In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss)
Abstract
:1. Introduction
2. Experimental Section
2.1. Animals and Treatment
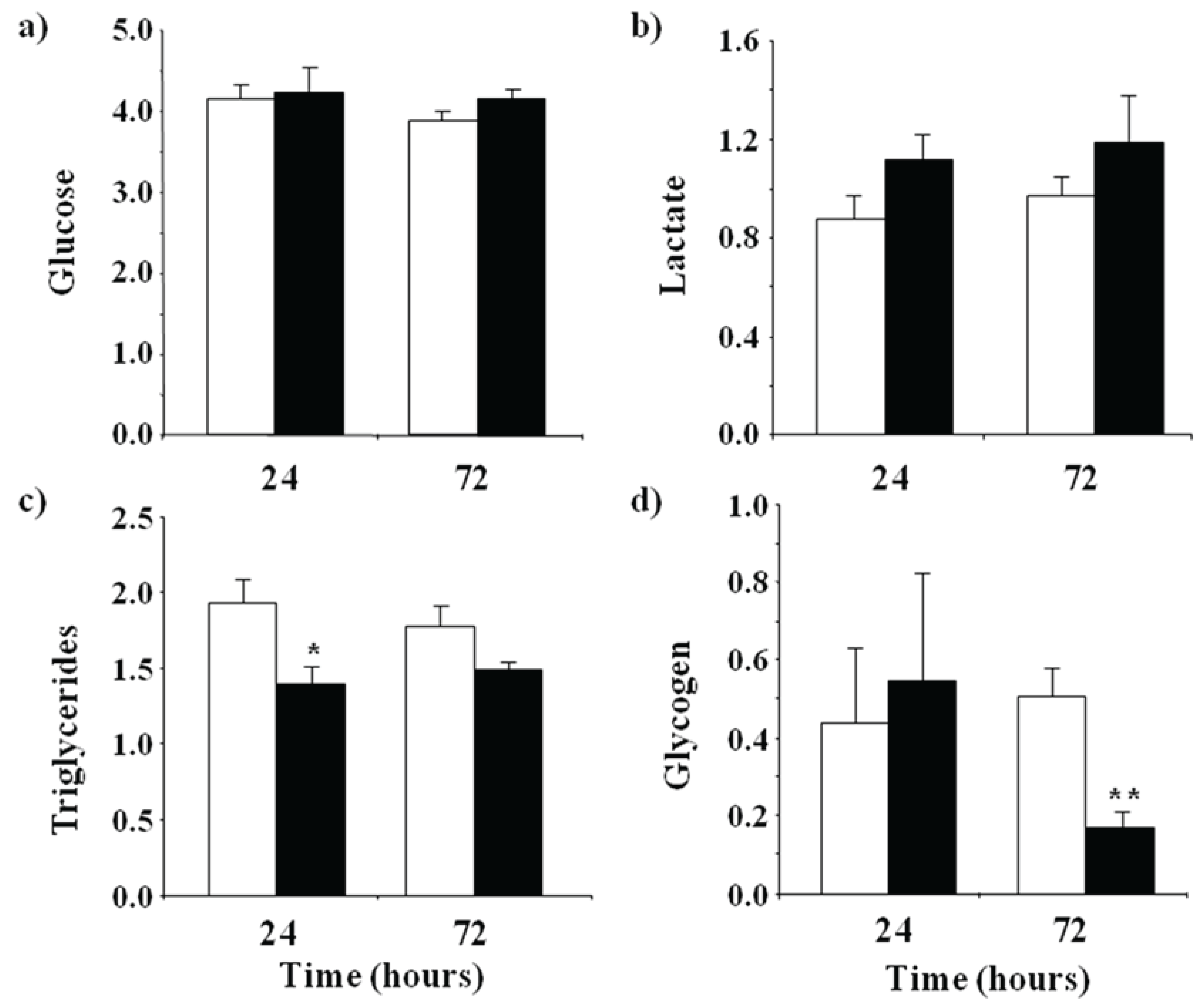
2.2. Biochemical Parameters
2.3. RNA Extraction and cDNA Synthesis
2.4. Microarray Analysis
2.5. Quantitative Real-Time PCR (qPCR)
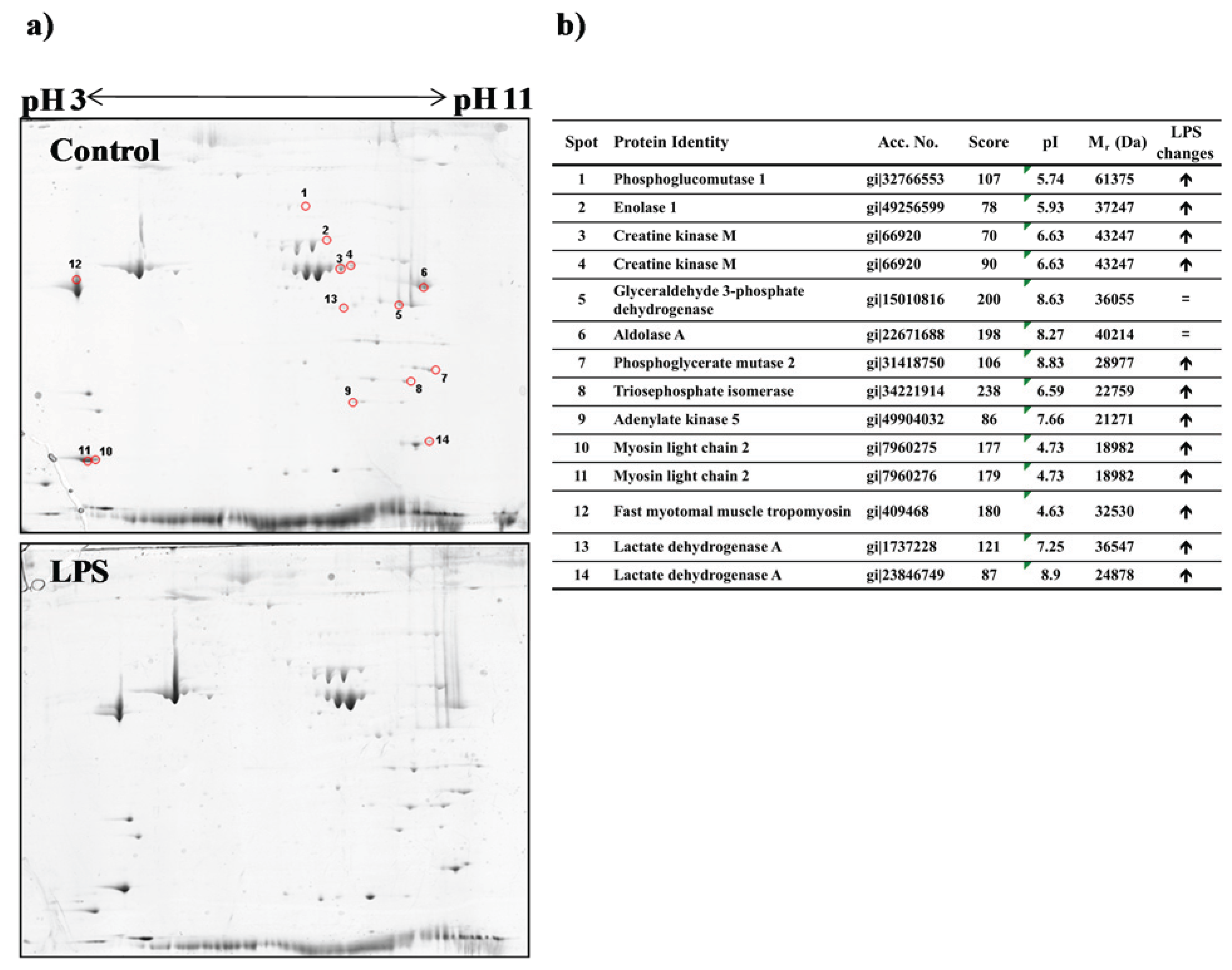
2.6. Protein Extraction, Separation and Visualization
| GeneBank ID | Clone Name | Slow Muscle | Fast Muscle | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | ||||||
| MA | qPCR | MA | qPCR | MA | qPCR | MA | qPCR | ||
| CB511095 | Glyceraldehyde-3-phosphate dehydrogenase-3 | −1.75 | −1.20 | −1.44 | −1.20 | ||||
| BX074486 | Heat shock protein HSP 90-beta-2 | 1.25 | 73.51 | ||||||
| CX146261 | Myosin light chain 1, skeletal muscle isoform | −3.78 | −14.32 | ||||||
| CU069718 | Parvalbumin alpha-3 | 1.82 | 4.59 | −74.54 | −592.22 | 1.99 | 5.86 | 2.38 | 5.54 |
| DY467707 | Serine protease-like protein-1 | 2.23 | 3.39 | −1.34 | −1.83 | ||||
| CA039449 | Tropomyosin alpha 3 chain-2 | −2.11 | −37.79 | ||||||
| ES325822 | Troponin I-4, fast skeletal muscle | 1.72 | 3.53 | −5.54 | −5.90 | 1.58 | 3.25 | 3.76 | 2.69 |
| CA371001 | Very-long-chain acyl-CoA synthetase | 1.27 | 22.32 | ||||||
2.7. Protein Identification by Mass Spectrometry (MS)
3. Results and Discussion
3.1. Metabolic Effects of LPS Administration in Rainbow Trout
3.2. Transcriptomic Analysis of Fast and Slow Skeletal Muscle in Response to LPS Administration in Rainbow Trout
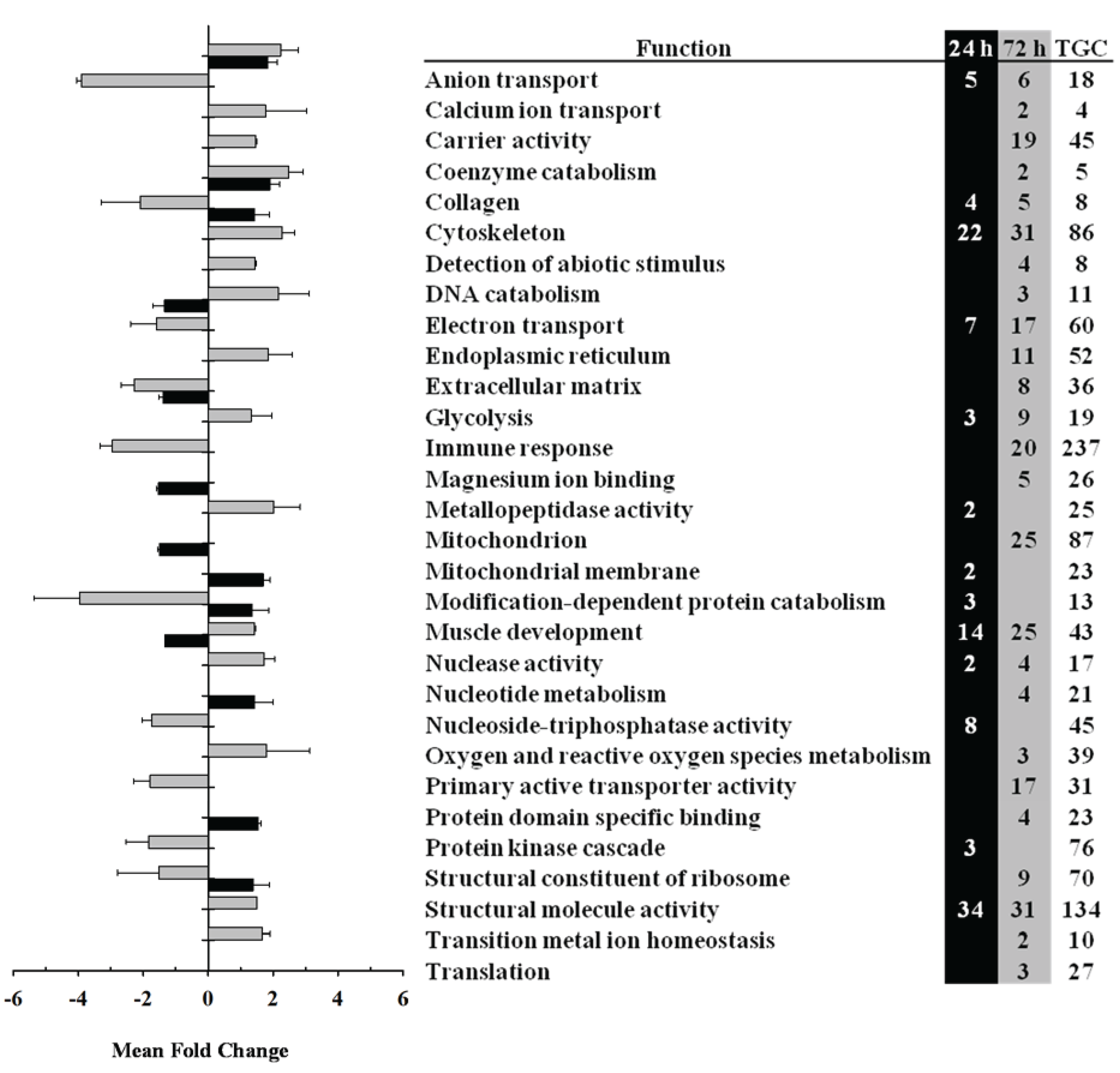

| Clone Name | Slow Muscle | Fast Muscle | ||
|---|---|---|---|---|
| 24 h | 72 h | 24 h | 72 h | |
| Carbohydrate metabolism | ||||
| Beta enolase-3 | −2.85 | 2.41 | ||
| 6-phosphofructokinase | −1.53 | −1.88 | ||
| Glyceraldehyde-3-phosphate dehydrogenase-5 | −1.84 | −1.61 | 1.66 | |
| Glycogen phosphorylase-2 | −1.99 | 1.51 | ||
| Fructose-1,6-bisphosphatase isozyme 2 | 1.62 | |||
| Glucose-6-phosphate isomerase-1 | 3.10 | |||
| Protein synthesis/catabolism/modification | ||||
| 40S ribosomal protein S9-3 | −1.51 | 1.77 | ||
