Genome-Wide Characterization and Identification of Auxin Response Factor (ARF) Gene Family Reveals the Regulation of RrARF5 in AsA Metabolism in Rosa roxburghii Tratt. Fruits
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of ARF Family Genes in R. roxburghii Genome
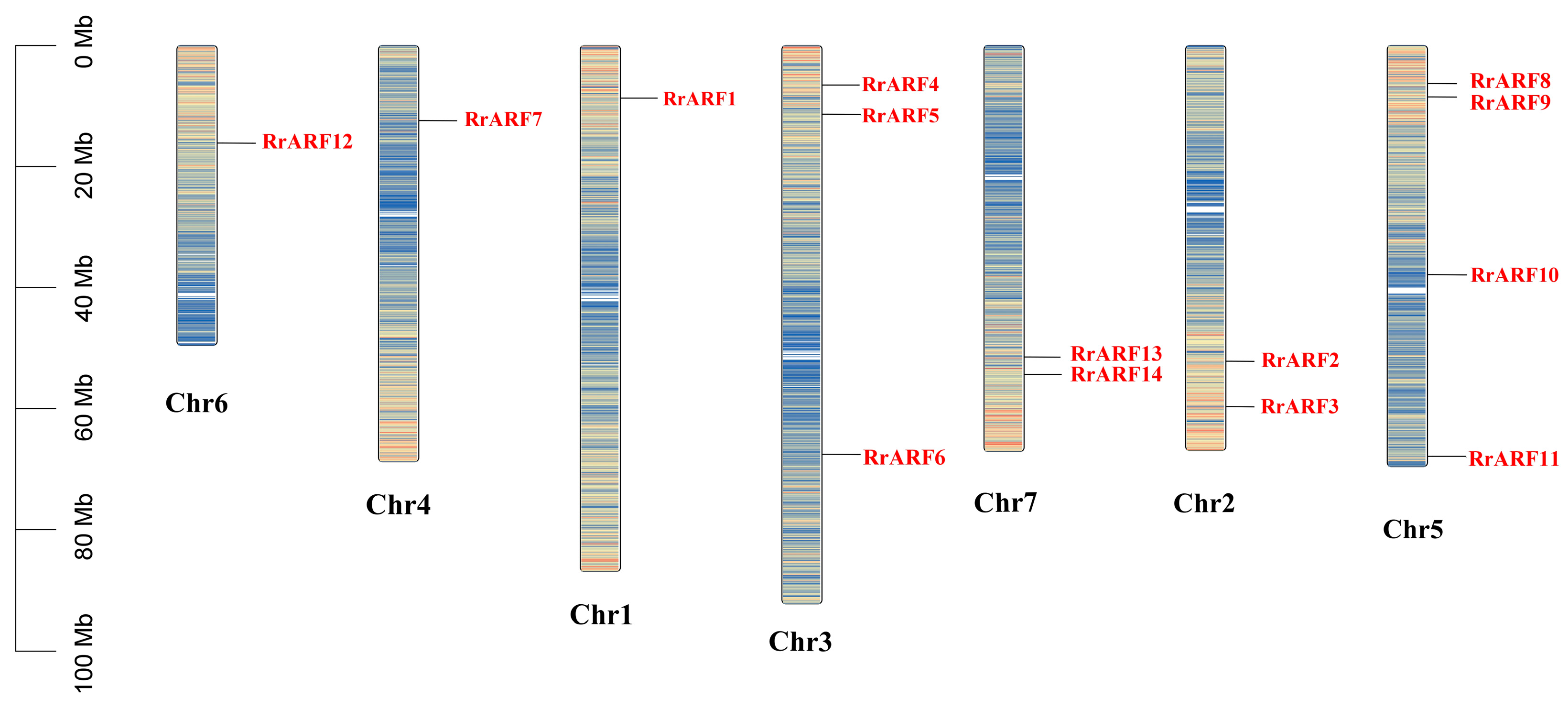
2.2. Chromosomal Distribution of RrARFs
2.3. Analysis of Cis-Acting Elements in RrARF Promoters
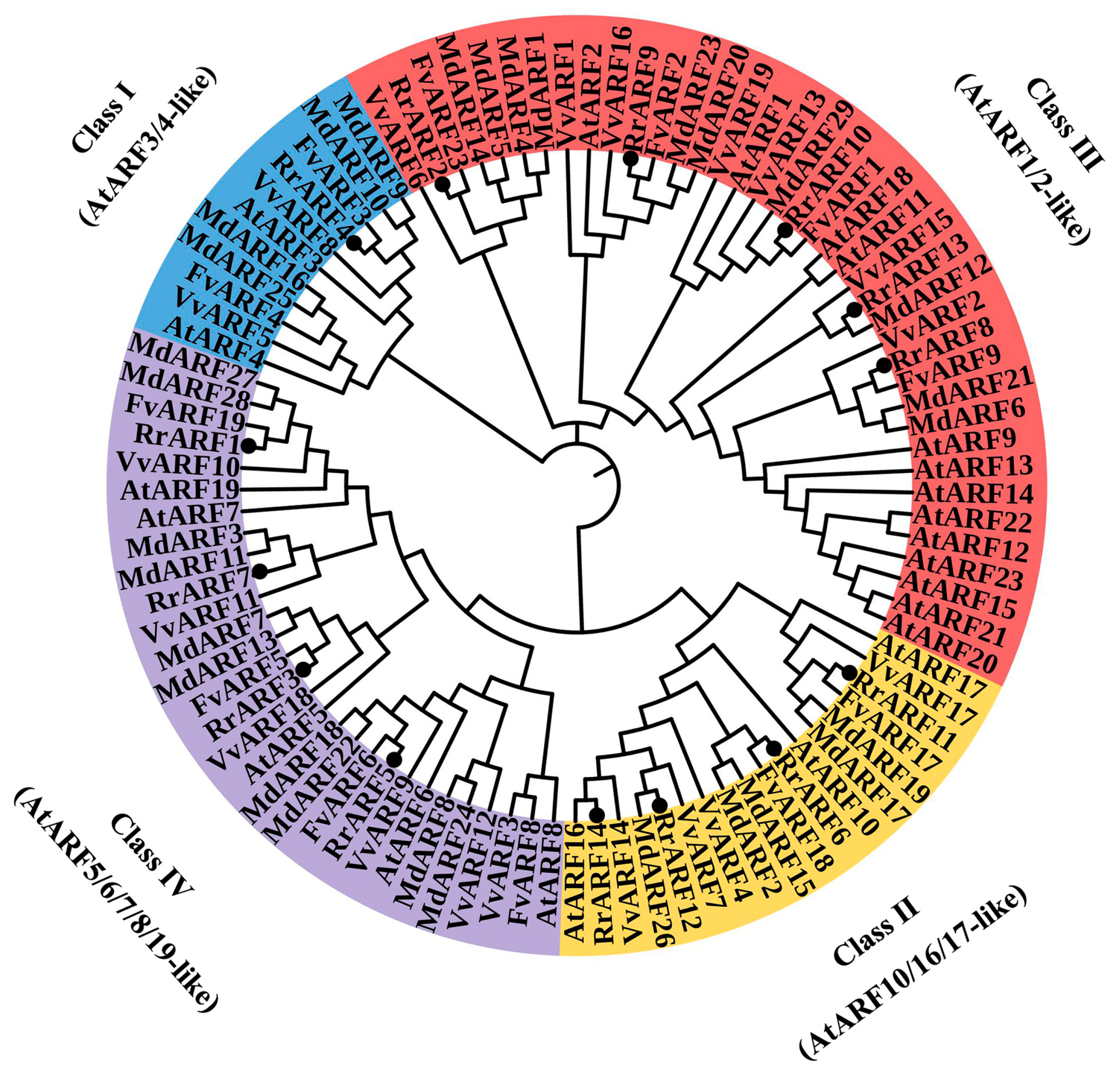
2.4. Phylogenetic Analysis of the R. roxburghii ARF Gene Family
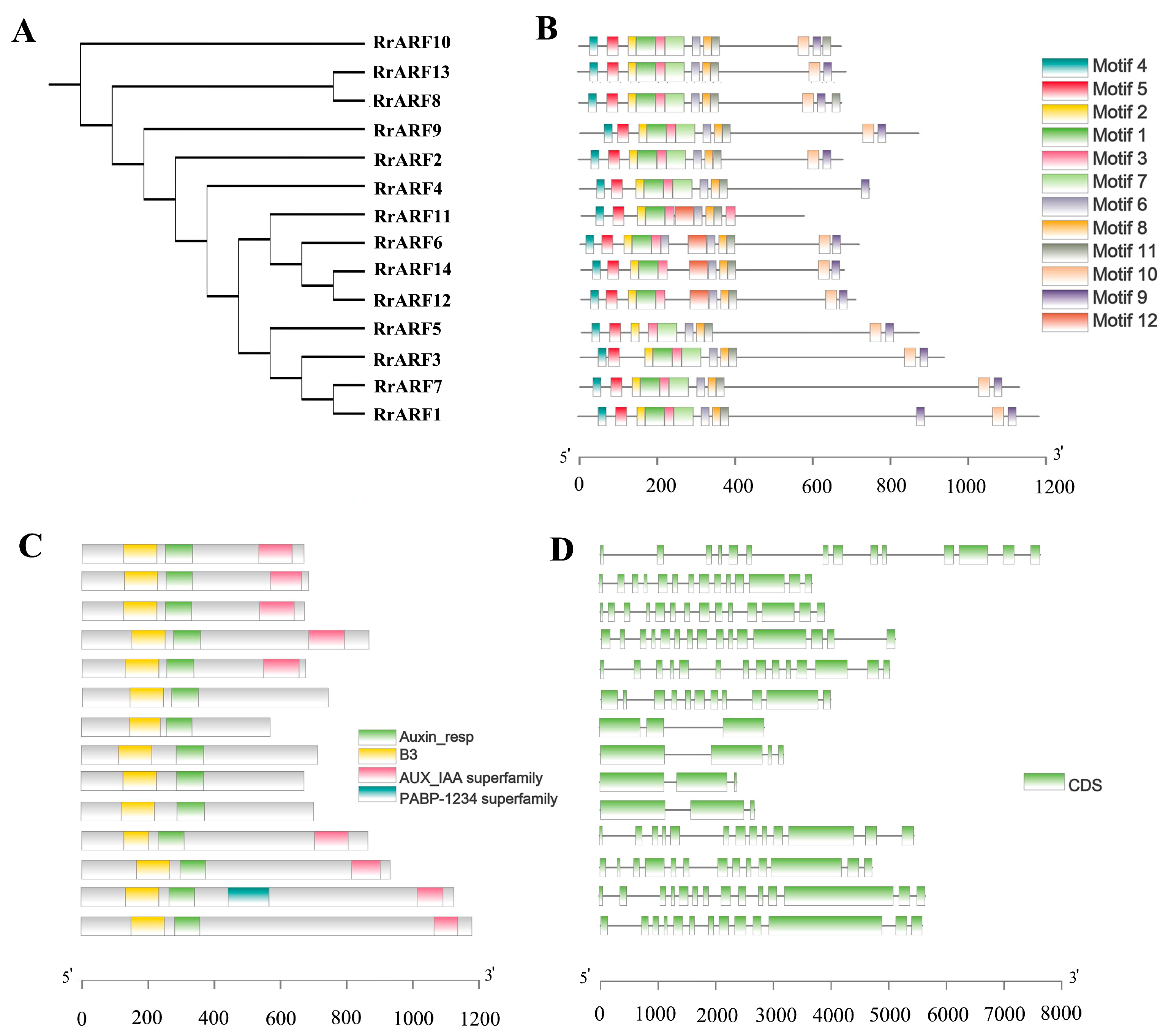
2.5. Gene Structure and Conserved Motif Characterization
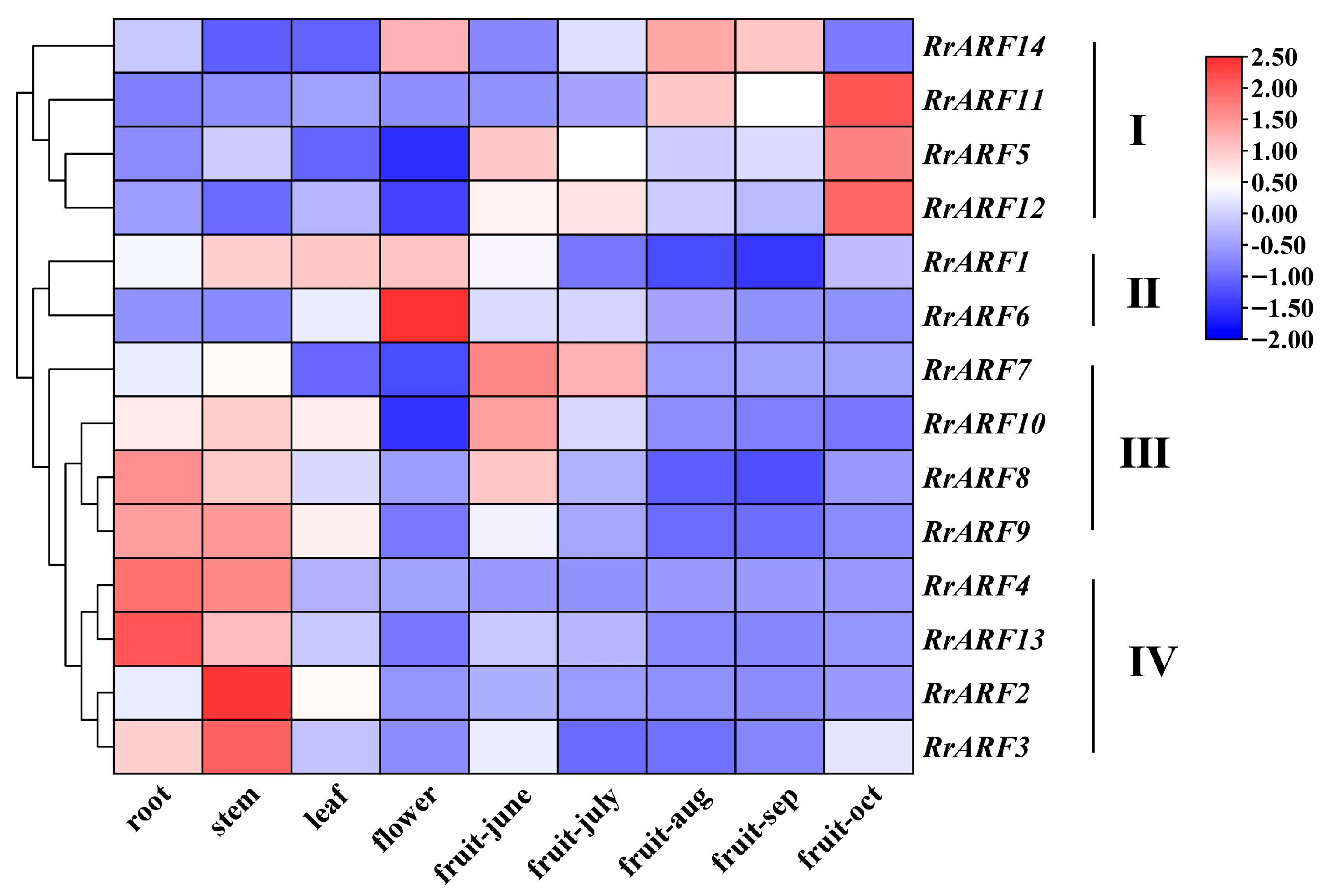
2.6. Gene Expression Analysis of ARF Family Genes
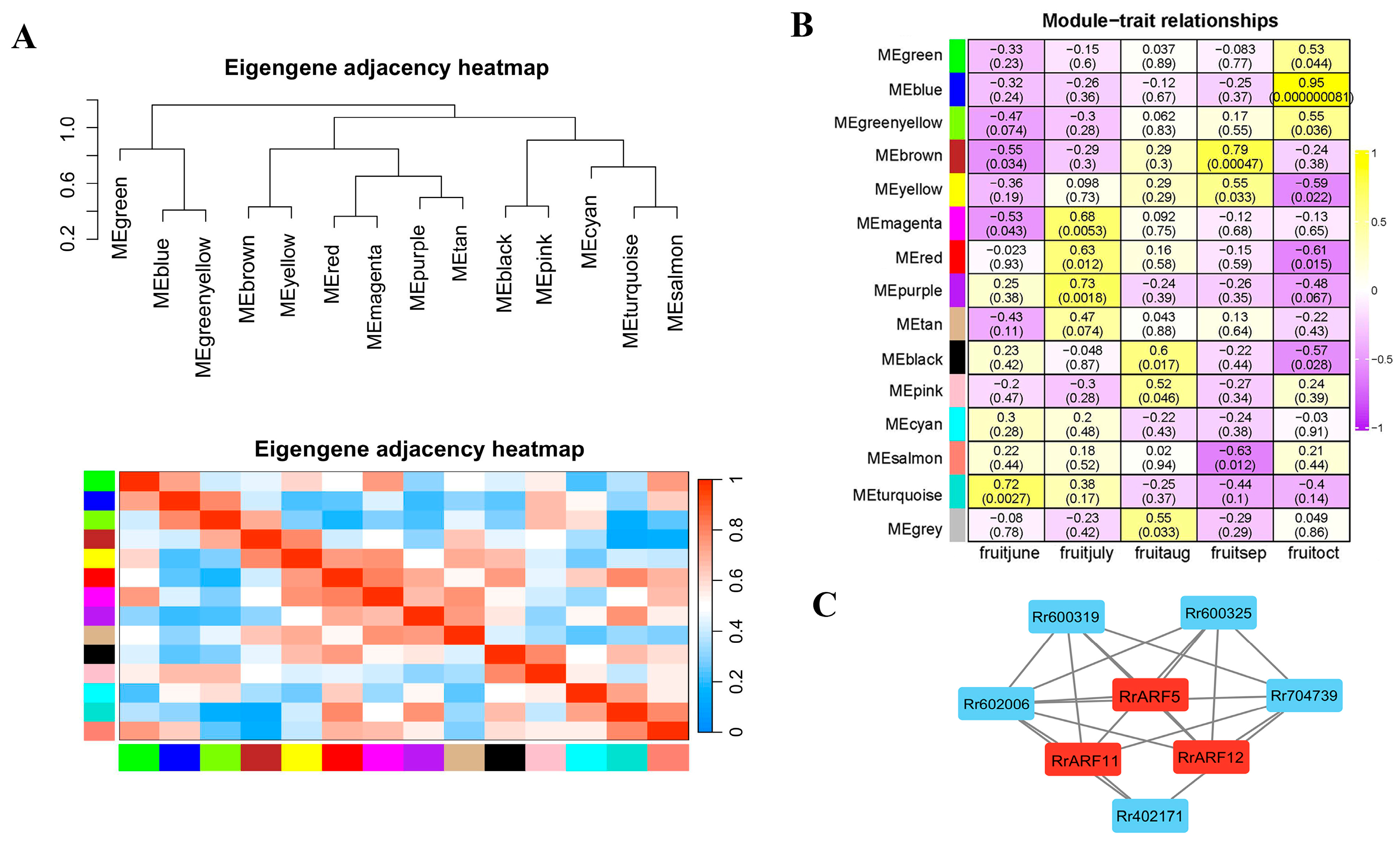
2.7. Weighted Correlation Network Analysis (WGCNA)
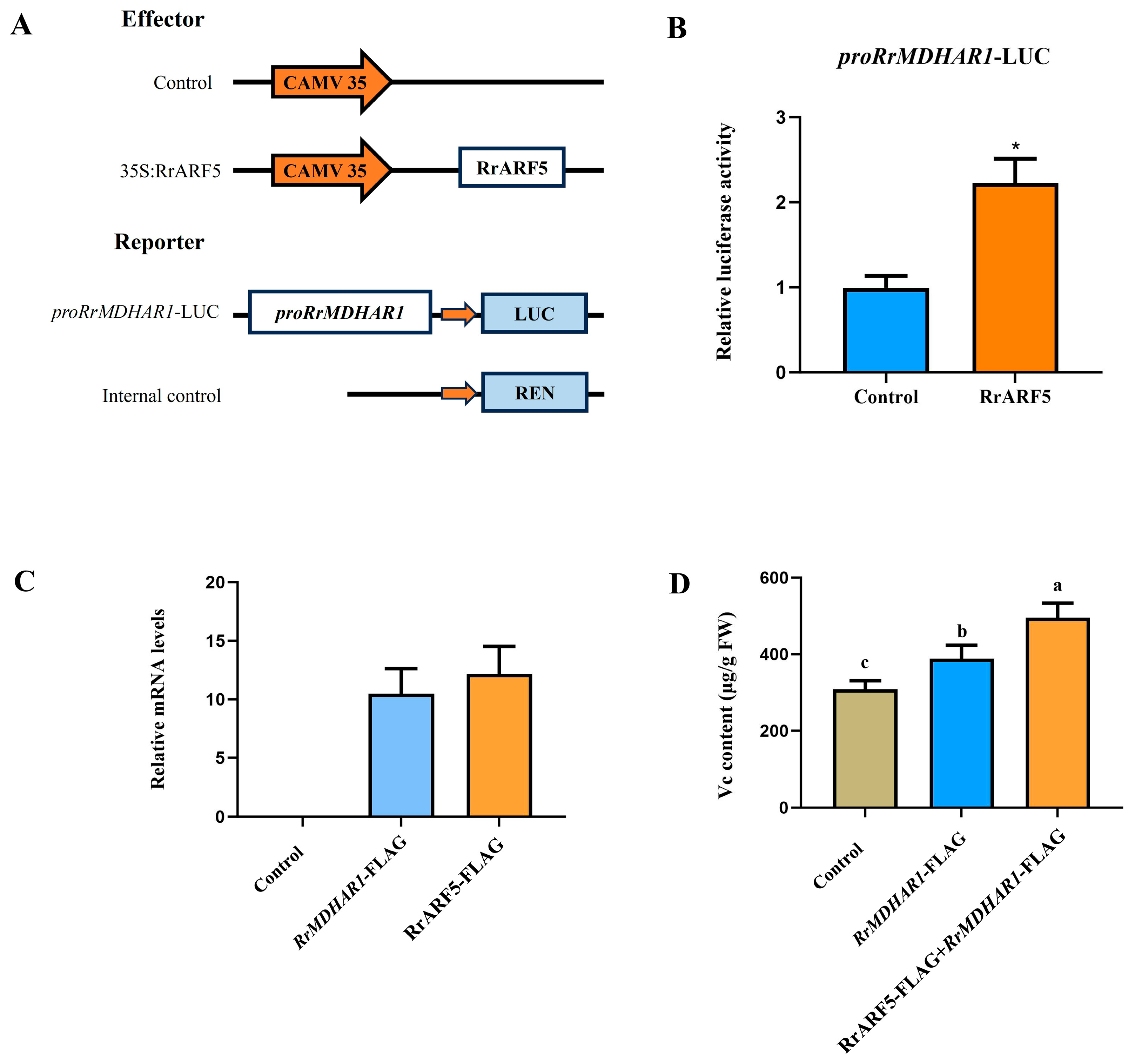
2.8. Dual-Luciferase (Dual-LUC) Reporter Assay
2.9. Accession Numbers
3. Results
3.1. Identification of ARF Family Genes in R. roxburghii Genome
3.2. Analysis of Conserved Structural Domains and Promoter Sequences of ARF Family Members in R. roxburghii
3.3. Expression Profiles of RrARFs in Different Tissues and Developmental Stages
3.4. Coexpressed Gene Networks of RrARFs
3.5. RrARF5 Involved in VC Biosynthesis by Activating RrMDHAR1 Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA Proteins Are Active Repressors, and Their Stability and Activity Are Modulated by Auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef][Green Version]
