Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits
Abstract
:1. Introduction
2. Methods
3. Results
3.1. Baseline Resting Hippocampal Histone Lysine Methylation Levels in Young and Aged Adults
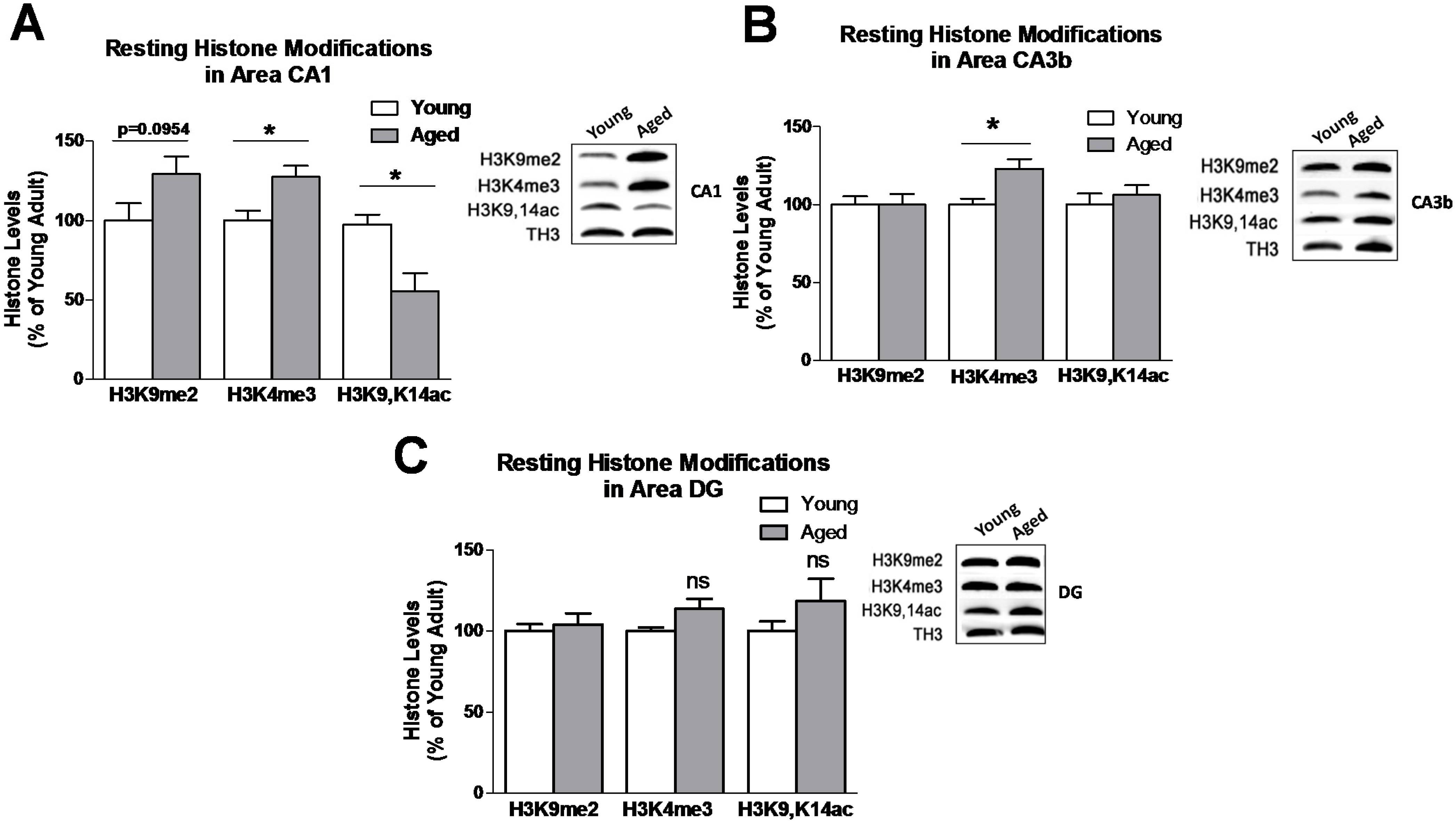
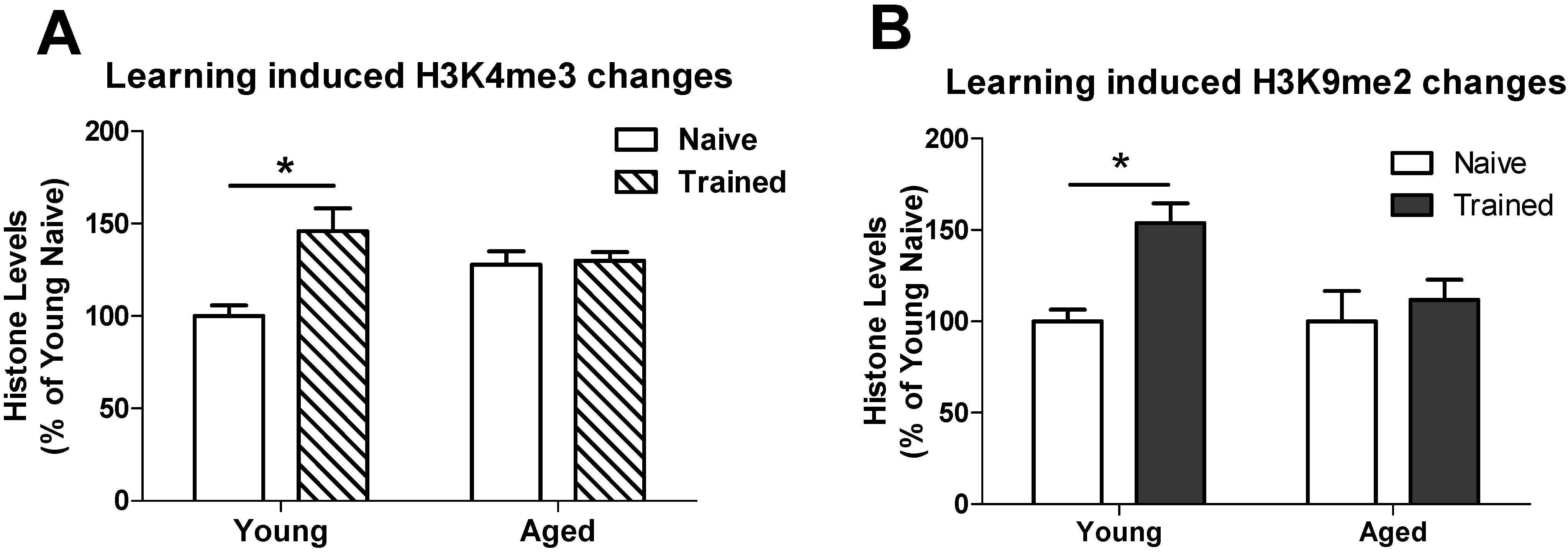
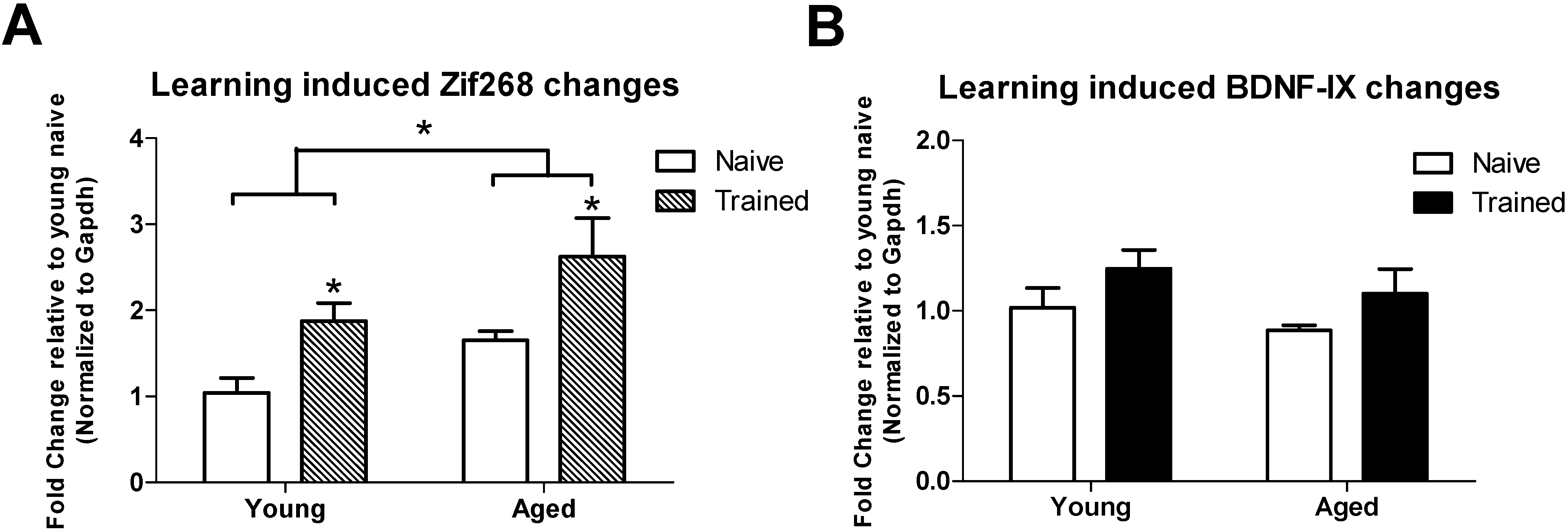
3.2. Learning induced Histone Lysine Methylation and Gene Expression Changes in the Young and Aged Hippocampus
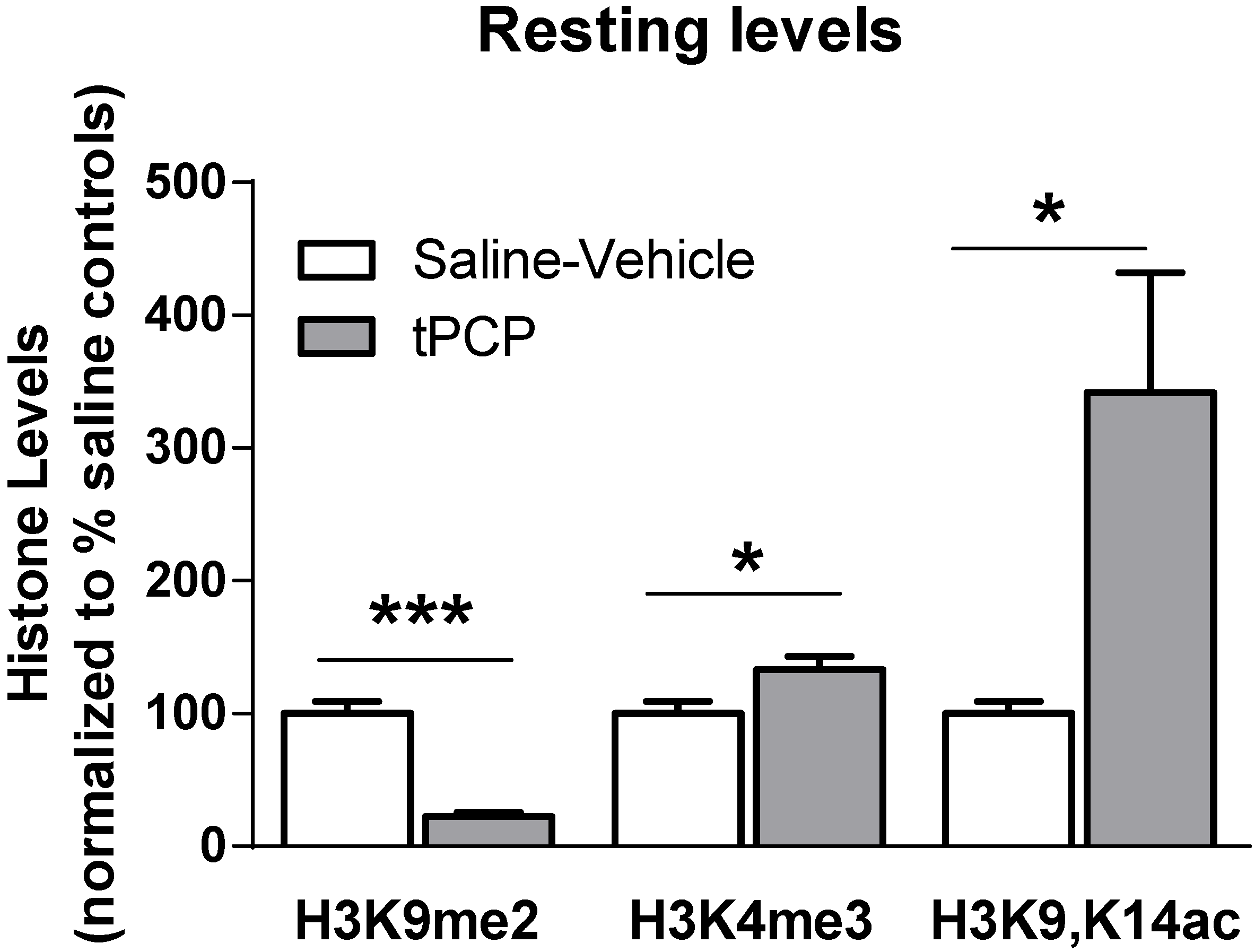
3.3. Inhibition of the LSD1 Histone Demethylase Mimics Age-Related Histone Lysine Methylation Changes and Memory Impairments in Young Adults
3.4. Environmental Enrichment Alters Age-Related H3K4me3 Levels at Gene Regions during Memory Formation

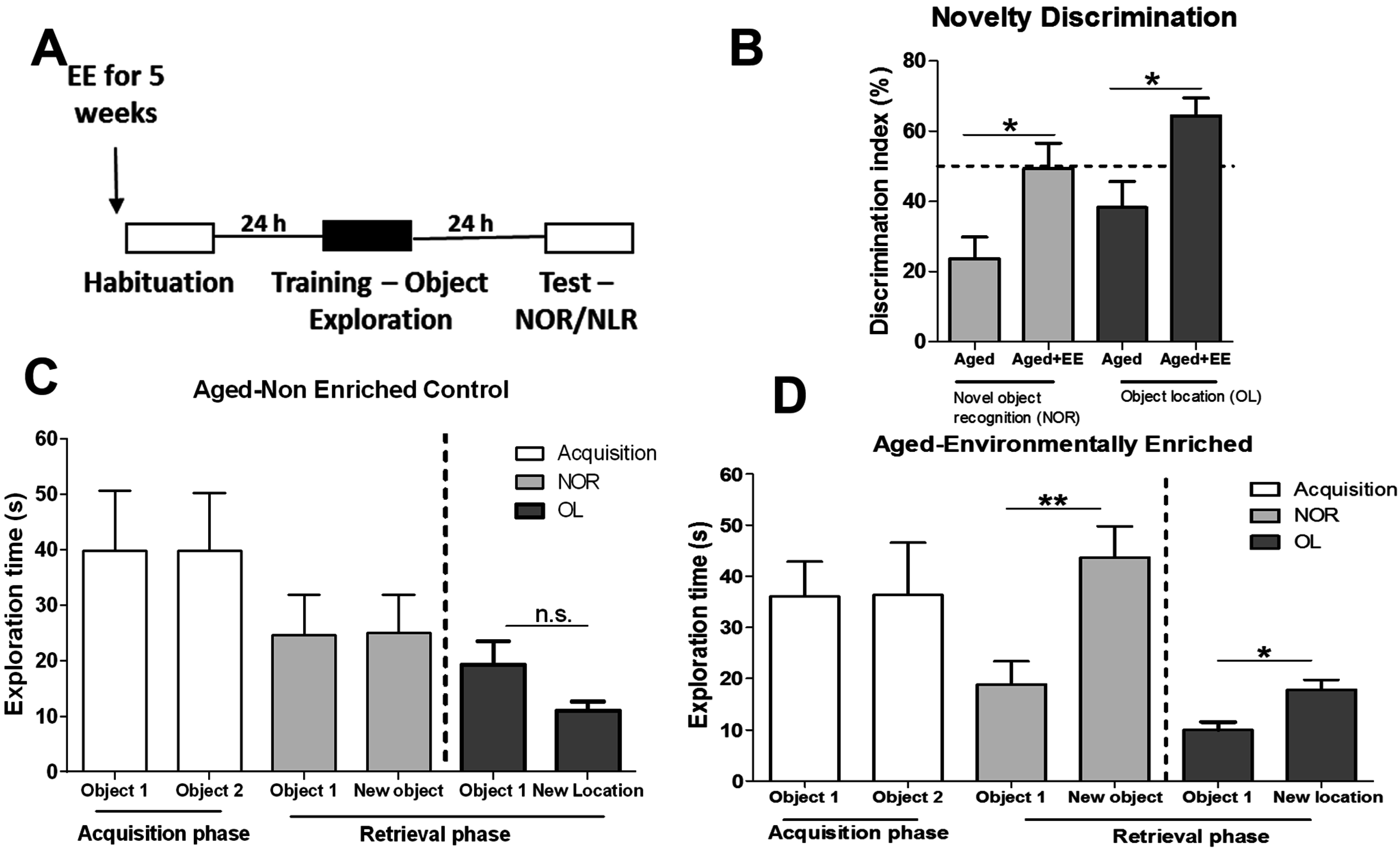
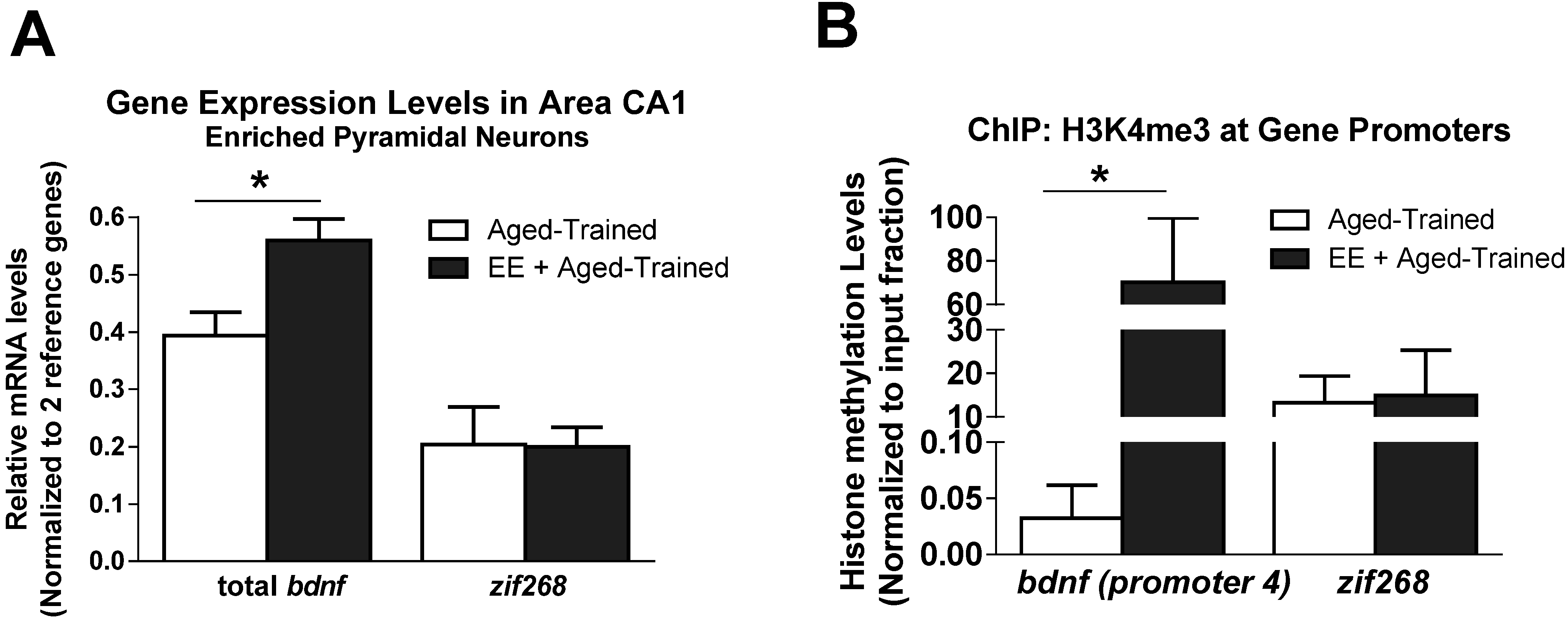
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bizon, J.L.; LaSarge, C.L.; Montgomery, K.S.; McDermott, A.N.; Setlow, B.; Griffith, W.H. Spatial reference and working memory across the lifespan of male fischer 344 rats. Neurobiol. Aging 2009, 30, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.; Rapp, P.R. The use of animal models to study the effects of aging on cognition. Annu. Rev. Psychol. 1997, 48, 339–370. [Google Scholar] [CrossRef] [PubMed]
