Molecular Regulation of Phenylpropanoid and Flavonoid Biosynthesis Pathways Based on Transcriptomic and Metabolomic Analyses in Oat Seedlings Under Sodium Selenite Treatment
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Treatments
2.2. Determination of Physiological and Biochemical Indices
2.3. Determination of Selenium Content
2.4. Metabolite Extraction and LC-MS/MS Analysis
2.5. cDNA Library Preparation and Transcriptome Sequencing
2.6. qRT-PCR Analysis
2.7. Statistical Analysis
3. Results
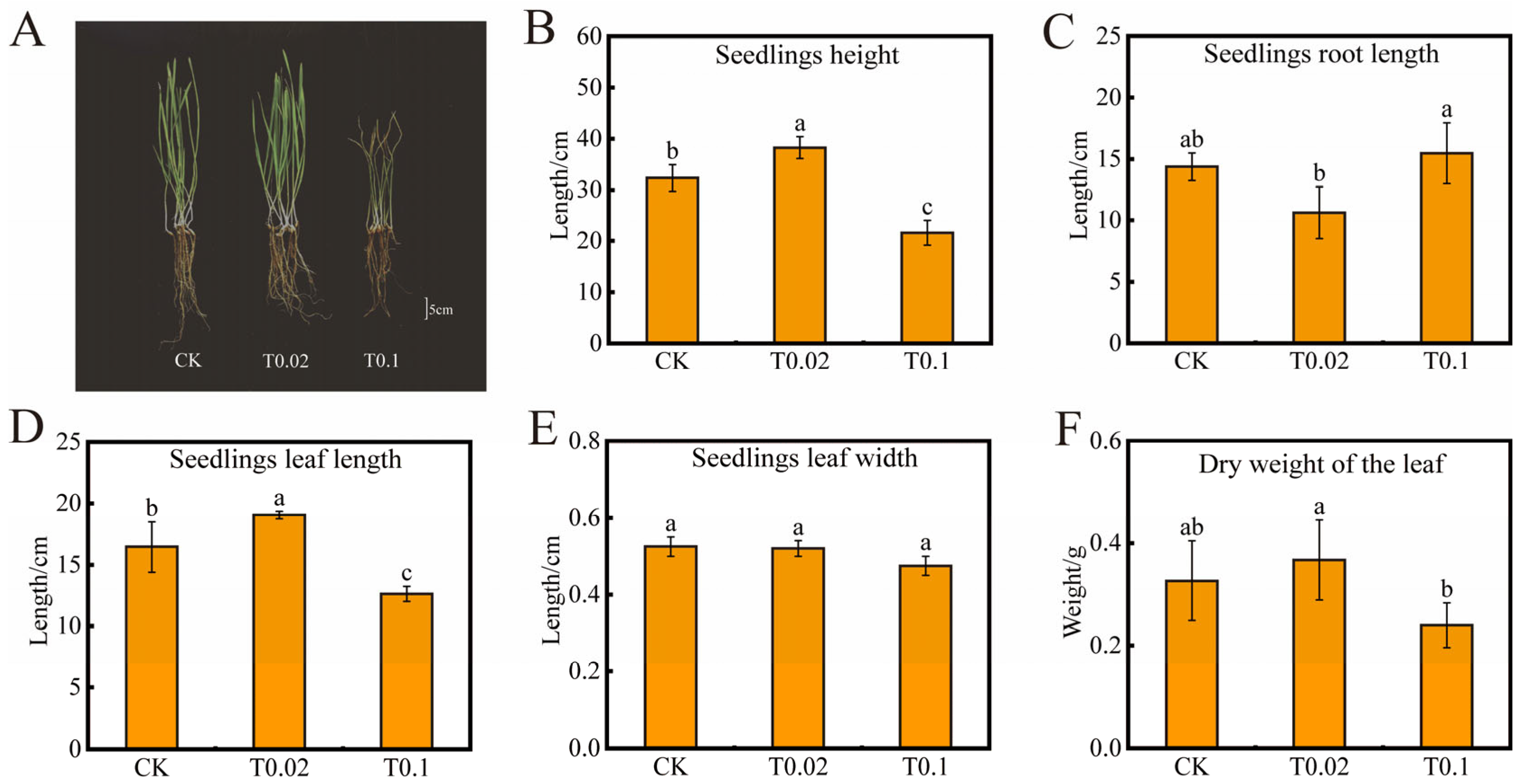
3.1. Effect of Oat Seedlings on Morphology and Physiological Indexes Under Na2SeO3 Treatment
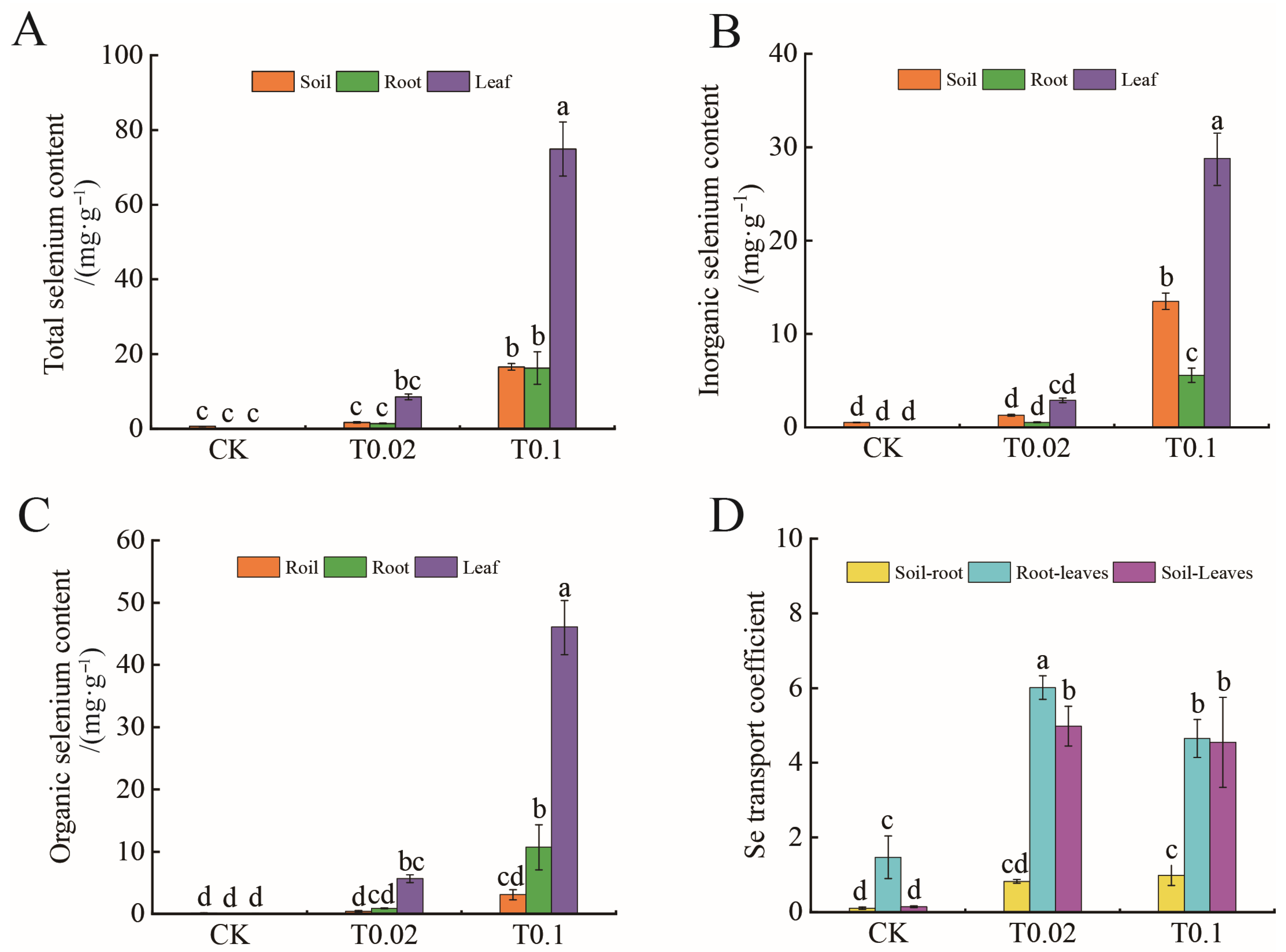
3.2. Selenium Content and Mobility Coefficient in Oat Seedlings Under Na2SeO3 Treatment
3.3. Metabolomic Analysis of Oat Seedlings Under Na2SeO3 Treatment
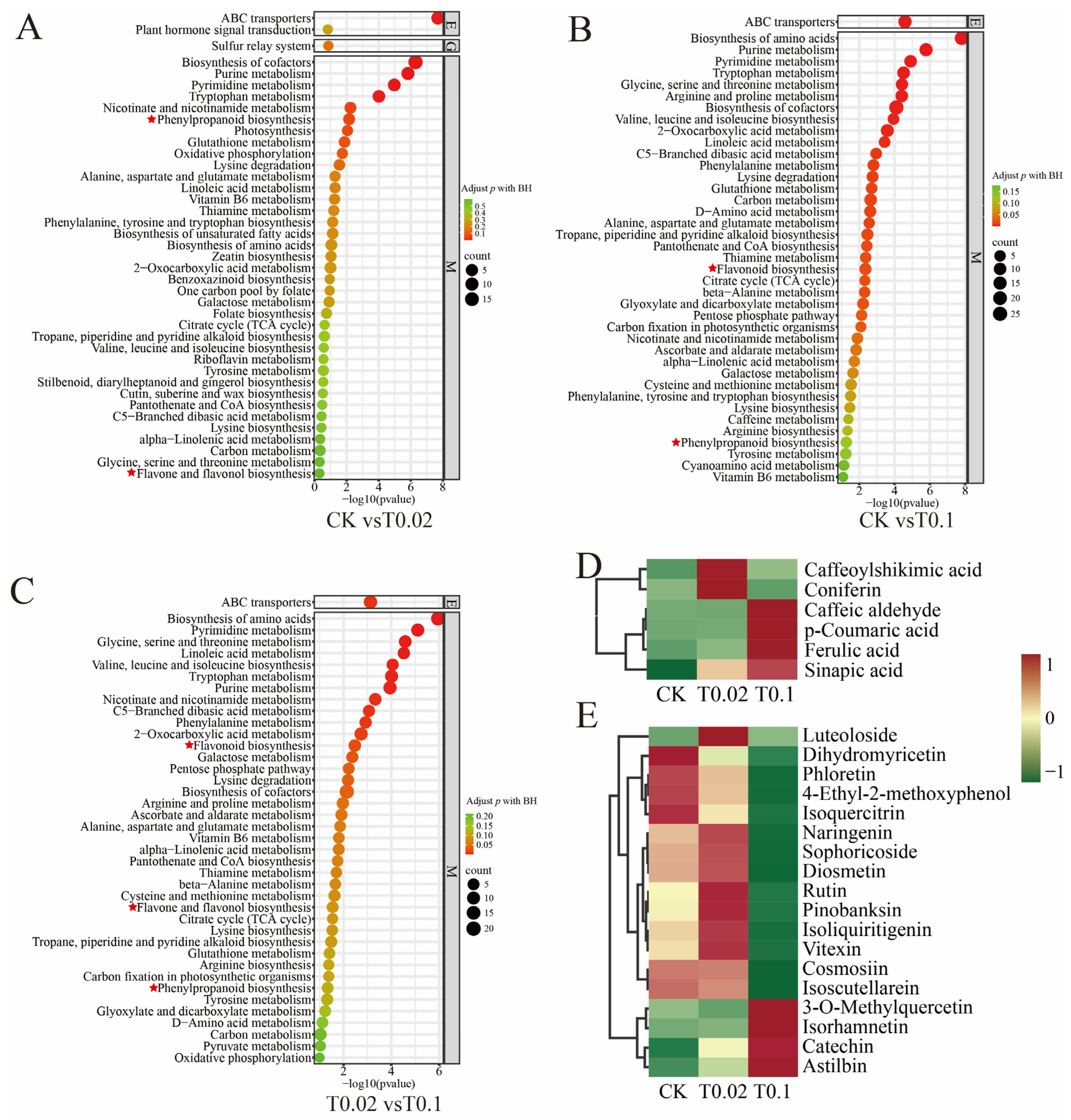
3.4. KEGG Pathway Analysis of DAMs in Oat Seedlings Under Na2SeO3 Treatment
3.5. Transcriptomic Analysis of Oat Seedlings Under Na2SeO3 Treatment
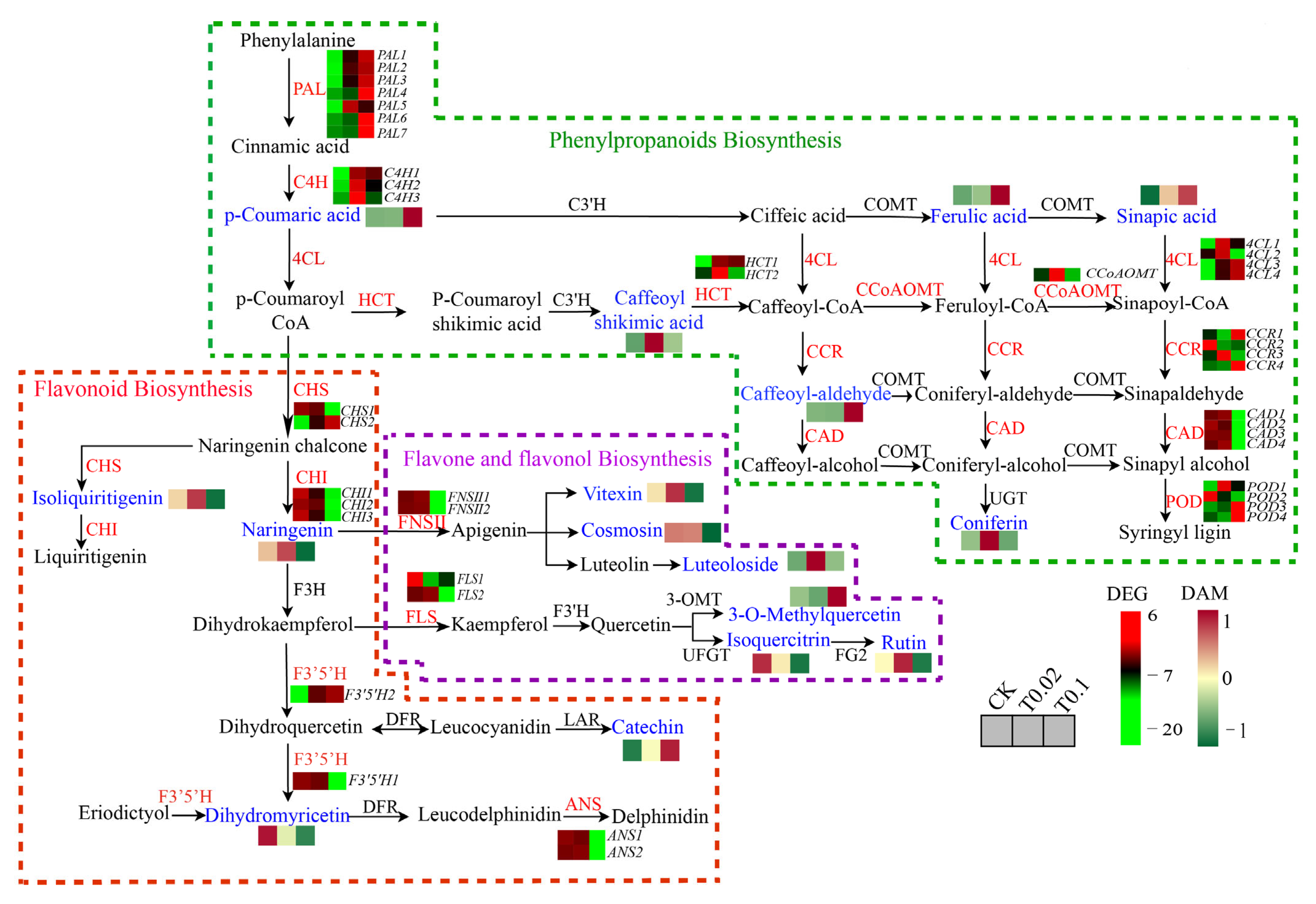
3.6. DEGs in the Phenylpropanoid and Flavonoid Biosynthesis in Oat Seedlings
3.7. Verification of DEGs in Phenylpropanoid and Flavonoid Metabolism in Oat Seedlings by RT-qPCR
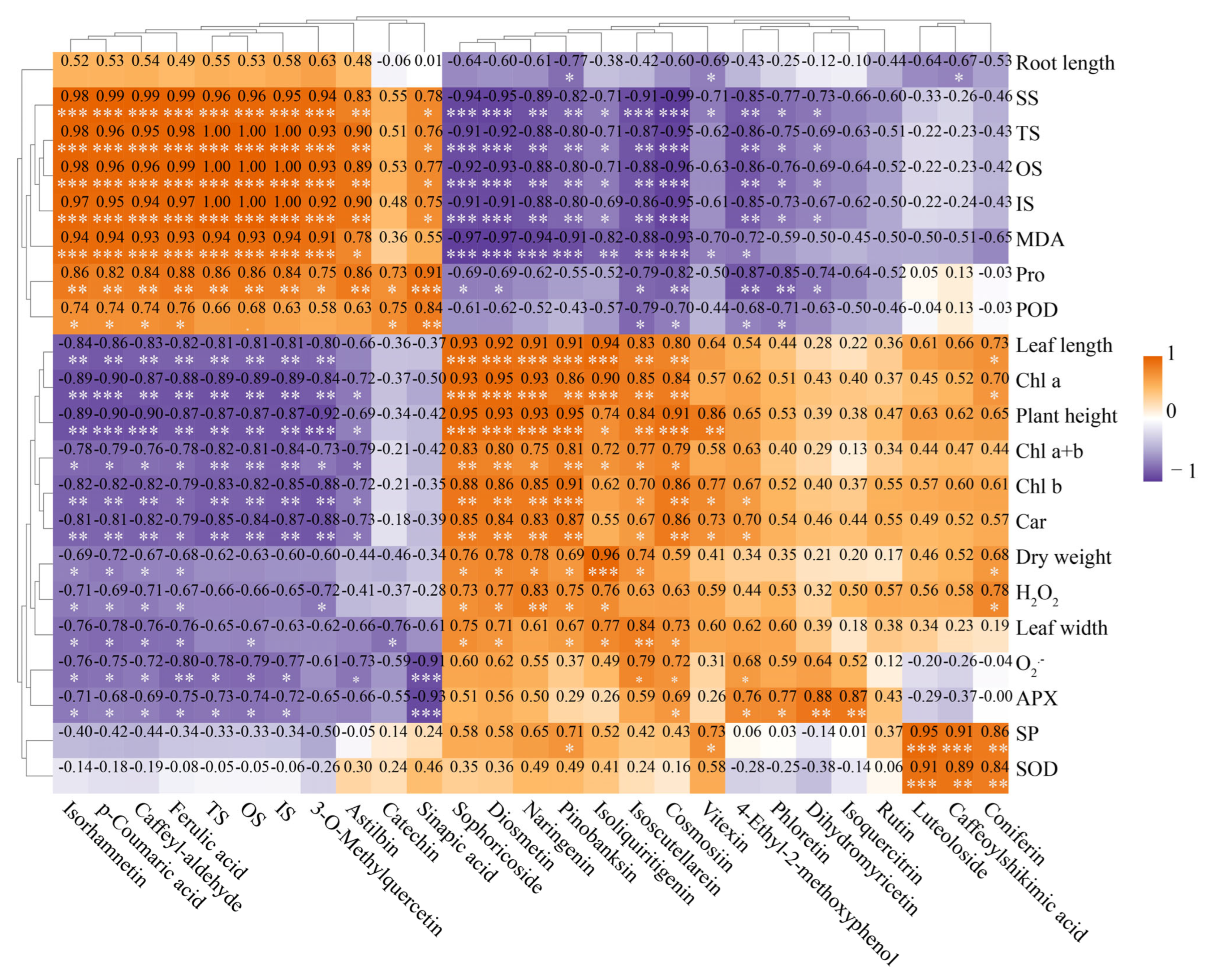
3.8. Correlation Analysis of Physiological and Biochemical Indicators, Gene Expression, Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TS | Total selenium content |
| OS | Organic selenium content |
| IS | Inorganic selenium content |
| MDA | Malondialdehyde |
| H2O2 | Hydrogen peroxide |
| APX | Ascorbate peroxidase |
| SOD | Superoxide dismutase |
| O2− | Superoxide anion radical |
| PAL | Phenylalanine ammonia-lyase |
| C4H | Trans-cinnamate 4-monooxygenase |
| 4CL | 4-coumarate--CoA ligase |
| CCR | Cinnamoyl-CoA reductase |
| CAD | Cinnamyl alcohol dehydrogenase |
| POD | Peroxidase |
| CCoAOMT | Caffeoyl-CoA O-methyltransferase |
| CHS | Chalcone synthase |
| CHI | Chalcone isomerase |
| ANS | Anthocyanidin synthase |
| F3′5′H | Flavonoid 3′,5′-hydroxylase |
| HCT | Hydroxycinnamoyl transferase |
| FLS | Flavonol synthase |
| FNSII | Flavone synthase II |
| DFR | Dihydroflavonol 4-reductase |
| COMT | Caffeic acid 3-O-methyltransferase |
| UGT | Uridine Diphosphate Glycosyltransferase |
