Stage-Specific Alternative Polyadenylation During Human Neural Differentiation Revealed by Integrated Long- and Short-Read Sequencing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. hESCs Culture
2.2. hESC Differentiation into Neural Stem Cells (NSCs)
2.3. hESC Differentiation into NPCs
2.4. Immunofluorescence Staining
2.5. Library Construction and Sequencing for ONT
2.6. ONT Transcriptome Sequencing and Data Processing
2.7. APA Analysis
2.8. Illumina RNA-Seq Data Analysis
2.9. Quality Control of PASs
2.10. Functional Enrichment Analysis
2.11. Quantitative Real-Time PCR
2.12. Target microRNA (miRNA) and RNA-Binding Protein (RBP) Prediction
3. Results
3.1. ONT Long-Read Sequencing Reveals the Transcriptomic Diversity in Human Neural Differentiation
3.2. Identification of PASs in All Gene Regions by Long-Read Sequencing
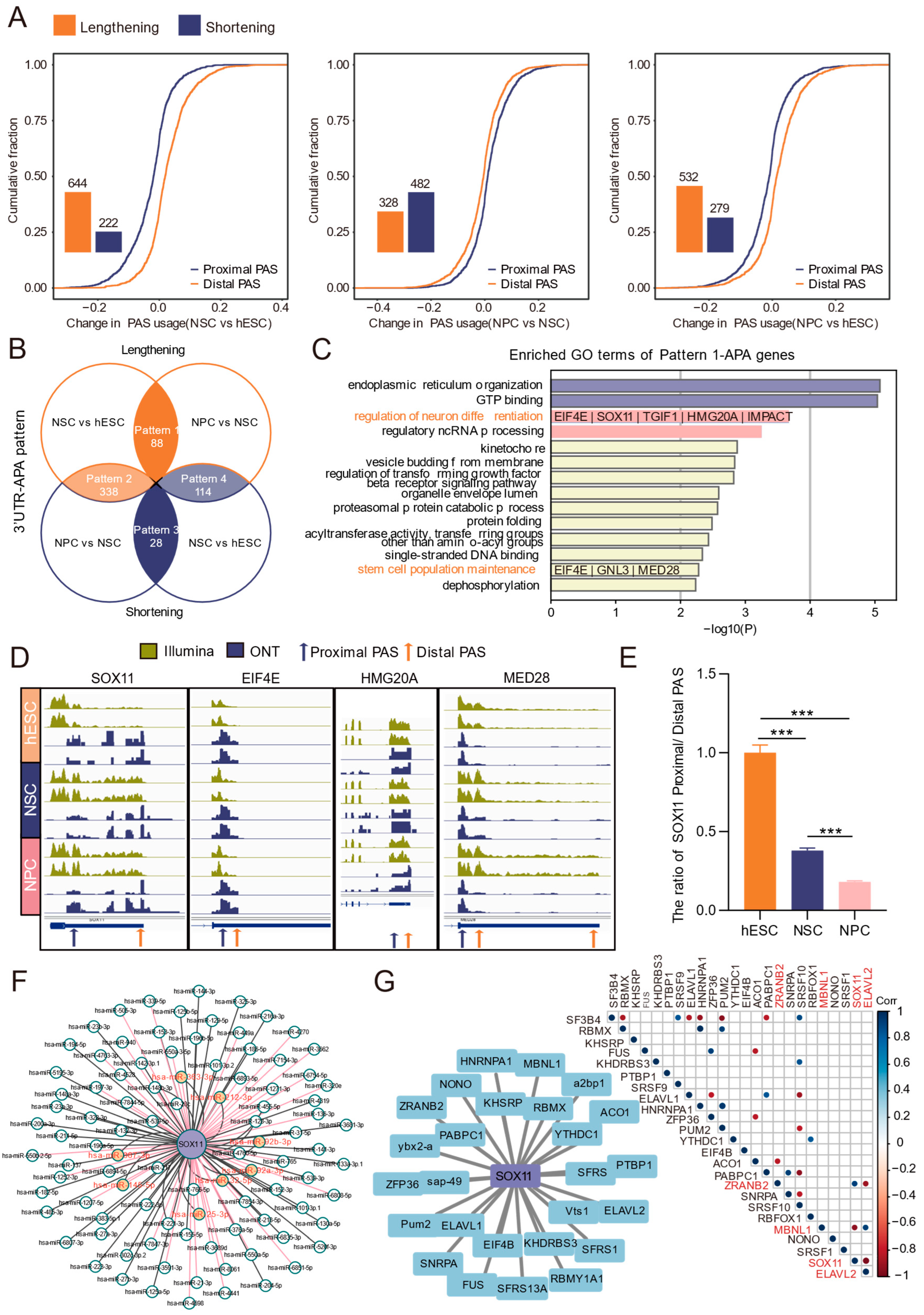
3.3. Dynamic Profiles of 3′ UTR-APA During Human Neural Differentiation
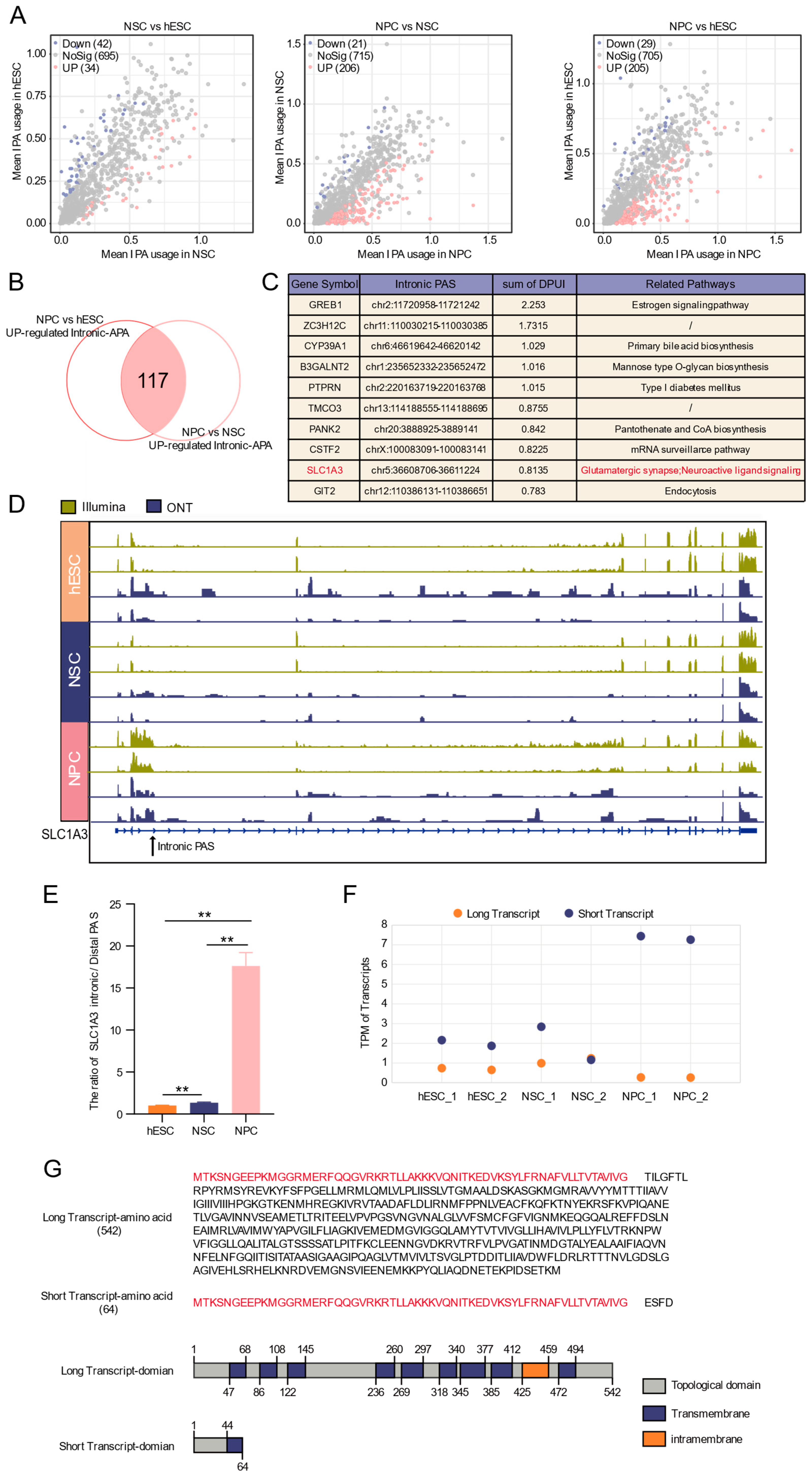
3.4. Widespread Intronic APA Events in the Neural Progenitor Cell Stage
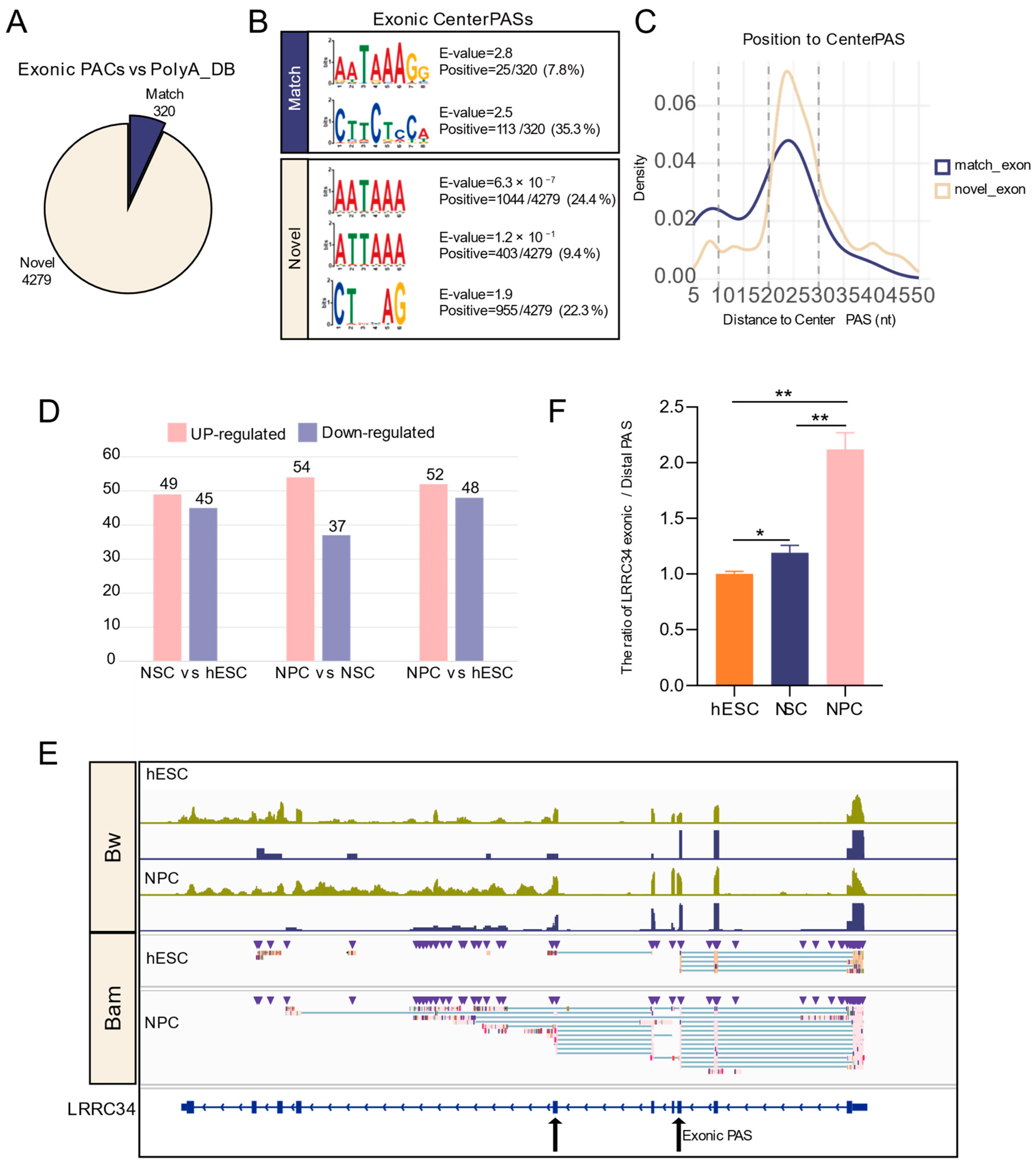
3.5. Accurate Identification of Exonic PASs by Long-Read Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Millevoi, S.; Vagner, S. Molecular Mechanisms of Eukaryotic Pre-mRNA 3′ End Processing Regulation. Nucleic Acids Res. 2010, 38, 2757–2774. [Google Scholar] [CrossRef]
- Hoque, M.; Ji, Z.; Zheng, D.; Luo, W.; Li, W.; You, B.; Park, J.Y.; Yehia, G.; Tian, B. Analysis of Alternative Cleavage and Polyadenylation by 3′ Region Extraction and Deep Sequencing. Nat. Methods 2013, 10, 133–139. [Google Scholar] [CrossRef]
- Tian, B. A Large-Scale Analysis of mRNA Polyadenylation of Human and Mouse Genes. Nucleic Acids Res. 2005, 33, 201–212. [Google Scholar] [CrossRef]
- Tian, B.; Manley, J.L. Alternative Polyadenylation of mRNA Precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Di Giammartino, D.C.; Nishida, K.; Manley, J.L. Mechanisms and Consequences of Alternative Polyadenylation. Mol. Cell 2011, 43, 853–866. [Google Scholar] [CrossRef]