- El Sharkawy, I.; Sherif, S.M.; Jones, B.; Mila, I.; Kumar, P.P.; Bouzayen, M.; Jayasankar, S. TIR1-like auxin-receptors are involved in the regulation of plum fruit development. J. Exp. Bot. 2014, 65, 5205–5215. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated Auxin Homeostasis Links Growth Regulation with Stress Adaptation Response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Ulmasov, T.; Hagen, G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 1998, 54, 619–627. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, J.; Wang, W.; Xu, L. ARF family identification in Tamarix chinensis reveals the salt responsive expression of TcARF6 targeted by miR167. PeerJ. 2020, 8, e8829. [Google Scholar] [CrossRef]
- Zhou, Z.; Schenke, D.; Miao, Y.; Cai, D. Investigation of the crosstalk between the flg22 and the UV-B-induced flavonol pathway in Arabidopsis thaliana seedlings. Plant Cell Environ. 2017, 40, 453–458. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana: Unique and Overlapping Functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef]
- Wang, S.X.; Shi, F.Y.; Dong, X.X.; Li, Y.X.; Zhang, Z.H.; Li, H. Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in strawberry (Fragaria vesca). J. Integr. Agric. 2019, 18, 1587–1603. [Google Scholar] [CrossRef]
- Wan, S.B.; Li, W.L.; Zhu, Y.Y.; Liu, Z.M.; Huang, W.D.; Zhan, J.C. Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 2014, 33, 1365–1375. [Google Scholar] [CrossRef]
- Li, S.B.; Ouyang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 119. [Google Scholar] [CrossRef]
