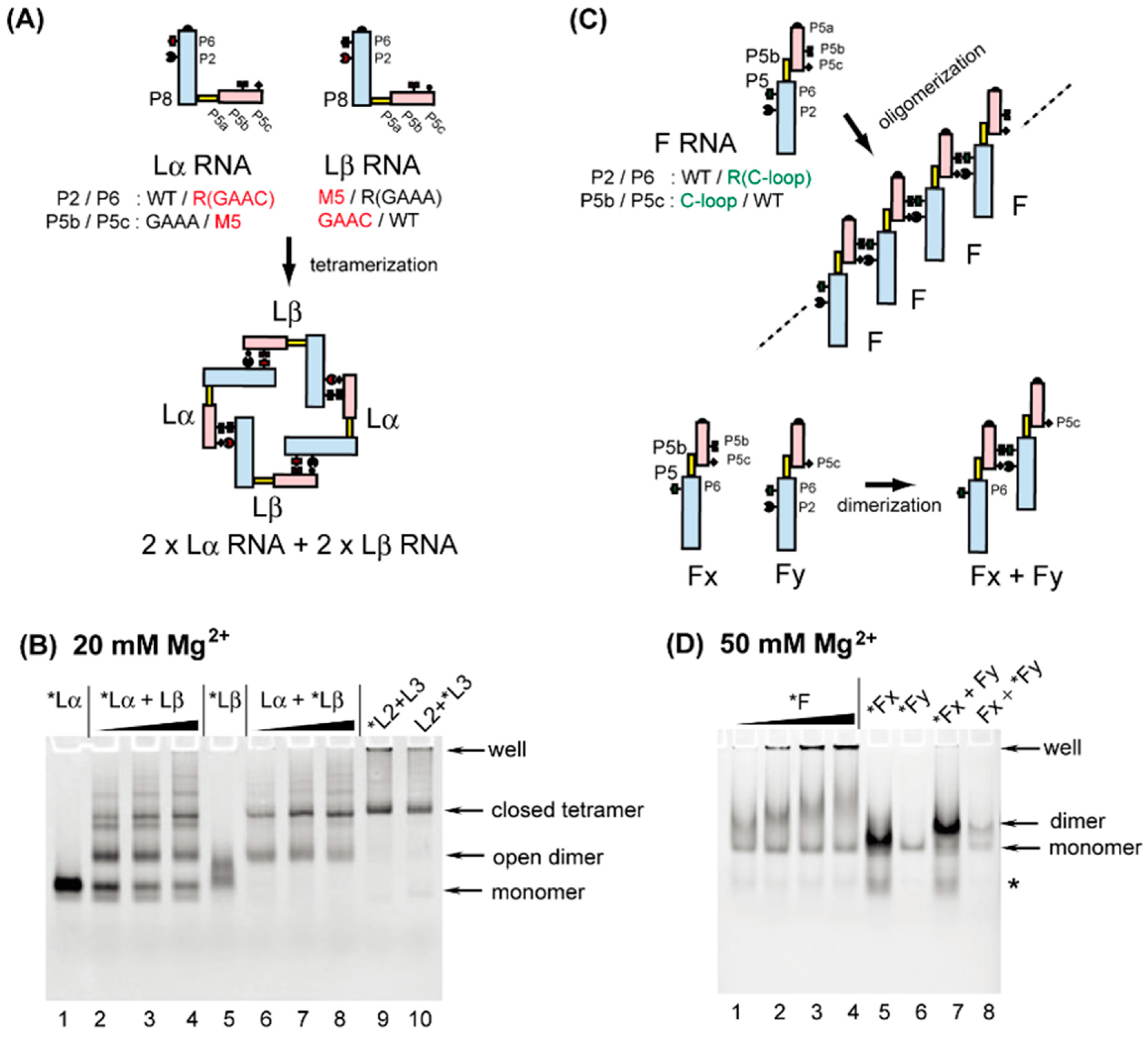
3.3. Formation of RNA Squares through Selective Assembly of Engineered Group I Ribozymes
3.4. Formation of Novel RNA 1D Array through Selective Oligomerization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Simmel, S.S.; Nickels, P.C.; Liedl, T. Wireframe and tensegrity DNA nanostructures. Acc. Chem. Res. 2014, 47, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, L.; Li, J.; Fan, C. Precisely tailored DNA nanostructures and their theranostic applications. Chem. Rec. 2017. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Jaeger, L. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014, 47, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for creating higher order structures. Chem. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Takeuchi, Y.; Emura, T.; Hidaka, K.; Sugiyama, H. Preparation of chemically modified RNA origami nanostructures. Chem. Eur. J. 2014, 20, 15330–15333. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.; Rothemund, P.W.; Andersen, E.S. RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 2014, 345, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Sparvath, S.L.; Geary, C.W.; Andersen, E.S. Computer-aided design of RNA origami structures. Methods Mol. Biol. 2017, 1500, 51–80. [Google Scholar] [PubMed]
- Kozyra, J.; Ceccarelli, A.; Torelli, E.; Lopiccolo, A.; Gu, J.Y.; Fellermann, H.; Stimming, U.; Krasnogor, N. Designing uniquely addressable bio-orthogonal synthetic scaffolds for DNA and RNA origami. ACS Synth. Biol. 2017, 6, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Chworos, A.; Severcan, I.; Koyfman, A.Y.; Weinkam, P.; Oroudjev, E.; Hansma, H.G.; Jaeger, L. Building programmable jigsaw puzzles with RNA. Science 2004, 306, 2068–2072. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.W.; Zakrevsky, P.; Afonin, K.A.; Chworos, A.; Shapiro, B.A.; Jaeger, L. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011, 11, 878–887. [Google Scholar] [CrossRef] [PubMed]
- Bui, M.N.; Brittany Johnson, M.; Viard, M.; Satterwhite, E.; Martins, A.N.; Li, Z.; Marriott, I.; Afonin, K.A.; Khisamutdinov, E.F. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomedicine 2017, 13, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.B.; Halman, J.R.; Satterwhite, E.; Zakharov, A.V.; Bui, M.N.; Benkato, K.; Goldsworthy, V.; Kim, T.; Hong, E.; Dobrovolskaia, M.A.; et al. Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small 2017. [Google Scholar] [CrossRef] [PubMed]
- Delebecque, C.J.; Lindner, A.B.; Silver, P.A.; Aldaye, F.A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 2011, 333, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Oi, H.; Fujita, D.; Suzuki, Y.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. Programmable formation of catalytic RNA triangles and squares by assembling modular RNA enzymes. J. Biochem. 2017, 161, 451–462. [Google Scholar] [PubMed]