| 60S ribosomal protein L13a-acute pahse protein | 2.14 | |||
| 60S ribosomal protein L32-2 | −1.88 | 2.08 | ||
| Cathepsin D | 1.66 | |||
| Proteasome activator complex subunit 2 | 1.60 | 1.83 | ||
| Proteasome subunit alpha type 7-2 | −3.84 | |||
| Proteasome subunit beta type 9 precursor | 2.01 | 1.49 | 1.88 | |
| Serine protease-like protein-1 | 2.23 | 1.78 | ||
| Ubiquitin | −3.73 | −265.03 | ||
| Muscle development/cytoskeleton | ||||
| Actin, alpha skeletal 3 | 1.73 | −3.53 | 1.87 | |
| Creatine kinase, M-3 | −2.75 | |||
| Myosin heavy chain, skeletal, adult 1-2 | −3.32 | −1.51 | ||
| Myosin light chain 2-2 | 1.77 | −5.94 | 3.20 | |
| Parvalbumin alpha-3 | 1.82 | −74.54 | 1.99 | 2.38 |
| Troponin I-4, fast skeletal muscle | 1.72 | −5.54 | 1.58 | 3.76 |
| Tropomyosin alpha 3 chain-2 | −2.11 | 2.55 | ||
| Immune response | ||||
| Beta-2-microglobulin-1 | 2.13 | 1.52 | 1.56 | |
| CC chemokine SCYA110-2 | −2.14 | −2.97 | ||
| Ferritin heavy chain-1 | 1.62 | |||
| Ig kappa chain V-IV region B17-2 | 1.58 | 3.41 | ||
| Lysozyme C precursor | 1.59 | 2.17 | ||
| Macrophage receptor MARCO | 1.60 | −1.59 | ||
| MHC class 1b antigen | 2.45 | 1.64 | 2.36 | |
| MHC class II invariant chain-like protein 1 | 1.60 | 2.19 | 2.06 | |
| Lipid metabolism | ||||
| Apolipoprotein A-I-1 | 4.56 | |||
| Apolipoprotein E-2 | 1.72 | |||
| Fatty acid-binding protein-1 | −1.62 | 1.68 | −1.64 | −2.17 |
| Acyl-Coenzyme A dehydrogenase, long chain | 2.46 | |||
| Mitochondrion/electron transport | ||||
| ATP synthase coupling factor 6, mitochondrial | 2.19 | |||
| Cytochrome b-1 | 1.73 | −1.71 | ||
| Cytochrome oxidase subunit III-2 | 2.89 | |||
| NADH dehydrogenase subunit 5-1 | 7.21 | 1.53 | 1.89 | |

3.2.1. Muscle Type-Specific Responses to LPS Administration
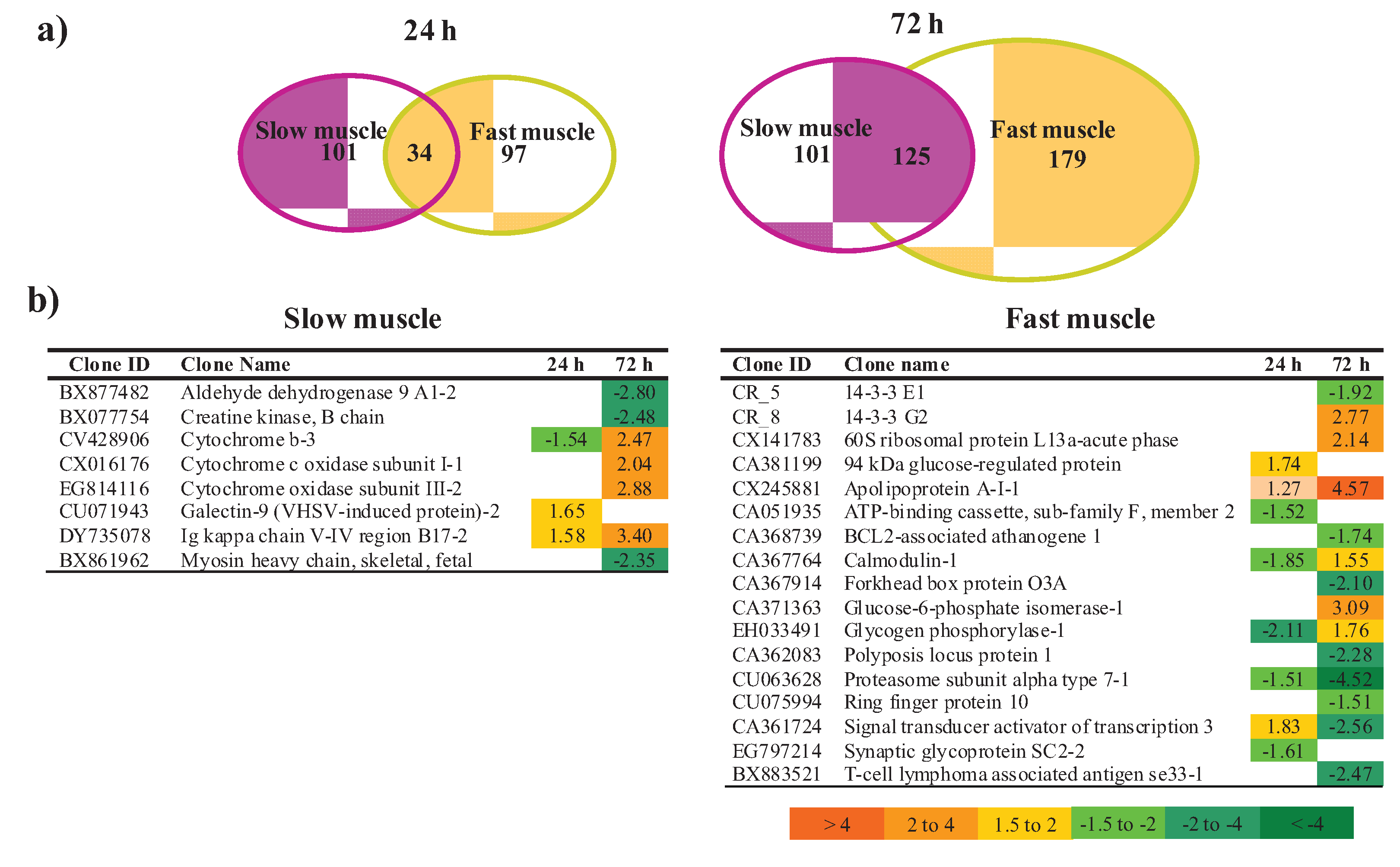
3.3. Proteomic Analysis of Fast Skeletal Muscle in Response to LPS Administration in Rainbow Trout