- Zhang, H.; Ning, C.; Chunjuan, D.; Shang, Q. Genome-wide Identification and Expression of ARF Gene Family during Adventitious Root Development in Hot Pepper (Capsicum annuum). Hortic. Plant J. 2017, 3, 151–164. [Google Scholar] [CrossRef]
- Weijers, D.; Benkova, E.; Jäger, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jürgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef]
- Wilmoth, J.C.; Wang, S.; Tiwari, S.B.; Joshi, A.D.; Hagen, G.; Guilfoyle, T.J.; Alonso, J.M.; Ecker, J.R.; Reed, J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005, 43, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Lee, I.C.; Kim, J.; Kim, H.J.; Ryu, J.S.; Woo, H.R.; Nam, H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010, 61, 1419–1430. [Google Scholar] [CrossRef]
- Wang, J.N.; Guo, L.Y.; Li, Y.; Wu, Y.Z.; Wang, M.M.; Zhao, S.C.; Xu, X.; Li, J.H. Genome-wide identification and expression pattern analysis of auxin response factor (ARF) genes in Chionanthus retusus and functional characterization of CrARF37 in terms of its effect on flower shape. BMC Plant Biol. 2025, 25, 1046. [Google Scholar] [CrossRef]
- Gambhir, P.; Raghuvanshi, U.; Kumar, R.; Sharma, A.K. Transcriptional regulation of tomato fruit ripening. Physiol. Mol. Biol. Plants 2024, 30, 289–303. [Google Scholar] [CrossRef]