- Neve, J.; Patel, R.; Wang, Z.; Louey, A.; Furger, A.M. Cleavage and Polyadenylation: Ending the Message Expands Gene Regulation. RNA Biol. 2017, 14, 865–890. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; You, B.; Hoque, M.; Zheng, D.; Luo, W.; Ji, Z.; Park, J.Y.; Gunderson, S.I.; Kalsotra, A.; Manley, J.L.; et al. Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3′ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation. PLoS Genet. 2015, 11, e1005166. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Pan, Z.; Lee, J.Y. Widespread mRNA Polyadenylation Events in Introns Indicate Dynamic Interplay between Polyadenylation and Splicing. Genome Res. 2007, 17, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Miura, P.; Shenker, S.; Andreu-Agullo, C.; Westholm, J.O.; Lai, E.C. Widespread and Extensive Lengthening of 3′ UTRs in the Mammalian Brain. Genome Res. 2013, 23, 812–825. [Google Scholar] [CrossRef]
- Ji, Z.; Lee, J.Y.; Pan, Z.; Jiang, B.; Tian, B. Progressive Lengthening of 3′ Untranslated Regions of mRNAs by Alternative Polyadenylation during Mouse Embryonic Development. Proc. Natl. Acad. Sci. USA 2009, 106, 7028–7033. [Google Scholar] [CrossRef]
- Lackford, B.; Yao, C.; Charles, G.M.; Weng, L.; Zheng, X.; Choi, E.-A.; Xie, X.; Wan, J.; Xing, Y.; Freudenberg, J.M.; et al. Fip1 Regulates mRNA Alternative Polyadenylation to Promote Stem Cell Self-Renewal. EMBO J. 2014, 33, 878–889. [Google Scholar] [CrossRef]
- Shepard, P.J.; Choi, E.-A.; Lu, J.; Flanagan, L.A.; Hertel, K.J.; Shi, Y. Complex and Dynamic Landscape of RNA Polyadenylation Revealed by PAS-Seq. RNA 2011, 17, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Tian, B. Reprogramming of 3′ Untranslated Regions of mRNAs by Alternative Polyadenylation in Generation of Pluripotent Stem Cells from Different Cell Types. PLoS ONE 2009, 4, e8419. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating Cells Express mRNAs with Shortened 3′ Untranslated Regions and Fewer MicroRNA Target Sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Bartel, D.P. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef]
- Papadimitriou, E.; Thomaidou, D. Post-Transcriptional Mechanisms Controlling Neurogenesis and Direct Neuronal Reprogramming. Neural Regen. Res. 2024, 19, 1929–1939. [Google Scholar] [CrossRef]