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Degens, H. The role of systemic inflammation in age-related muscle weakness and wasting. Scand. J. Med. Sci. Sports 2010, 20, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Rectenwald, J.E.; Moldawer, L.L. Skeletal muscle and cytokines in sepsis and severe injury. In Skeletal Muscle: Pathology, Diagnosis and Management of Disease; Preedy, V.R., Peters, T.J., Eds.; Cambridge University Press: Cambridge, UK, 2002; pp. 301–310. [Google Scholar]
- Frost, R.A.; Lang, C.H. Regulation of muscle growth by pathogen associated molecules. J. Anim. Sci. 2008, 86, 84–93. [Google Scholar] [CrossRef]
- Wiendl, H.; Hohlfeld, R.; Kieseier, B. Immunobiology of muscle: Advances in understanding an immunological microenvironment. Trends Immunol. 2005, 26, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, K. Immunological capabilities of skeletal muscle cells. Acta Physiol. Scand. 2001, 171, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Read, R.C.; Wyllie, D.H. Toll receptors and sepsis. Curr. Opin. Crit. Care 2001, 7, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Michie, H.R. Metabolism of sepsis and multiple organ failure. World J. Surg. 1996, 20, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Iliev, D.B.; Roach, J.C.; MacKenzie, S.; Planas, J.V.; Goetz, F.W. Endotoxin recognition: In fish or not in fish? FEBS Lett. 2005, 579, 6519–6528. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y. Toll-like receptors in bony fish: From genomics to function. Dev. Comp. Immunol. 2011, 35, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.; Iliev, D.; Liarte, C.; Koskinen, H.; Planas, J.V.; Goetz, F.W.; Mölsä, H.; Krasnov, A.; Tort, L. Transcriptional analysis of LPS-stimulated activation of trout (Oncorhynchus mykiss) monocyte/macrophage cells in primary culture treated with cortisol. Mol. Immunol. 2006, 43, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.; Balasch, J.C.; Novoa, B.; Ribas, L.; Roher, N.; Krasnov, A.; Figueras, A. Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Swain, P.; Nayak, S.K.; Nanda, P.K.; Dash, S. Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: A review. Fish Shellfish Immunol. 