- Shamy, J.L.; Buonocore, M.H.; Makaron, L.M.; Amaral, D.G.; Barnes, C.A.; Rapp, P.R. Hippocampal volume is preserved and fails to predict recognition memory impairment in aged rhesus monkeys (macaca mulatta). Neurobiol. Aging 2006, 27, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Thomas, K.L.; Everitt, B.J. Rapid and selective induction of Bdnf expression in the hippocampus during contextual learning. Nat. Neurosci. 2000, 3, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Blalock, E.M.; Chen, K.C.; Sharrow, K.; Herman, J.P.; Porter, N.M.; Foster, T.C.; Landfield, P.W. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 2003, 23, 3807–3819. [Google Scholar] [PubMed]
- Bramham, C.R. Control of synaptic consolidation in the dentate gyrus: Mechanisms, functions, and therapeutic implications. Prog. Brain Res. 2007, 163, 453–471. [Google Scholar] [PubMed]
- Burger, C. Region-specific genetic alterations in the aging hippocampus: Implications for cognitive aging. Front. Aging Neurosci. 2010, 2, 140. [Google Scholar] [CrossRef] [PubMed]
- Poirier, R.; Cheval, H.; Mailhes, C.; Charnay, P.; Davis, S.; Laroche, S. Paradoxical role of an EGR transcription factor family member, egr2/krox20, in learning and memory. Front. Behav. Neurosci. 2007, 1, 6. [Google Scholar] [PubMed]
- Rowe, W.B.; Blalock, E.M.; Chen, K.C.; Kadish, I.; Wang, D.; Barrett, J.E.; Thibault, O.; Porter, N.M.; Rose, G.M.; Landfield, P.W. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J. Neurosci. 2007, 27, 3098–3110. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D.; Roth, T.L.; Sweatt, J.D. Epigenetic regulation of Bdnf gene transcription in the consolidation of fear memory. J. Neurosci. 2008, 28, 10576–10586. [Google Scholar] [CrossRef] [PubMed]
