Integrating Multi-Domain Approach for Identification of Neo Anti-DHPS Inhibitors Against Pathogenic Stenotrophomonas maltophilia
Simple Summary
Abstract
1. Introduction
2. Methodology
2.1. Target Identification and Energy Minimization Phase
2.2. Compound Library Selection and Preparation Phase
2.3. Molecular Docking Step
2.4. ADME Property Prediction Step
2.5. Molecular Dynamics (MD) Simulation
2.6. Hydrogen Bonding Analysis
2.7. Salt Bridges Analysis
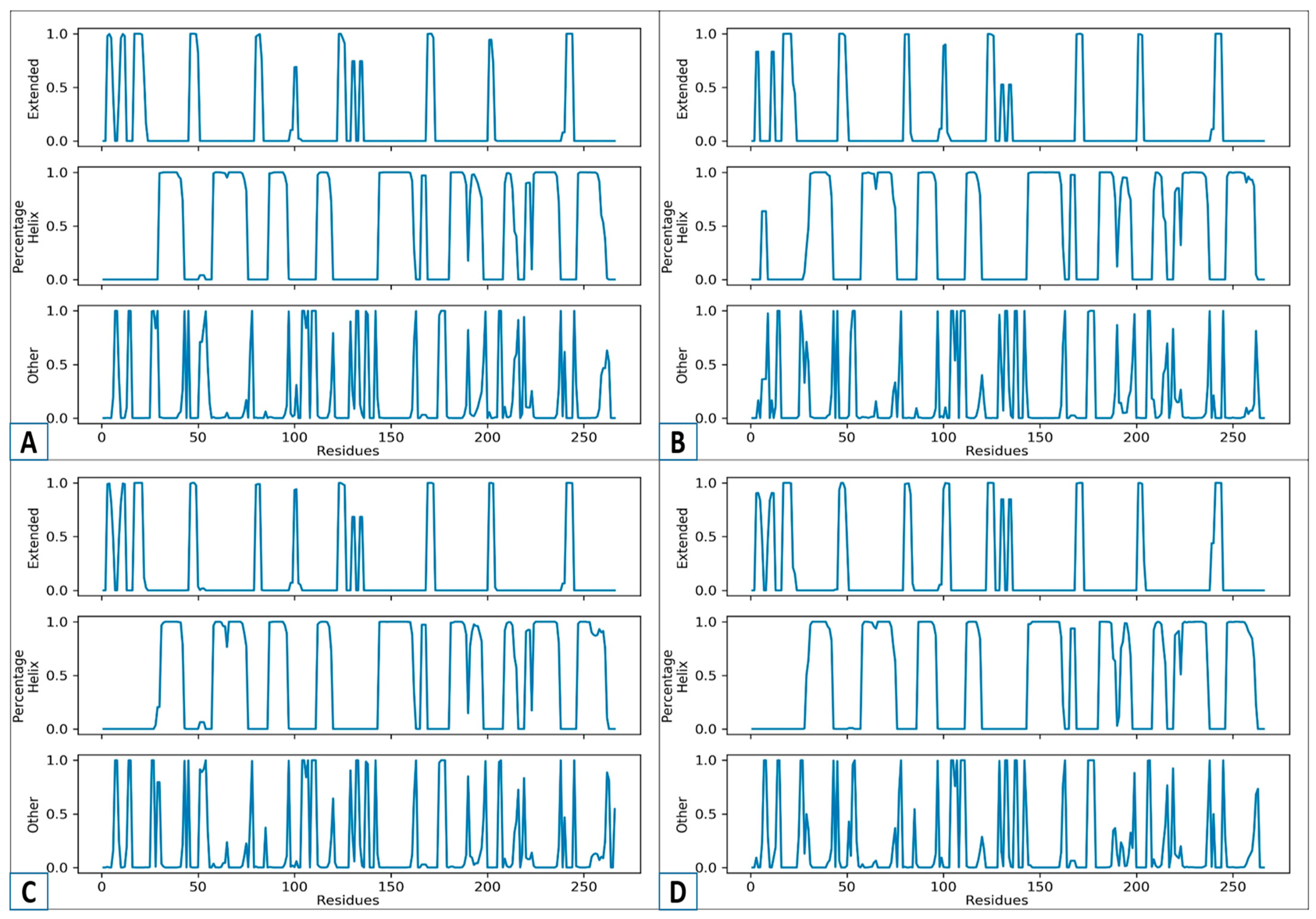
2.8. Secondary Structure Analysis
2.9. MMPB/GBSA Calculations
2.10. Entropy Energy Calculations
3. Results
3.1. Target Receptor Identification and Preparation Phase
3.2. Molecular Docking Findings and Binding Affinity Score of Compounds Against DHPS
3.3. Molecular Interaction Between DHPS and Top-3 Hits
3.4. Molecular Docking Analysis of the Control as a Baseline Compound
3.5. ADME Properties and Lipinski’s Rule of Five Profile of Screened Compounds
3.6. Molecular Dynamics (MD) Simulation
3.7. Hydrogen Bonding Analysis
3.8. Salt Bridge Formation and Electrostatic Interaction
3.9. An Analysis of the Secondary Structure Transitions upon Ligand Binding
3.10. MMPBS/GBSA Calculations
3.11. Entropy Energy Estimation
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooke, J.S. Advances in the Microbiology of Stenotrophomonas Maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef]
- Agri, H. An Epidemiological Study on Stenotrophomonas maltophilia from Clinical and Environmental Samples of Animals, and Milk Samples from Cattle and Buffalo. Ph.D. Thesis, Deemed University, 2024. [Google Scholar]
- García, G.; Girón, J.A.; Yañez, J.A.; Cedillo, M.L. Stenotrophomonas Maltophilia and Its Ability to Form Biofilms. Microbiol. Res. 2022, 14, 1–20. [Google Scholar] [CrossRef]
- Parija, S.C. Pseudomonas, Burkholderia and Acinetobacter. In Textbook of Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2023; pp. 553–561. [Google Scholar]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial Resistance: Impacts, Challenges, and Future Prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Hafiz, T.A.; Aldawood, E.; Albloshi, A.; Alghamdi, S.S.; Mubaraki, M.A.; Alyami, A.S.; Aldriwesh, M.G. Stenotrophomonas Maltophilia Epidemiology, Resistance Characteristics, and Clinical Outcomes: Understanding of the Recent Three Years’ Trends. Microorganisms 2022, 10, 2506. [Google Scholar] [CrossRef]