| F3′H | Flavonoid 3′-monooxygenase |
| F3H | Naringenin 3-dioxygenase |
| 3-OMT | Flavonol 3-O-methyltransferase |
| UFGT | Flavonol 3-O-glucosyltransferase |
| FG2 | Flavonol-3-O-glucoside L-rhamnosyltransferase |
| LAR | Leucoanthocyanidin reductase |
References
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Sarkar, S.; Ahmad, Z.; Vemuri, H.; Garai, S.; Mondal, M.; Bhatt, R.; et al. Selenium Biofortification: Roles, Mechanisms, Responses and Prospects. Molecules 2021, 26, 881. [Google Scholar] [CrossRef]
- Wang, Y.-S.; Liang, S.-S.; Ren, J.-J.; Wang, Z.-Y.; Deng, X.-X.; Liu, W.-D.; Yan, Y.-L.; Song, G.-H.; Li, X.-X. The Effects of Selenium Supplementation in the Treatment of Autoimmune Thyroiditis: An Overview of Systematic Reviews. Nutrients 2023, 15, 3194. [Google Scholar] [CrossRef]
- Zhang, F.; Li, X.; Wei, Y. Selenium and Selenoproteins in Health. Biomolecules 2023, 13, 799. [Google Scholar] [CrossRef]
- Morán-Serradilla, C.; Angulo-Elizari, E.; Henriquez-Figuereo, A.; Sanmartín, C.; Sharma, A.K.; Plano, D. Seleno-Metabolites and Their Precursors: A New Dawn for Several Illnesses? Metabolites 2022, 12, 874. [Google Scholar] [CrossRef]
- Hariharan, S.; Dharmaraj, S. Selenium and Selenoproteins: It’s Role in Regulation of Inflammation. Inflammopharmacology 2020, 28, 667–695. [Google Scholar] [CrossRef] [PubMed]
- Rua, R.M.; Nogales, F.; Carreras, O.; Ojeda, M.L. Selenium, Selenoproteins and Cancer of the Thyroid. J. Trace Elem. Med. Biol. 2023, 76, 127115. [Google Scholar] [CrossRef]
- Fairweather-Tait, S.J.; Filippini, T.; Vinceti, M. Selenium Status and Immunity. Proc. Nutr. Soc. 2023, 82, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Thounaojam, T.C.; Chowdhury, D.; Upadhyaya, H. The Role of Selenium and Nano Selenium on Physiological Responses in Plant: A Review. Plant Growth Regul. 2023, 100, 409–433. [Google Scholar] [CrossRef]
- Ikram, S.; Li, Y.; Lin, C.; Yi, D.; Heng, W.; Li, Q.; Tao, L.; Hongjun, Y.; Weijie, J. Selenium in Plants: A Nexus of Growth, Antioxidants, and Phytohormones. J. Plant Physiol. 2024, 296, 154237. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Yang, X.; Rao, S.; Zhu, Z.; Cong, X.