- Kadish, I.; Thibault, O.; Blalock, E.M.; Chen, K.C.; Gant, J.C.; Porter, N.M.; Landfield, P.W. Hippocampal and cognitive aging across the lifespan: A bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J. Neurosci. 2009, 29, 1805–1816. [Google Scholar] [CrossRef]
- Gupta, S.; Kim, S.Y.; Artis, S.; Molfese, D.L.; Schumacher, A.; Sweatt, J.D.; Paylor, R.E.; Lubin, F.D. Histone methylation regulates memory formation. J. Neurosci. 2010, 30, 3589–3599. [Google Scholar] [CrossRef] [PubMed]
- Levenson, J.M.; O’Riordan, K.J.; Brown, K.D.; Trinh, M.A.; Molfese, D.L.; Sweatt, J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004, 279, 40545–40559. [Google Scholar] [CrossRef]
- Fischer, A.; Sananbenesi, F.; Wang, X.; Dobbin, M.; Tsai, L.H. Recovery of learning and memory is associated with chromatin remodelling. Nature 2007, 447, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Peleg, S.; Sananbenesi, F.; Zovoilis, A.; Burkhardt, S.; Bahari-Javan, S.; Agis-Balboa, R.C.; Cota, P.; Wittnam, J.L.; Gogol-Doering, A.; Opitz, L.; et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010, 328, 753–756. [Google Scholar] [CrossRef]