- Majumdar, R.; Karthikeyan, H.; Senthilnathan, V.; Sugumar, S. Review on Stenotrophomonas Maltophilia: An Emerging Multidrug-Resistant Opportunistic Pathogen. Recent Pat. Biotechnol. 2022, 16, 329–354. [Google Scholar] [CrossRef]
- Kunz Coyne, A.J.; Herbin, S.; Caniff, K.; Rybak, M.J. Steno-sphere: Navigating the Enigmatic World of Emerging Multidrug-resistant Stenotrophomonas maltophilia. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2023, 43, 833–846. [Google Scholar] [CrossRef]
- Ramasco, F.; Méndez, R.; de la Rica, A.S.; de Castro, R.G.; Maseda, E. Sepsis Stewardship: The Puzzle of Antibiotic Therapy in the Context of Individualization of Decision Making. J. Pers. Med. 2024, 14, 106. [Google Scholar] [CrossRef] [PubMed]
- AlFonaisan, M.K.; Mubaraki, M.A.; Althawadi, S.I.; Obeid, D.A.; Al-Qahtani, A.A.; Almaghrabi, R.S.; Alhamlan, F.S. Temporal Analysis of Prevalence and Antibiotic-Resistance Patterns in Stenotrophomonas maltophilia Clinical Isolates in a 19-Year Retrospective Study. Sci. Rep. 2024, 14, 14459. [Google Scholar] [CrossRef]
- Ochoa-Sánchez, L.E.; Martínez, J.L.; Gil-Gil, T. Evolution of Resistance against Ciprofloxacin, Tobramycin, and Trimethoprim/Sulfamethoxazole in the Environmental Opportunistic Pathogen Stenotrophomonas maltophilia. Antibiotics 2024, 13, 330. [Google Scholar] [CrossRef] [PubMed]
- Gil-Gil, T.; Martínez, J.L. Fosfomycin Resistance Evolutionary Pathways of Stenotrophomonas maltophilia in Different Growing Conditions. Int. J. Mol. Sci. 2022, 23, 1132. [Google Scholar] [CrossRef]
- Sehrawat, R.; Rathee, P.; Khatkar, S.; Akkol, E.; Khayatkashani, M.; Nabavi, S.M.; Khatkar, A. Dihydrofolate Reductase (DHFR) Inhibitors: A Comprehensive Review. Curr. Med. Chem. 2024, 31, 799–824. [Google Scholar] [CrossRef]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of Clinical Drug Development Fails and How to Improve It? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Macalino, S.J.Y.; Billones, J.B.; Organo, V.G.; Carrillo, M.C.O. In Silico Strategies in Tuberculosis Drug Discovery. Molecules 2020, 25, 665. [Google Scholar] [CrossRef]