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; Xu, F. Advances in Physiological Mechanisms of Selenium to Improve Heavy Metal Stress Tolerance in Plants. Plant Biol. 2022, 24, 913–919. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Pan, Y.; Ma, L.; Fang, Y.; Pan, C.; Qiang, Y.; Cao, X.; Xu, H. Nano-Selenium Promotes the Product Quality and Plant Defense of Salvia Miltiorrhiza by Inducing Tanshinones and Salvianolic Acids Accumulation. Ind. Crops Prod. 2023, 195, 116436. [Google Scholar] [CrossRef]
- Yi, Q.; Sun, X.; Tang, S.; Xu, P.; Pang, Y.; Huang, X.; Huang, Q.; Huang, J.; Zhang, M. Comparation of Se Accumulation and Distribution of Two Rice (Oryza sativa L.) Cultivars with High- and Low- Se Efficiency as Affected by Exogenous Application of Selenite. J. Cereal Sci. 2022, 105, 103475. [Google Scholar] [CrossRef]
- Eiche, E.; Nothstein, A.K.; Göttlicher, J.; Steininger, R.; Dhillon, K.S.; Neumann, T. The Behaviour of Irrigation Induced Se in the Groundwater-Soil-Plant System in Punjab, India. Environ. Sci. Process. Impacts 2019, 21, 957–969. [Google Scholar] [CrossRef]
- Li, L.; Yu, J.; Li, L.; Rao, S.; Wu, S.; Wang, S.; Cheng, S.; Cheng, H. Treatment of Ginkgo Biloba with Exogenous Sodium Selenite Affects Its Physiological Growth, Changes Its Phytohormones, and Synthesizes Its Terpene Lactones. Molecules 2022, 27, 7548. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liao, X.; Yu, L.; Rao, S.; Chen, Q.; Zhu, Z.; Cong, X.; Zhang, W.; Ye, J.; Cheng, S.; et al. Combined Metabolome and Transcriptome Analysis Reveal the Mechanism of Selenate Influence on the Growth and Quality of Cabbage (Brassica oleracea var. capitata L.). Food Res. Int. 2022, 156, 111135. [Google Scholar] [CrossRef]
- Ulhassan, Z.; Gill, R.A.; Ali, S.; Mwamba, T.M.; Ali, B.; Wang, J.; Huang, Q.; Aziz, R.; Zhou, W. Dual Behavior of Selenium: Insights into Physio-Biochemical, Anatomical and Molecular Analyses of Four Brassica napus Cultivars. Chemosphere 2019, 225, 329–341. [Google Scholar] [CrossRef]
- Zhou, W.; Duan, Y.; Zhang, Y.; Wang, H.; Huang, D.; Zhang, M. Effects of Foliar Selenium Application on Growth and Rhizospheric Soil Micro-Ecological Environment of Atractylodes Macrocephala Koidz. S. Afr. J. Bot 2021, 137, 98–109. [Google Scholar] [CrossRef]