- Penner, M.R.; Roth, T.L.; Chawla, M.K.; Hoang, L.T.; Roth, E.D.; Lubin, F.D.; Sweatt, J.D.; Worley, P.F.; Barnes, C.A. Age-related changes in arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging 2010, 32, 2198–2210. [Google Scholar] [CrossRef] [PubMed]
- Castellano, J.F.; Fletcher, B.R.; Kelley-Bell, B.; Kim, D.H.; Gallagher, M.; Rapp, P.R. Age-related memory impairment is associated with disrupted multivariate epigenetic coordination in the hippocampus. PLOS ONE 2012, 7, e33249. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D. Epigenetic gene regulation in the adult mammalian brain: Multiple roles in memory formation. Neurobiol. Learn. Mem. 2011, 96, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Jarome, T.J.; Lubin, F.D. Histone lysine methylation: Critical regulator of memory and behavior. Rev. Neurosci. 2013, 24, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Gupta-Agarwal, S.; Franklin, A.V.; Deramus, T.; Wheelock, M.; Davis, R.L.; McMahon, L.L.; Lubin, F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012, 32, 5440–5453. [Google Scholar] [CrossRef] [PubMed]
- Mora-Gallegos, A.; Rojas-Carvajal, M.; Salas, S.; Saborio-Arce, A.; Fornaguera-Trias, J.; Brenes, J.C. Age-dependent effects of environmental enrichment on spatial memory and neurochemistry. Neurobiol. Learn. Mem. 2014, 118C, 96–104. [Google Scholar] [CrossRef]