- Serrani, J.C.; Fos, M.; Atarés, A.; García-Martínez, J.L. Effect of Gibberellin and Auxin on Parthenocarpic Fruit Growth Induction in the cv Micro-Tom of Tomato. J. Plant Growth Regul. 2007, 26, 211–221. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Feron, R.; Maretto, F.; Keijman, J.; Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008, 177, 60–76. [Google Scholar] [CrossRef]
- Yi, S.N.; Mao, J.X.; Zhang, X.Y.; Li, X.M.; Zhang, Z.H.; Li, H. FveARF2 negatively regulates fruit ripening and quality in strawberry. Front. Plant Sci. 2022, 13, 1023739. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.J.; Xu, X.; Gong, Z.H.; Tang, Y.W.; Wu, M.B.; Yan, F.; Zhang, X.L.; Zhang, Q.; Yang, F.Q.; Hu, X.W.; et al. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 85. [Google Scholar] [CrossRef]
- Yuan, Y.J.; Mei, L.H.; Wu, M.B.; Wei, W.W.; Shan, W.; Gong, Z.H.; Zhang, Q.; Yang, F.Q.; Yan, F.; Zhang, Q.; et al. SlARF10, an auxin response factor, is involved in chlorophyll and sugar accumulation during tomato fruit development. J. Exp. Bot. 2018, 69, 5507–5518. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.T.; Lu, Q.; Liu, Z.; Lv, T.X.; Li, X.Y.; Bu, H.D.; Liu, W.T.; Xu, Y.X.; Yuan, H.; Wang, A. Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 2020, 226, 1781–1795. [Google Scholar] [CrossRef]
- Zhang, T.; Li, W.; Xie, R.X.; Xu, L.; Zhou, Y.; Li, H.L.; Yuan, C.C.; Zheng, X.L.; Xiao, L.T.; Liu, K.D. CpARF2 and CpEIL1 interact to mediate auxin–ethylene interaction and regulate fruit ripening in papaya. Plant J. 2020, 103, 1318–1337. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; An, H.M.; Li, L.L. Genome Survey Sequencing for the Characterization of the Genetic Background of Rosa roxburghii Tratt and Leaf Ascorbate Metabolism Genes. PLoS ONE 2016, 11, e0147530. [Google Scholar] [CrossRef]