- Gallicchio, L.; Olivares, G.H.; Berry, C.W.; Fuller, M.T. Regulation and Function of Alternative Polyadenylation in Development and Differentiation. RNA Biol. 2023, 20, 908–925. [Google Scholar] [CrossRef]
- Langhnoja, J.; Buch, L.; Pillai, P. Potential Role of NGF, BDNF, and Their Receptors in Oligodendrocytes Differentiation from Neural Stem Cell: An in Vitro Study. Cell Biol. Int. 2021, 45, 432–446. [Google Scholar] [CrossRef]
- Langhnoja, J.; Buch, L.; Chruvattil, R.; Gupta, S.; Pillai, P. Insulin Receptor Regulates Neurotrophin and Neurotrophin Receptor Expression in the Differentiation of Neural Stem Cells: In Vitro Study. J. Biochem. Mol. Toxicol. 2025, 39, e70198. [Google Scholar] [CrossRef]
- An, J.J.; Gharami, K.; Liao, G.-Y.; Woo, N.H.; Lau, A.G.; Vanevski, F.; Torre, E.R.; Jones, K.R.; Feng, Y.; Lu, B.; et al. Distinct Role of Long 3′ UTR BDNF mRNA in Spine Morphology and Synaptic Plasticity in Hippocampal Neurons. Cell 2008, 134, 175–187. [Google Scholar] [CrossRef]
- Rodrigues, D.C.; Kim, D.-S.; Yang, G.; Zaslavsky, K.; Ha, K.C.H.; Mok, R.S.F.; Ross, P.J.; Zhao, M.; Piekna, A.; Wei, W.; et al. MECP2 Is Post-Transcriptionally Regulated during Human Neurodevelopment by Combinatorial Action of RNA-Binding Proteins and miRNAs. Cell Rep. 2016, 17, 720–734. [Google Scholar] [CrossRef]
- Coutinho, A.M.; Oliveira, G.; Katz, C.; Feng, J.; Yan, J.; Yang, C.; Marques, C.; Ataíde, A.; Miguel, T.S.; Borges, L.; et al. MECP2 Coding Sequence and 3′UTR Variation in 172 Unrelated Autistic Patients. Am. J. Med. Genet. Pt. B Neuropsychiatr. Genet. 2007, 144B, 475–483. [Google Scholar] [CrossRef]
- Alser, M.; Rotman, J.; Deshpande, D.; Taraszka, K.; Shi, H.; Baykal, P.I.; Yang, H.T.; Xue, V.; Knyazev, S.; Singer, B.D.; et al. Technology Dictates Algorithms: Recent Developments in Read Alignment. Genome Biol. 2021, 22, 249. [Google Scholar] [CrossRef]