- Xue, Z.; Wang, B.; Qu, C.; Tao, M.; Wang, Z.; Zhang, G.; Zhao, M.; Zhao, S. Response of Salt Stress Resistance in Highland Barley (Hordeum vulgare L. var. nudum) through Phenylpropane Metabolic Pathway. PLoS ONE 2023, 18, e0286957. [Google Scholar] [CrossRef]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Chen, X.; Su, W.; Zhang, H.; Zhan, Y.; Zeng, F. Fraxinus Mandshurica 4-Coumarate-CoA Ligase 2 Enhances Drought and Osmotic Stress Tolerance of Tobacco by Increasing Coniferyl Alcohol Content. Plant Physiol. Biochem. 2020, 155, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.-P.; Zhang, L.; Yan, P.; Ahammed, G.J.; Han, W.-Y. Methyl Salicylate Enhances Flavonoid Biosynthesis in Tea Leaves by Stimulating the Phenylpropanoid Pathway. Molecules 2019, 24, 362. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Cheng, Y.; Gao, H.; Chen, X. A Review of Classification, Biosynthesis, Biological Activities and Potential Applications of Flavonoids. Molecules 2023, 28, 4982. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Q.; Zhang, L.; Ren, F.; Zhang, Z.; Jia, Q. Integrated Metabolomics and Transcriptomics Analysis of Flavonoid Biosynthesis Pathway in Polygonatum Cyrtonema Hua. Molecules 2024, 29, 2248. [Google Scholar] [CrossRef]
- Zhuang, W.-B.; Li, Y.-H.; Shu, X.-C.; Pu, Y.-T.; Wang, X.-J.; Wang, T.; Wang, Z. The Classification, Molecular Structure and Biological Biosynthesis of Flavonoids, and Their Roles in Biotic and Abiotic Stresses. Molecules 2023, 28, 3599. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant Flavonoids: Classification, Distribution, Biosynthesis, and Antioxidant Activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, R.; Hu, Y.; Deng, H.; Pei, X.; Liu, F.; Chen, C. Impact of Oat (Avena sativa L.) on Metabolic Syndrome and Potential Physiological Mechanisms of Action: A Current Review. J. Agric. Food Chem. 2023, 71, 14838–14852. [Google Scholar] [CrossRef]
- Huang, C.-T.; Klos, K.E.; Huang, Y.-F. Genome-Wide Association Study Reveals the Genetic Architecture of Seed Vigor in Oats. G3 Genes|Genomes|Genet. 2020, 10, 4489–4503. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, Y.; Huo, P.; Zhang, Q.; Chen, X.; Zhang, Z. Transcriptome Analysis of Hexaploid Hulless Oat in Response to Salinity Stress. PLoS ONE 2017, 12, e0171451. [Google Scholar] [CrossRef]