- Yang, H.; Hu, J.W.; Huang, X.F.; Zhou, C.; Li, L.Y.; Fan, M.Y. Risk Assessment of Heavy Metals Pollution for Rosa sterilis and Soil from Planting Bases Located in Karst Areas of Guizhou Province. Appl. Mech. Mater. 2015, 700, 475–481. [Google Scholar] [CrossRef]
- Liu, M.H.; Zhang, Q.; Zhang, Y.H.; Lu, X.Y.; Fu, W.M.; He, J.Y. Chemical Analysis of Dietary Constituents in Rosa roxburghii and Rosa sterilis Fruits. Molecules 2016, 21, 1204. [Google Scholar] [CrossRef]
- An, H.M.; Fan, W.G.; Chen, L.G.; Asghar, S.; Liu, Q.L. Molecular characterisation and expression of L-galactono-1,4-lactone dehydrogenase and L-ascorbic acid accumulation during fruit development in Rosa roxburghii. J. Hortic. Sci. Biotechnol. 2007, 82, 627–635. [Google Scholar] [CrossRef]
- Liao, G.L.; Chen, L.; He, Y.Q.; Li, X.S.; Lv, Z.X.; Yi, S.Y.; Zhong, M.; Huang, C.H.; Jia, D.F.; Qu, X.Y.; et al. Three metabolic pathways are responsible for the accumulation and maintenance of high AsA content in kiwifruit (Actinidia eriantha). BMC Genom. 2021, 22, 13. [Google Scholar] [CrossRef]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.L.; Xu, Q.; Allan, A.C.; Xu, X.B. L-Ascorbic acid metabolism and regulation in fruit crops. Plant Physiol. 2023, 192, 1684–1695. [Google Scholar] [CrossRef]