- Freret, T.; Billard, J.M.; Schumann-Bard, P.; Dutar, P.; Dauphin, F.; Boulouard, M.; Bouet, V. Rescue of cognitive aging by long-lasting environmental enrichment exposure initiated before median lifespan. Neurobiol. Aging 2012, 33, 1005.e1–1005.e10. [Google Scholar] [CrossRef]
- Bouet, V.; Freret, T.; Dutar, P.; Billard, J.M.; Boulouard, M. Continuous enriched environment improves learning and memory in adult nmri mice through theta burst-related-LTP independent mechanisms but is not efficient in advanced aged animals. Mech. Ageing Dev. 2011, 132, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Leal-Galicia, P.; Castaneda-Bueno, M.; Quiroz-Baez, R.; Arias, C. Long-term exposure to environmental enrichment since youth prevents recognition memory decline and increases synaptic plasticity markers in aging. Neurobiol. Learn. Mem. 2008, 90, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Frick, K.M.; Fernandez, S.M. Enrichment enhances spatial memory and increases synaptophysin levels in aged female mice. Neurobiol. Aging 2003, 24, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Van Praag, H.; Christie, B.R.; Sejnowski, T.J.; Gage, F.H. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc. Natl. Acad. Sci. USA 1999, 96, 13427–13431. [Google Scholar] [CrossRef] [PubMed]
- Vaynman, S.; Ying, Z.; Gomez-Pinilla, F. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience 2003, 122, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Vaynman, S.; Ying, Z.; Gomez-Pinilla, F. Hippocampal Bdnf mediates the efficacy of exercise on synaptic plasticity and cognition. Eur. J. Neurosci. 2004, 20, 2580–2590. [Google Scholar] [CrossRef] [PubMed]