2008, 25, 191–201. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.; Planas, J.V.; Goetz, F.W. LPS-Stimulated expression of a tumor necrosis factor-a mRNA in primary trout monocytes and in vitro differentiated macrophages. Dev. Comp. Immunol. 2003, 27, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Iliev, D.B.; Liarte, C.Q.; MacKenzie, S.; Goetz, F.W. Activation of rainbow trout (Oncorhynchus mykiss) mononuclear phagocytes by different pathogen associated molecular pattern (PAMP) bearing agents. Mol. Immunol. 2005, 42, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Moyes, C.D.; West, T.G. Exercise metabolism of fish. In Metabolic Biochemistry; Hochachka, P.W., Mommsen, T.P., Hochachka, P.W., Mommsen, T.P., Eds.; Elsevier Science: Amsterdam, the Netherlands, 1995; Volume 4, pp. 368–392. [Google Scholar]
- Bone, Q. Locomotor muscle. In Fish Physiology; Hoar, W.S., Randall, D.J., Eds.; Academic Press: Oxford, UK, 1978; Volume 7, pp. 361–424. [Google Scholar]
- Altringham, J.D.; Ellerby, D.J. Fish swimming: patterns in muscle function. J. Exp. Biol. 1999, 202, 3397–3403. [Google Scholar] [PubMed]
- Kaitetzidou, E.; Crespo, D.; Vraskou, Y.; Antonopoulou, E.; Planas, J.V. Transcriptomic response of skeletal muscle to lipopolysaccharide in the gilthead seabream (Sparus aurata). Mar. Biotechnol. 2012, 14, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Magnoni, L.J.; Crespo, D.; Ibarz, A.; Blasco, J.; Fernández-Borràs, J.; Planas, J.V. Effects of sustained swimming on the red and white muscle transcriptome of rainbow trout (Oncorhynchus mykiss) fed a carbohydrate-rich diet. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 166, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hassid, W.Z.; Abraham, S. Chemical procedures for the analysis of polysaccharides. In Methods in Enzymology; Colowick, S.P., Kaplan, N.O., Eds.; Academic: New York, NY, USA, 1957; Volume 3, pp. 34–50. [Google Scholar]