- Zong, D.; Liu, H.; Gan, P.; Ma, S.; Liang, H.; Yu, J.; Li, P.; Jiang, T.; Sahu, S.K.; Yang, Q. Chromosomal-scale genomes of two Rosa species provide insights into genome evolution and ascorbate accumulation. Plant J. 2024, 117, 1264–1280. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.G.; Xue, Q.H.; Shu, P.; Xu, W.J.; Du, X.F.; Wu, M.B.; Liu, K.D.; Pirrello, J.; Bouzayen, M.; Hong, Y.G.; et al. Bifunctional transcription factors SlERF.H5 and H7 activate cell wall and repress gibberellin biosynthesis genes in tomato via a conserved motif. Dev. Cell 2024, 59, 1345–1359. [Google Scholar] [CrossRef]
- Luo, X.C.; Sun, M.H.; Xu, R.R.; Shu, H.R.; Wang, J.W.; Zhang, S.Z. Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 2014, 93, 785–797. [Google Scholar] [CrossRef]
- Xing, H.Y.; Pudake, R.N.; Guo, G.G.; Xing, G.F.; Hu, Z.R.; Zhang, Y.R.; Sun, Q.X.; Ni, Z.F. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef]
- Zouine, M.; Fu, Y.Y.; Chateigner-Boutin, A.L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.P.; Bouzayen, M. Characterization of the Tomato ARF Gene Family Uncovers a Multi-Level Post-Transcriptional Regulation Including Alternative Splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Liu, K.D.; Yuan, C.C.; Li, H.L.; Lin, W.H.; Yang, Y.J.; Shen, C.J.; Zheng, X.L. Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics 2015, 16, 901. [Google Scholar] [CrossRef]