- Ha, K.C.H.; Blencowe, B.J.; Morris, Q. QAPA: A New Method for the Systematic Analysis of Alternative Polyadenylation from RNA-Seq Data. Genome Biol. 2018, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio Sequencing and Its Applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Bayega, A.; Fahiminiya, S.; Oikonomopoulos, S.; Ragoussis, J. Current and Future Methods for mRNA Analysis: A Drive Toward Single Molecule Sequencing. In Gene Expression Analysis; Raghavachari, N., Garcia-Reyero, N., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1783, pp. 209–241. ISBN 978-1-4939-7833-5. [Google Scholar]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso-Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S.N. A Survey of the Sorghum Transcriptome Using Single-Molecule Long Reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Zhang, Z.; Bae, B.; Cuddleston, W.H.; Miura, P. Coordination of Alternative Splicing and Alternative Polyadenylation Revealed by Targeted Long Read Sequencing. Nat. Commun. 2023, 14, 5506. [Google Scholar] [CrossRef] [PubMed]
- Foord, C.; Prjibelski, A.D.; Hu, W.; Michielsen, L.; Vandelli, A.; Narykov, O.; Evans, B.; Hsu, J.; Belchikov, N.; Jarroux, J.; et al. A Spatial Long-Read Approach at near-Single-Cell Resolution Reveals Developmental Regulation of Splicing and Polyadenylation Sites in Distinct Cortical Layers and Cell Types. Nat. Commun. 2025, 16, 8093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Glatt-Deeley, H.; Zou, L.; Song, D.; Miura, P. APALORD: An R-Based Tool for Differential Alternative Polyadenylation Analysis of Long-Read RNA-Seq Data. bioRxiv 2025, 2025.06.11.658931. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Ye, C.; Zhao, D.; Ye, W.; Wu, X.; Ji, G.; Li, Q.Q.; Lin, J. QuantifyPoly(A): Reshaping Alternative Polyadenylation Landscapes of Eukaryotes with Weighted Density Peak Clustering. Brief. Bioinform. 2021, 22, bbab268. [Google Scholar] [CrossRef]
- Trivedi, U.H.; Cézard, T.; Bridgett, S.; Montazam, A.; Nichols, J.; Blaxter, M.; Gharbi, K. Quality Control of Next-Generation Sequencing Data without a Reference. Front. Genet. 2014, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Xia, Z.; Donehower, L.A.; Cooper, T.A.; Neilson, J.R.; Wheeler, D.A.; Wagner, E.J.; Li, W. Dynamic Analyses of Alternative Polyadenylation from RNA-Seq Reveal a 3′-UTR Landscape across Seven Tumour Types. Nat. Commun. 