- Koskinen, H.; Pehkonen, P.; Vehniainen, E.; Krasnov, A.; Rexroad, C.; Afanasyev, S.; Molsa, H.; Oikari, A. Response of rainbow trout transcriptome to model chemical contaminants. Biochem. Biophys. Res. Commun. 2004, 320, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Koskinen, H.; Pehkonen, P.; Rexroad, C.; Afanasyev, S.; Molsa, H. Gene expression in the brain and kidney of rainbow trout in response to handling stress. BMC Genomics 2005, 6, 3–14. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, S.; Montserrat, N.; Mas, M.; Acerete, L.; Tort, L.; Krasnov, A.; Goetz, F.W.; Planas, J.V. Bacterial lipopolysaccharide induces apoptosis in the trout ovary. Reprod. Biol. Endocrinol. 2006, 4. [Google Scholar] [CrossRef]
- Jorgensen, S.M.; Hetland, D.L.; Press, C.M.; Grimholt, U.; Gjøen, T. Effect of early infectious salmon anaemia virus (ISAV) infection on expression of MHC pathway genes and type I and II interferon in Atlantic salmon (Salmo salar L.) tissues. Fish Shellfish Immunol. 2007, 23, 576–588. [Google Scholar] [CrossRef]
- Schiotz, B.L.; Jorgensen, S.M.; Rexroad, C.; Gjoen, T.; Krasnov, A. Transcriptomic analysis of responses to infectious salmon anemia virus infection in macrophage-like cells. Virus Res. 2008, 136, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Skugor, S.; Glover, K.A.; Nilsen, F.; Krasnov, A. Local and systemic gene expression responses of Atlantic salmon (Salmo salar L.) to infection with the salmon louse (Lepeophtheirus salmonis). BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Djordjevic, B.; Skugor, S.; Jorgensen, S.M.; Overland, M.; Mydland, L.T.; Krasnov, A. Modulation of splenic immune responses to bacterial lipopolysaccharide in rainbow trout (Oncorhynchus mykiss) fed lentinan, a beta-glucan from mushroom Lentinula edodes. Fish Shellfish Immunol. 2009, 26, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Goetz, F.W.; Iliev, D.B.; McCauley, L.A.R.; Liarte, C.Q.; Tort, L.B.; Planas, J.V.; MacKenzie, S. Analysis of genes isolated from lipopolysaccharide-stimulated rainbow trout (Oncorhynchus mykiss) macrophages. Mol. Immunol. 2004, 41, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, S.A.; Elwyn, D.H. The effects of injury and sepsis on fuel utilization. Annu. Rev. Nutr. 1989, 9, 445–473. [Google Scholar] [CrossRef] [PubMed]