- Peng, Y.; Fang, T.; Zhang, Y.Y.; Zhang, M.Y.; Zeng, L.H. Genome-Wide Identification and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Longan (Dimocarpus longan L.). Plants 2020, 9, 221. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, F.Y.; Cheng, L.; Kong, F.L.; Peng, Z.; Liu, S.Y.; Yu, X.L.; Lu, G. Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 2011, 30, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.W.; Ma, Y.J.; Chang, Y.T.; Zhang, W.B.; Deng, Y.Y.; Zhang, N.; Zhang, X.; Fan, K.K.; Hu, X.M.; Wang, S.H.; et al. Identification and transcriptome data analysis of ARF family genes in five Orchidaceae species. Plant Mol. Biol. 2023, 112, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.; Page, D.; Gouble, B.; Garchery, C.; Zamir, D.; Causse, M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ. 2008, 31, 1086–1096. [Google Scholar] [CrossRef]
| Gene | Gene | CDS | Chromosome | Protein | ||
|---|---|---|---|---|---|---|
| Name | ID | Size | Location | Length (aa) | MW (KDa) | pI |
| RrARF1 | Rr100998 | 3549 | Chr1 | 1182 | 132.8 | 6.15 |
| RrARF2 | Rr203848 | 2031 | Chr2 | 676 | 75.5 | 5.83 |
| RrARF3 | Rr204729 | 2799 | Chr2 | 932 | 102.6 | 5.25 |
| RrARF4 | Rr300761 | 2232 | Chr3 | 743 | 81.5 | 6.31 |
| RrARF5 | Rr301243 | 2598 | Chr3 | 865 | 95.3 | 6.09 |
| RrARF6 | Rr304962 | 2145 | Chr3 | 714 | 78.7 | 8.56 |
| RrARF7 | Rr400968 | 3384 | Chr4 | 1127 | 124.7 | 6.24 |
| RrARF8 | Rr500747 | 2019 | Chr5 | 672 | 74.6 | 6.23 |
| RrARF9 | Rr500973 | 2607 | Chr5 | 868 | 96.5 | 6.07 |
| RrARF10 | Rr503508 | 2016 | Chr5 | 671 | 74.4 | 6.10 |
| RrARF11 | Rr505360 | 1713 | Chr5 | 570 | 62.6 | 6.13 |
| RrARF12 | Rr601745 | 2112 | Chr6 | 703 | 77.3 | 6.57 |