- Anwar, A.; She, M.; Wang, K.; Riaz, B.; Ye, X. Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 3681. [Google Scholar] [CrossRef]
- Liu, K.; Ju, Z.; Jia, Z.; Liang, G.; Ma, X.; Liu, W. Genome-Wide Identification and Characterization of the Oat (Avena sativa L.) WRKY Transcription Factor Family. Genes 2022, 13, 1918. [Google Scholar] [CrossRef]
- Jinqiu, Y.; Bing, L.; Tingting, S.; Jinglei, H.; Zelai, K.; Lu, L.; Wenhua, H.; Tao, H.; Xinyu, H.; Zengqing, L.; et al. Integrated Physiological and Transcriptomic Analyses Responses to Altitude Stress in Oat (Avena sativa L.). Front. Genet. 2021, 12, 638683. [Google Scholar] [CrossRef]
- Gao, Y.; Jin, Y.; Guo, W.; Xue, Y.; Yu, L. Metabolic and Physiological Changes in the Roots of Two Oat Cultivars in Response to Complex Saline-Alkali Stress. Front. Plant Sci. 2022, 13, 835414. [Google Scholar] [CrossRef]
- Kim, I.-S.; Hwang, C.-W.; Yang, W.-S.; Kim, C.-H. Multiple Antioxidative and Bioactive Molecules of Oats (Avena sativa L.) in Human Health. Antioxidants 2021, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, T.H.; Jeong, Y.J.; Park, S.H.; Park, S.C.; Lee, J.; Yang, K.Y.; Jeong, J.C.; Kim, C.Y. Synergistic Effect of Methyl Jasmonate and Abscisic Acid Co-Treatment on Avenanthramide Production in Germinating Oats. Int. J. Mol. Sci. 2021, 22, 4779. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into Plant Salt Stress Signaling and Tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Zhou, Y.; Feng, C.; Wang, Y.; Yun, C.; Zou, X.; Cheng, N.; Zhang, W.; Jing, Y.; Li, H. Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding. Int. J. Mol. Sci. 2024, 25, 10940. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pan, Y.; Li, J.; Chen, J.; Yang, S.; Zhao, M.; Xue, Y. Transcriptome Sequencing Analysis of Root in Soybean Responding to Mn Poisoning. Int. J. Mol. Sci. 2023, 24, 12727. [Google Scholar] [CrossRef]
- Li, J.; Zhou, X.; Zhou, J.; Shang, R.; Wang, Y.; Jing, P. Comparative Study on Several Determination Methods of Chlorophyll Content in Plants. IOP Conf. Ser. Mater. Sci. Eng. 2020, 730, 012066. [Google Scholar] [CrossRef]