- Marik, P.E.; Raghavan, M. Stress-hyperglycemia, insulin and immunomodulation in sepsis. Intens. Care Med. 2004, 30, 748–756. [Google Scholar] [CrossRef]
- Crespo, D.; Bonnet, E.; Roher, N.; Mackenzie, S.A.; Krasnov, A.; Goetz, F.W.; Bobe, J.; Planas, J.V. Cellular and molecular evidence for a role of tumor necrosis factor alpha in the ovulatory mechanism of trout. Reprod. Biol. Endocrinol. 2010, 8. [Google Scholar] [CrossRef] [PubMed]
- Callahan, L.A.; Supinski, G.S. Downregulation of diaphragm electron electron transport chain and glycolitic enzyme expression in sepsis. J. Appl. Physiol. 2004, 99, 1120–1126. [Google Scholar] [CrossRef]
- Macallan, D.C.; Cook, E.B.; Preedy, V.R.; Griffin, G.E. The effect of endotoxin on skeletal muscle protein gene expression in the rat. J. Biochem. Cell Biol. 1996, 28, 511–520. [Google Scholar] [CrossRef]
- Van den Thillart, G. Energy metabolism of swimming trout (Salmo gairdneri). J. Comp. Physiol. B 1986, 156, 511–520. [Google Scholar] [CrossRef]
- Salem, M.; Kenney, P.B.; Rexroad, C.E.; Yao, J. Microarray gene expression analysis in atrophying rainbow trout muscle: A unique nonmammalian muscle degradation model. Physiol. Genom. 2006, 28, 33–45. [Google Scholar] [CrossRef]
- Johansen, K.A.; Sealey, W.M.; Overturf, K. The effects of chronic immune stimulation on muscle growth in rainbow trout. Comp. Biochem. Physiol. B 2006, 144, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Arnold, L.; Henry, A.; Poron, F.; Baba-Amer, Y.; van Rooijen, N.; Plonquet, A.; Gherardi, R.K.; Chazaud, B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007, 204, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- McLennan, I.S. Degenerating and regenerating skeletal muscles contain several subpopulations of macrophages with distinct spatial and temporal distributions. J. Anat. 1996, 188, 17–28. [Google Scholar] [PubMed]