- Van der Horst, G.; Christian, A.; Inoue, T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. USA. 1991, 88, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Herschlag, D.; Doudna, J.A. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry 1999, 38, 2982–2990. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Furuta, H.; Ikawa, Y. Installation of orthogonality to the interface that assembles two modular domains in the Tetrahymena group I ribozyme. J. Biosci. Bioeng. 2014, 117, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.; Baudrey, S.; Jaeger, L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008, 36, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Fujita, Y.; Maeda, Y.; Furuta, H.; Ikawa, Y. GNRA/receptor interacting modules: Versatile modular units for natural and artificial RNA architectures. Methods 2011, 54, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Shiraishi, H.; Inoue, T. P5abc of the Tetrahymena ribozyme consists of three functionally independent elements. RNA 1998, 4, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, Y.; Moriyama, S.; Furuta, H. Facile syntheses of BODIPY derivatives for fluorescent labeling of the 3′ and 5′ ends of RNAs. Anal. Biochem. 2008, 378, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, M.A.; Doherty, E.A.; Knitt, D.S.; Doudna, J.A.; Herschlag, D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 2000, 39, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S.P.; Ikawa, Y.; Nakamura, Y. Selection of a novel class of RNA-RNA interaction motifs based on the ligase ribozyme with defined modular architecture. Nucleic Acids Res. 2008, 36, 3600–3607. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Matsumura, S.; Jaeger, L.; Inoue, T.; Furuta, H.; Ikawa, Y. Rational optimization of the DSL ligase ribozyme with GNRA/receptor interacting modules. Arch. Biochem. Biophys. 2009, 490, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Furuta, H.; Ikawa, Y. An in vitro-selected RNA receptor for the GAAC loop: Modular receptor for non-GNRA-type tetraloop. Nucleic Acids Res. 2013, 41, 3748–3759. [Google Scholar] [CrossRef] [PubMed]
- Serra, M.J.; Baird, J.D.; Dale, T.; Fey, B.L.; Retatagos, K.; Westhof, E. Effects of magnesium ions on the stabilization of RNA oligomers of defined structures. RNA 2002, 8, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.J.; Chen, S.J. Importance of diffuse metal ion binding to RNA. Met. Ions Life Sci. 2011, 9, 101–124. [Google Scholar] [PubMed]
- Lehnert, V.; Jaeger, L.; Michel, F.; Westhof, E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996, 3, 993–1009. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hassan, B.H.; Mirzoyan, M.G.; Leontis, N.B. Engineering cooperative tecto-RNA complexes having programmable stoichiometries. Nucleic Acids Res. 2011, 39, 2903–2917. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Westhof, E.; Leontis, N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001, 29, 455–463. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.M.; Matsumura, S.; Ikawa, Y. Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology 2017, 6, 37. https://doi.org/10.3390/biology6040037
Rahman MM, Matsumura S, Ikawa Y. Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology. 2017; 6(4):37. https://doi.org/10.3390/biology6040037
Chicago/Turabian StyleRahman, Md. Motiar, Shigeyoshi Matsumura, and Yoshiya Ikawa. 2017. "Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design" Biology 6, no. 4: 37. https://doi.org/10.3390/biology6040037
APA StyleRahman, M. M., Matsumura, S., & Ikawa, Y. (2017). Artificial RNA Motifs Expand the Programmable Assembly between RNA Modules of a Bimolecular Ribozyme Leading to Application to RNA Nanostructure Design. Biology, 6(4), 37. https://doi.org/10.3390/biology6040037