- Mas-Bargues, C.; Escrivá, C.; Dromant, M.; Borrás, C.; Viña, J. Lipid Peroxidation as Measured by Chromatographic Determination of Malondialdehyde. Human Plasma Reference Values in Health and Disease. Arch. Biochem. Biophys. 2021, 709, 108941. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Z.; Zhou, Q.; Wang, X.; Song, S.; Dong, S. Physiological Response of Soybean Plants to Water Deficit. Front. Plant Sci. 2021, 12, 809692. [Google Scholar] [CrossRef]
- Ghaffari, H.; Tadayon, M.R.; Nadeem, M.; Cheema, M.; Razmjoo, J. Proline-Mediated Changes in Antioxidant Enzymatic Activities and the Physiology of Sugar Beet under Drought Stress. Acta Physiol. Plant 2019, 41, 23. [Google Scholar] [CrossRef]
- Hadwan, M.H.; Hussein, M.J.; Mohammed, R.M.; Hadwan, A.M.; Saad Al-Kawaz, H.; Al-Obaidy, S.S.M.; Al Talebi, Z.A. An Improved Method for Measuring Catalase Activity in Biological Samples. Biol. Methods Protoc. 2024, 9, bpae015. [Google Scholar] [CrossRef]
- Sadak, M.S.; Abd El-Hameid, A.R.; Zaki, F.S.A.; Dawood, M.G.; El-Awadi, M.E. Physiological and Biochemical Responses of Soybean (Glycine Max L.) to Cysteine Application under Sea Salt Stress. Bull. Natl. Res. Cent. 2019, 44, 1. [Google Scholar] [CrossRef]
- Hosseini, F.; Mosaddeghi, M.R.; Dexter, A.R.; Sepehri, M. Maize Water Status and Physiological Traits as Affected by Root Endophytic Fungus Piriformospora Indica under Combined Drought and Mechanical Stresses. Planta 2018, 247, 1229–1245. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, M.; Zhou, W.; Ai, L.; You, J.; Liu, H.; You, J.; Wang, H.; Wassie, M.; Wang, M.; et al. Transcriptome Analysis Reveals Novel Insights into the Continuous Cropping Induced Response in Codonopsis Tangshen, a Medicinal Herb. Plant Physiol. Biochem. 2019, 141, 279–290. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, P.; Zhang, T.; Li, Y.; Wang, Y.; Gao, C. Comprehensive Analysis of BpHSP Genes and Their Expression under Heat Stresses in Betula platyphylla. Environ. Exp. Bot. 2018, 152, 167–176. [Google Scholar] [CrossRef]
- Ni, X.; Li, X.; Shen, T.-L.; Qian, W.-J.; Xian, M. A Sweet H2S/H2O2 Dual Release System and Specific Protein S-Persulfidation Mediated by Thioglucose/Glucose Oxidase. J. Am. Chem. Soc. 2021, 143, 13325–13332. [Google Scholar] [CrossRef] [PubMed]
- GB/T 5009.93-2017; Determination of Selenium in Foods. Standardization Administration of China: Beijing, China, 2017.