| RrARF13 | Rr703521 | 2061 | Chr7 | 686 | 76.0 | 6.56 |
| RrARF14 | Rr703808 | 2025 | Chr7 | 674 | 74.4 | 6.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, T.; Sun, Z.; Liu, M.; Zhao, H.; Zhang, Y.; Garcia-Caparros, P.; Yang, B.; Yang, Y. Genome-Wide Characterization and Identification of Auxin Response Factor (ARF) Gene Family Reveals the Regulation of RrARF5 in AsA Metabolism in Rosa roxburghii Tratt. Fruits. Biology 2025, 14, 1156. https://doi.org/10.3390/biology14091156
Feng T, Sun Z, Liu M, Zhao H, Zhang Y, Garcia-Caparros P, Yang B, Yang Y. Genome-Wide Characterization and Identification of Auxin Response Factor (ARF) Gene Family Reveals the Regulation of RrARF5 in AsA Metabolism in Rosa roxburghii Tratt. Fruits. Biology. 2025; 14(9):1156. https://doi.org/10.3390/biology14091156
Chicago/Turabian StyleFeng, Tu, Zhengliang Sun, Mingchun Liu, Hong Zhao, Yizhong Zhang, Pedro Garcia-Caparros, Bin Yang, and Yingdie Yang. 2025. "Genome-Wide Characterization and Identification of Auxin Response Factor (ARF) Gene Family Reveals the Regulation of RrARF5 in AsA Metabolism in Rosa roxburghii Tratt. Fruits" Biology 14, no. 9: 1156. https://doi.org/10.3390/biology14091156
APA StyleFeng, T., Sun, Z., Liu, M., Zhao, H., Zhang, Y., Garcia-Caparros, P., Yang, B., & Yang, Y. (2025). Genome-Wide Characterization and Identification of Auxin Response Factor (ARF) Gene Family Reveals the Regulation of RrARF5 in AsA Metabolism in Rosa roxburghii Tratt. Fruits. Biology, 14(9), 1156. https://doi.org/10.3390/biology14091156