2014, 5, 5274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xu, Q.; Wei, R.; Wang, W.; Ding, D.; Yang, Y.; Yao, J.; Zhang, L.; Hu, Y.-Q.; Wei, G.; et al. Cancer-Associated Dynamics and Potential Regulators of Intronic Polyadenylation Revealed by IPAFinder Using Standard RNA-Seq Data. Genome Res. 2021, 31, 2095–2106. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Nambiar, R.; Zheng, D.; Tian, B. PolyA_DB 3 Catalogs Cleavage and Polyadenylation Sites Identified by Deep Sequencing in Multiple Genomes. Nucleic Acids Res. 2018, 46, D315–D319. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Tsang, S.M.; Oliemuller, E.; Howard, B.A. Regulatory Roles for SOX11 in Development, Stem Cells and Cancer. Semin. Cancer Biol. 2020, 67, 3–11. [Google Scholar] [CrossRef]
- Matsugami, T.R.; Tanemura, K.; Mieda, M.; Nakatomi, R.; Yamada, K.; Kondo, T.; Ogawa, M.; Obata, K.; Watanabe, M.; Hashikawa, T.; et al. Indispensability of the Glutamate Transporters GLAST and GLT1 to Brain Development. Proc. Natl. Acad. Sci. USA 2006, 103, 12161–12166. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, D.; Gao, Q.; Tan, P.; Wang, Y.; Cai, X.; Li, A.; Zhao, Y.; Thurman, A.L.; Malekpour, S.A.; et al. Improving Gene Isoform Quantification with miniQuant. Nat. Biotechnol. 2025. [Google Scholar] [CrossRef]
- Glinos, D.A.; Garborcauskas, G.; Hoffman, P.; Ehsan, N.; Jiang, L.; Gokden, A.; Dai, X.; Aguet, F.; Brown, K.L.; Garimella, K.; et al. Transcriptome Variation in Human Tissues Revealed by Long-Read Sequencing. Nature 2022, 608, 353–359. [Google Scholar] [CrossRef]
- Sun, Q.; Han, Y.; He, J.; Wang, J.; Ma, X.; Ning, Q.; Zhao, Q.; Jin, Q.; Yang, L.; Li, S.; et al. Long-Read Sequencing Reveals the Landscape of Aberrant Alternative Splicing and Novel Therapeutic Target in Colorectal Cancer. Genome Med. 2023, 15, 76. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Hu, J.; Lutz, C.S.; Wilusz, J.; Tian, B. Bioinformatic Identification of Candidate Cis -Regulatory Elements Involved in Human mRNA Polyadenylation. RNA 2005, 11, 1485–1493. [Google Scholar] [CrossRef]