- Zhou, X.; Yi, D.; Ma, L.; Wang, X. Genome-Wide Analysis and Expression of the Aquaporin Gene Family in Avena sativa L. Front. Plant Sci. 2023, 14, 1305299. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, M.; Cui, L.; Liu, L. Deciphering the Dynamic Gene Expression Patterns of Pollen Abortion in a Male Sterile Line of Avena sativa through Transcriptome Analysis at Different Developmental Stages. BMC Plant Biol. 2021, 21, 101. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, G.; Qin, H.; Guan, Y.; Wang, T.; Ni, W.; Xie, H.; Xing, Y.; Tian, G.; Lyu, M.; et al. Metabolomics Combined with Physiology and Transcriptomics Reveal Key Metabolic Pathway Responses in Apple Plants Exposure to Different Selenium Concentrations. J. Hazard. Mater. 2024, 464, 132953. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, J.; Li, F.; Zeng, X.; Wu, Z.; Huang, Y.; Xue, Y.; Wang, Y. High Concentrations of Se Inhibited the Growth of Rice Seedlings. Plants 2024, 13, 1580. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, J.; Hu, H.; Li, Y.; Xiang, M.; Wang, D. Integrated Eco-Physiological, Biochemical, and Molecular Biological Analyses of Selenium Fortification Mechanism in Alfalfa. Planta 2022, 256, 114. [Google Scholar] [CrossRef] [PubMed]
- Hawrylak-Nowak, B.; Matraszek, R.; Pogorzelec, M. The Dual Effects of Two Inorganic Selenium Forms on the Growth, Selected Physiological Parameters and Macronutrients Accumulation in Cucumber Plants. Acta Physiol. Plant 2015, 37, 41. [Google Scholar] [CrossRef]
- Cabral Gouveia, G.C.; Galindo, F.S.; Dantas Bereta Lanza, M.G.; Caroline da Rocha Silva, A.; Pereira de Brito Mateus, M.; Souza da Silva, M.; Rimoldi Tavanti, R.F.; Tavanti, T.R.; Lavres, J.; Reis, A.R.D. Selenium Toxicity Stress-Induced Phenotypical, Biochemical and Physiological Responses in Rice Plants: Characterization of Symptoms and Plant Metabolic Adjustment. Ecotoxicol. Environ. Saf. 2020, 202, 110916. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Shi, J.; Pan, Y.; Yang, S.; Xue, Y. Combined Metabolome and Transcriptome Analysis Reveals the Key Pathways Involved in the Responses of Soybean Plants to High Se Stress. Ecotoxicol. Environ. Saf. 2024, 287, 117262. [Google Scholar] [CrossRef]
- Morales, M.; Munné-Bosch, S. Malondialdehyde: Facts and Artifacts. Plant Physiol. 2019, 180, 1246–1250. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Devi, D.D.; Shanker, A.K.; Sheeba, J.A.; Bangarusamy, U. Selenium—an Antioxidative Protectant in Soybean during Senescence. Plant Soil. 2005, 272, 77–86. [Google Scholar] [CrossRef]
- Khalofah, A.; Migdadi, H.; El-Harty, E. Antioxidant Enzymatic Activities and Growth Response of Quinoa (Chenopodium quinoa Willd) to Exogenous Selenium Application. Plants 2021, 10, 719. [Google Scholar] [CrossRef]
- Jin, W.; Cheng, L.; Liu, C.; Liu, H.; Jiao, Q.; Wang, H.; Deng, Z.; Seth, C.S.; Guo, H.; Shi, Y. Cadmium Negatively Affects the Growth and Physiological Status and the Alleviation Effects by Exogenous Selenium in Silage Maize (Zea mays L.). Environ. Sci. Pollut. Res. Int. 2024, 31, 21646–21658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Q.; Wang, Y.; Xu, Y.; Li, J.; Zhao, S.; Wang, D.; Ma, Z.; Yan, F.; Liu, Y. Combined Transcriptomic and Metabolomic Analysis Reveals the Role of Phenylpropanoid Biosynthesis Pathway in the Salt Tolerance Process of Sophora Alopecuroides. Int. J. Mol. Sci. 2021, 22, 2399. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, L.; Chao, W.; Xiang, J.; Yang, X.; Ye, J.; Liao, X.; Zhou, X.; Rao, S.; Cheng, S.; et al. Comparative Physiological and Transcriptome Analysis Reveals the Potential Mechanism of Selenium Accumulation and Tolerance to Selenate Toxicity of Broussonetia Papyrifera. Tree Physiol. 2022, 42, 2578–2595. [Google Scholar] [CrossRef]
- Xiang, J.; Rao, S.; Chen, Q.; Zhang, W.; Cheng, S.; Cong, X.; Zhang, Y.; Yang, X.; Xu, F. Research Progress on the Effects of Selenium on the Growth and Quality of Tea Plants. Plants 2022, 11, 2491. [Google Scholar] [CrossRef]
- Liu, P.; Song, L.; Hao, S.; Qin, J.; Yang, C.; Yang, W.; Feng, M.; Zhang, M.; Wang, C.; Song, X. Effects of Selenium Application Concentration, Period and Method on the Selenium Content and Grain Yield of Tartary Buckwheat of Different Varieties. J. Sci. Food Agric. 2022, 102, 6868–6876. [Google Scholar] [CrossRef]