- Ennaceur, A.; Cavoy, A.; Costa, J.C.; Delacour, J. A new one-trial test for neurobiological studies of memory in rats. II: Effects of piracetam and pramiracetam. Behav. Brain Res. 1989, 33, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Bevins, R.A.; Besheer, J. Object recognition in rats and mice: A one-trial non-matching-to-sample learning task to study “recognition memory”. Nat. Protoc. 2006, 1, 1306–1311. [Google Scholar] [CrossRef]
- Ryley Parrish, R.; Albertson, A.J.; Buckingham, S.C.; Hablitz, J.J.; Mascia, K.L.; Davis Haselden, W.; Lubin, F.D. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and nmda glutamate receptor grin2b DNA methylation levels in the hippocampus. Neuroscience 2013, 248, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Neelamegam, R.; Ricq, E.L.; Malvaez, M.; Patnaik, D.; Norton, S.; Carlin, S.M.; Hill, I.T.; Wood, M.A.; Haggarty, S.J.; Hooker, J.M. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chem. Neurosci. 2012, 3, 120–128. [Google Scholar] [CrossRef]
- Yu, J.L.; Ma, L.; Ma, L.; Tao, Y.Z. Voluntary wheel running enhances cell proliferation and expression levels of Bdnf, IGF1 and WNT4 in dentate gyrus of adult mice. Sheng Li Xue Bao 2014, 66, 559–568. [Google Scholar]