- Pimorady-Esfahani, A.; Grounds, M.D.; McMenamin, P.G. Macrophages and dendritic cells in normal and regenerating murine skeletal muscle. Muscle Nerve 1997, 20, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.H.; Silvis, C.; Deshpande, N.; Nystrom, G.; Frost, R.A. Endotoxin stimulates in vivo expression of inflammatory cytokines tumor necrosis factor alpha, interleukin-1[beta], -6, and high-mobility-group protein-1 in skeletal muscle. Shock 2003, 19, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.A.; Nystrom, G.J.; Lang, C.H. Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R698–R709. [Google Scholar] [PubMed]
- Grunfeld, C.; Feingold, K.R. Tumor necrosis factor, interleukin, and interferon induced changes in lipid metabolism as part of host defense. Proc. Soc. Exp. Biol. Med. 1992, 200, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Tracey, K.J.; Cerami, A. Tumor necrosis factor, other cytokines and disease. Annu. Rev. Cell Biol. 1993, 9, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Hardardóttir, I.; Grunfeld, C.; Feingold, K.R. Effects of endotoxin and cytokines on lipid metabolism. Curr. Opin. Lipidol. 1994, 5, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kong, X.; Zhou, C.; Li, L.; Nie, G.; Li, X. Toll-Like receptor recognition of bacteria in fish: Ligand specificity and signal pathways. Fish Shellfish Immunol. 2014, 41, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Roher, N.; Callol, A.; Planas, J.V.; Goetz, F.W.; Mackenzie, S.A. Endotoxin recognition in fish results in inflammatory cytokine secretion not gene expression. Innate Immun. 2011, 17, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Albalat, A.; Liarte, C.; MacKenzie, S.; Tort, L.; Planas, J.V.; Navarro, I. Control of adipose tissue lipid metabolism by tumor necrosis factor alpha in rainbow trout (Oncorhynchus mykiss). J. Endocrinol. 2005, 184, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Barcia, A.M.; Harris, H.W. Triglyceride-rich lipoproteins as agents of innate immunity. Clin. Infect. Dis. 2005, 41, S498–S503. [Google Scholar] [CrossRef] [PubMed]