- Sock, E.; Rettig, S.D.; Enderich, J.; Bösl, M.R.; Tamm, E.R.; Wegner, M. Gene Targeting Reveals a Widespread Role for the High-Mobility-Group Transcription Factor Sox11 in Tissue Remodeling. Mol. Cell. Biol. 2004, 24, 6635–6644. [Google Scholar] [CrossRef]
- Kato, K.; Bhattaram, P.; Penzo-Méndez, A.; Gadi, A.; Lefebvre, V. SOXC Transcription Factors Induce Cartilage Growth Plate Formation in Mouse Embryos by Promoting Noncanonical WNT Signaling. J. Bone Miner. Res. 2015, 30, 1560–1571. [Google Scholar] [CrossRef]
- Bergsland, M.; Ramsköld, D.; Zaouter, C.; Klum, S.; Sandberg, R.; Muhr, J. Sequentially Acting Sox Transcription Factors in Neural Lineage Development. Genes Dev. 2011, 25, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Kavyanifar, A.; Turan, S.; Lie, D.C. SoxC Transcription Factors: Multifunctional Regulators of Neurodevelopment. Cell Tissue Res. 2018, 371, 91–103. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Geng, X.; Guo, X.; Wang, T.; Xu, J.; Jiang, L.; Zhen, H. microRNA-125b-5p Alleviated CCI-induced Neuropathic Pain and Modulated Neuroinflammation via Targeting SOX11. Synapse 2024, 78, e22306. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, J.; Wang, H.; Zhang, L.; Guo, J.; Jin, G. GAS5, a Long Noncoding RNA, Contributes to Annulus Fibroblast Osteogenic Differentiation and Apoptosis in Intervertebral Disk Degeneration via the miR-221-3p/SOX11 Axis. Aging 2024, 16, 3896–3914. [Google Scholar] [CrossRef]
- Struebing, F.L.; Wang, J.; Li, Y.; King, R.; Mistretta, O.C.; English, A.W.; Geisert, E.E. Differential Expression of Sox11 and Bdnf mRNA Isoforms in the Injured and Regenerating Nervous Systems. Front. Mol. Neurosci. 2017, 10, 354. [Google Scholar] [CrossRef] [PubMed]
- Kainov, Y.A.; Makeyev, E.V. A Transcriptome-Wide Antitermination Mechanism Sustaining Identity of Embryonic Stem Cells. Nat. Commun. 2020, 11, 361. [Google Scholar] [CrossRef]
- La Rosa, P.; Bielli, P.; Compagnucci, C.; Cesari, E.; Volpe, E.; Farioli Vecchioli, S.; Sette, C. Sam68 Promotes Self-Renewal and Glycolytic Metabolism in Mouse Neural Progenitor Cells by Modulating Aldh1a3 Pre-mRNA 3′-End Processing. eLife 2016, 5, e20750. [Google Scholar] [CrossRef] [PubMed]