- Boehme, F.; Gil-Mohapel, J.; Cox, A.; Patten, A.; Giles, E.; Brocardo, P.S.; Christie, B.R. Voluntary exercise induces adult hippocampal neurogenesis and Bdnf expression in a rodent model of fetal alcohol spectrum disorders. Eur. J. Neurosci. 2011, 33, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J.; Zhao, X.; van Praag, H.; Wodtke, K.; Gage, F.H.; Christie, B.R. Effects of voluntary exercise on synaptic plasticity and gene expression in the dentate gyrus of adult male sprague-dawley rats in vivo. Neuroscience 2004, 124, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D.; Gupta, S.; Parrish, R.R.; Grissom, N.M.; Davis, R.L. Epigenetic mechanisms: Critical contributors to long-term memory formation. Neuroscientist 2011, 17, 616–632. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, A.; Tsai, L.H. Epigenetic modifications in the nervous system and their impact upon cognitive impairments. Neuropharmacology 2014, 80, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Jarome, T.J.; Lubin, F.D. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn. Mem. 2014, 115C, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Alzayady, K.; Stewart, R.; Ye, P.; Yang, S.; Li, W.; Shi, Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010, 30, 1997–2005. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.G.; Hillenmeyer, S.; Lawrence, C.; Chang, C.; Hosier, S.; Lightfoot, W.; Mukherjee, E.; Jiang, N.; Schorl, C.; Brodsky, A.S.; et al. Chromatin remodeling in the aging genome of drosophila. Aging Cell 2010, 9, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Kuzumaki, N.; Ikegami, D.; Tamura, R.; Hareyama, N.; Imai, S.; Narita, M.; Torigoe, K.; Niikura, K.; Takeshima, H.; Ando, T.; et al. Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus 2011, 21, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015, 8, 199–209. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morse, S.J.; Butler, A.A.; Davis, R.L.; Soller, I.J.; Lubin, F.D. Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits. Biology 2015, 4, 298-313. https://doi.org/10.3390/biology4020298
Morse SJ, Butler AA, Davis RL, Soller IJ, Lubin FD. Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits. Biology. 2015; 4(2):298-313. https://doi.org/10.3390/biology4020298
Chicago/Turabian StyleMorse, Sarah J., Anderson A. Butler, Robin L. Davis, Ian J. Soller, and Farah D. Lubin. 2015. "Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits" Biology 4, no. 2: 298-313. https://doi.org/10.3390/biology4020298
APA StyleMorse, S. J., Butler, A. A., Davis, R. L., Soller, I. J., & Lubin, F. D. (2015). Environmental Enrichment Reverses Histone Methylation Changes in the Aged Hippocampus and Restores Age-Related Memory Deficits. Biology, 4(2), 298-313. https://doi.org/10.3390/biology4020298