- Ge, X.; Zhang, J.; He, L.; Yu, N.; Pan, C.; Chen, Y. Integration of Metabolomics and Transcriptomics Analyses Reveals the Mechanism of Nano-Selenium Treated to Activate Phenylpropanoid Metabolism and Enhance the Antioxidant Activity of Peach. J. Food Sci. 2023, 88, 4529–4543. [Google Scholar] [CrossRef]
- Dong, Q.; Li, D.; Wu, Y.; Zhou, C.; Lin, Y.; Miao, P.; Li, J.; Pan, C. Exogenous Nanoselenium Alleviates Imidacloprid-Induced Oxidative Stress Toxicity by Improving Phenylpropanoid Metabolism and Antioxidant Defense System in Perilla frutescens (L.) Britt. J. Plant Physiol. 2023, 289, 154095. [Google Scholar] [CrossRef]
- Li, D.; An, Q.; Wu, Y.; Li, J.-Q.; Pan, C. Foliar Application of Selenium Nanoparticles on Celery Stimulates Several Nutrient Component Levels by Regulating the α-Linolenic Acid Pathway. ACS Sustain. Chem. Eng. 2020, 8, 10502–10510. [Google Scholar] [CrossRef]
- Shiriaev, A.; Brizzolara, S.; Sorce, C.; Meoni, G.; Vergata, C.; Martinelli, F.; Maza, E.; Djari, A.; Pirrello, J.; Pezzarossa, B.; et al. Selenium Biofortification Impacts the Tomato Fruit Metabolome and Transcriptional Profile at Ripening. J. Agric. Food Chem. 2023, 71, 13554–13565. [Google Scholar] [CrossRef]
- Dong, B.; Tang, H.; Zhu, D.; Yao, Q.; Han, H.; He, K.; Ding, X. Benzothiazole Treatment Regulates the Reactive Oxygen Species Metabolism and Phenylpropanoid Pathway of Rosa roxburghii Fruit to Delay Senescence During Low Temperature Storage. Front. Plant Sci. 2021, 12, 753261. [Google Scholar] [CrossRef]
- Dong, B.; Da, F.; Chen, Y.; Ding, X. Melatonin Treatment Maintains the Quality of Fresh-Cut Gastrodia Elata under Low-Temperature Conditions by Regulating Reactive Oxygen Species Metabolism and Phenylpropanoid Pathway. Int. J. Mol. Sci. 2023, 24, 14284. [Google Scholar] [CrossRef]
- Wang, G.; Wu, L.; Zhang, H.; Wu, W.; Zhang, M.; Li, X.; Wu, H. Regulation of the Phenylpropanoid Pathway: A Mechanism of Selenium Tolerance in Peanut (Arachis hypogaea L.) Seedlings. J. Agric. Food Chem. 2016, 64, 3626–3635. [Google Scholar] [CrossRef]
- Guo, G.; Wang, Y.; Zhang, B.; Yu, H.; Li, L.; Cao, G.; Chen, B.; Li, C.; Bu, F.; Teng, S.; et al. Comparative Transcriptomic and Metabolomic Analysis Reveals Mechanisms of Selenium-Regulated Anthocyanin Synthesis in Waxy Maize (Zea mays L.). Front. Plant Sci. 2024, 15, 1466756. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhou, C.; Ma, J.; Wu, Y.; Kang, L.; An, Q.; Zhang, J.; Deng, K.; Li, J.-Q.; Pan, C. Nanoselenium Transformation and Inhibition of Cadmium Accumulation by Regulating the Lignin Biosynthetic Pathway and Plant Hormone Signal Transduction in Pepper Plants. J. Nanobiotechnol. 2021, 19, 316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, X.; Wu, Q.; Lu, P.; Kang, Q.; Zhao, M.; Wang, A.; Dong, Q.; Sun, M.; Yang, Z.; et al. Selenium Application Enhances the Accumulation of Flavones and Anthocyanins in Bread Wheat (Triticum aestivum L.) Grains. J. Agric. Food Chem. 2022, 70, 13431–13444. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Wu, X.; Xie, H.; Geng, G.; Qiao, F. Molecular Regulation of Phenylpropanoid and Flavonoid Biosynthesis Pathways Based on Transcriptomic and Metabolomic Analyses in Oat Seedlings Under Sodium Selenite Treatment. Biology 2025, 14, 1131. https://doi.org/10.3390/biology14091131
Ma J, Wu X, Xie H, Geng G, Qiao F. Molecular Regulation of Phenylpropanoid and Flavonoid Biosynthesis Pathways Based on Transcriptomic and Metabolomic Analyses in Oat Seedlings Under Sodium Selenite Treatment. Biology. 2025; 14(9):1131. https://doi.org/10.3390/biology14091131
Chicago/Turabian StyleMa, Jianxia, Xiaozhuo Wu, Huichun Xie, Guigong Geng, and Feng Qiao. 2025. "Molecular Regulation of Phenylpropanoid and Flavonoid Biosynthesis Pathways Based on Transcriptomic and Metabolomic Analyses in Oat Seedlings Under Sodium Selenite Treatment" Biology 14, no. 9: 1131. https://doi.org/10.3390/biology14091131
APA StyleMa, J., Wu, X., Xie, H., Geng, G., & Qiao, F. (2025). Molecular Regulation of Phenylpropanoid and Flavonoid Biosynthesis Pathways Based on Transcriptomic and Metabolomic Analyses in Oat Seedlings Under Sodium Selenite Treatment. Biology, 14(9), 1131. https://doi.org/10.3390/biology14091131