- Llorens-Bobadilla, E.; Zhao, S.; Baser, A.; Saiz-Castro, G.; Zwadlo, K.; Martin-Villalba, A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells That Become Activated upon Brain Injury. Cell Stem Cell 2015, 17, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Rieskamp, J.D.; Rosado-Burgos, I.; Christofi, J.E.; Ansar, E.; Einstein, D.; Walters, A.E.; Valentini, V.; Bruno, J.P.; Kirby, E.D. Excitatory Amino Acid Transporter 1 Supports Adult Hippocampal Neural Stem Cell Self-Renewal. iScience 2023, 26, 107068. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, C.; Nuriel, Y.; Lazar, D.; Kohl, A.; Muir, E.; Genin, O.; Cinnamon, Y.; Benyamini, H.; Nevo, Y.; Sela-Donenfeld, D. Hindbrain Boundaries as Niches of Neural Progenitor and Stem Cells Regulated by the Extracellular Matrix Proteoglycan Chondroitin Sulphate. Development 2024, 151, dev201934. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Lou, Z.; Zeng, X.; Jiang, T.; Du, P.; Rao, J.; Dai, X.; Lin, H.; Zhu, Y. Stage-Specific Alternative Polyadenylation During Human Neural Differentiation Revealed by Integrated Long- and Short-Read Sequencing. Biology 2026, 15, 24. https://doi.org/10.3390/biology15010024
Lou Z, Zeng X, Jiang T, Du P, Rao J, Dai X, Lin H, Zhu Y. Stage-Specific Alternative Polyadenylation During Human Neural Differentiation Revealed by Integrated Long- and Short-Read Sequencing. Biology. 2026; 15(1):24. https://doi.org/10.3390/biology15010024
Chicago/Turabian StyleLou, Zheqi, Xianyan Zeng, Tinghui Jiang, Peizhen Du, Jiyao Rao, Xinyan Dai, Haishuang Lin, and Yong Zhu. 2026. "Stage-Specific Alternative Polyadenylation During Human Neural Differentiation Revealed by Integrated Long- and Short-Read Sequencing" Biology 15, no. 1: 24. https://doi.org/10.3390/biology15010024
APA StyleLou, Z., Zeng, X., Jiang, T., Du, P., Rao, J., Dai, X., Lin, H., & Zhu, Y. (2026). Stage-Specific Alternative Polyadenylation During Human Neural Differentiation Revealed by Integrated Long- and Short-Read Sequencing. Biology, 15(1), 24. https://doi.org/10.3390/biology15010024