- Tessier, J.-P.; Thurner, B.; Jungling, E.; Luckhoff, A.; Fischer, Y. Impairment of glucose metabolism in hearts from rats treated with endotoxin. Cardiovasc. Res. 2003, 60, 119–130. [Google Scholar] [CrossRef]
- Celis, J.E.; Kruhøffer, M.; Gromova, I.; Frederiksen, C.; Ostergaard, M.; Thykjaer, T.; Gromov, P.; Yu, J.; Pálsdóttir, H.; Magnusson, N.; et al. Gene expression profiling: Monitoring transcription and translation products using DNA microarrays and proteomics. FEBS Lett. 2000, 480, 2–16. [Google Scholar]
- Forné, I.; Castellana, B.; Marín-Juez, R.; Cerdà, J.; Abián, J.; Planas, J.V. Transcriptional and proteomic profiling of flatfish (Solea senegalensis) spermatogenesis. Proteomics 2011, 11, 2195–2211. [Google Scholar] [CrossRef] [PubMed]
- Bosworth, C.; Chou, C.W.; Cole, R.B.; Rees, B.B. Protein expression patterns in zebrafish skeletal muscle: Initial characterization and the effects of hypoxic exposure. Proteomics 2005, 5, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Ton, C.; Stamatiou, D.; Liew, C.-C. Gene expression profile of zebrafish exposed to hypoxia during development. Physiol. Genomics 2003, 13, 97–106. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magnoni, L.J.; Roher, N.; Crespo, D.; Krasnov, A.; Planas, J.V. In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss). Biology 2015, 4, 67-87. https://doi.org/10.3390/biology4010067
Magnoni LJ, Roher N, Crespo D, Krasnov A, Planas JV. In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss). Biology. 2015; 4(1):67-87. https://doi.org/10.3390/biology4010067
Chicago/Turabian StyleMagnoni, Leonardo J., Nerea Roher, Diego Crespo, Aleksei Krasnov, and Josep V. Planas. 2015. "In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss)" Biology 4, no. 1: 67-87. https://doi.org/10.3390/biology4010067
APA StyleMagnoni, L. J., Roher, N., Crespo, D., Krasnov, A., & Planas, J. V. (2015). In Vivo Molecular Responses of Fast and Slow Muscle Fibers to Lipopolysaccharide in a Teleost Fish, the Rainbow Trout (Oncorhynchus mykiss). Biology, 4(1), 67-87. https://doi.org/10.3390/biology4010067