- dos Santos Nascimento, I.J.; da Silva Rodrigues, É.E.; da Silva, M.F.; de Araújo-Júnior, J.X.; de Moura, R.O. Advances in Computational Methods to Discover New NS2B-NS3 Inhibitors Useful Against Dengue and Zika Viruses. Curr. Top. Med. Chem. 2022, 22, 2435–2462. [Google Scholar] [CrossRef]
- Yadav, M.; Abdalla, M.; Madhavi, M.; Chopra, I.; Bhrdwaj, A.; Soni, L.; Shaheen, U.; Prajapati, L.; Sharma, M.; Sikarwar, M.S.; et al. Structure-Based Virtual Screening, Molecular Docking, Molecular Dynamics Simulation and Pharmacokinetic Modelling of Cyclooxygenase-2 (COX-2) Inhibitor for the Clinical Treatment of Colorectal Cancer. Mol. Simul. 2022, 48, 1081–1101. [Google Scholar] [CrossRef]
- Jejurikar, B.L.; Rohane, S.H. Drug Designing in Discovery Studio. Asian J. Res. Chem. 2021, 14, 135–138. [Google Scholar]
- Richard, A.M.; Williams, C.R. Distributed Structure-Searchable Toxicity (DSSTox) Public Database Network: A Proposal. Mutat. Res. Mol. Mech. Mutagen. 2002, 499, 27–52. [Google Scholar] [CrossRef]
- Akash, S.; Bayıl, I.; Rahman, M.A.; Mukerjee, N.; Maitra, S.; Islam, M.R.; Rajkhowa, S.; Ghosh, A.; Al-Hussain, S.A.; Zaki, M.E.A.; et al. Target Specific Inhibition of West Nile Virus Envelope Glycoprotein and Methyltransferase Using Phytocompounds: An in Silico Strategy Leveraging Molecular Docking and Dynamics Simulation. Front. Microbiol. 2023, 14, 1189786. [Google Scholar] [CrossRef] [PubMed]
- Bharatam, P.V. Computer-Aided Drug Design. In Drug Discovery and Development: From Targets and Molecules to Medicines; Springer: Berlin/Heidelberg, Germany, 2021; pp. 137–210. [Google Scholar]
- Egieyeh, S.A.; Syce, J.; Malan, S.F.; Christoffels, A. Prioritization of Anti-Malarial Hits from Nature: Chemo-Informatic Profiling of Natural Products with in Vitro Antiplasmodial Activities and Currently Registered Anti-Malarial Drugs. Malar. J. 2016, 15, 50. [Google Scholar] [CrossRef]
- Yao, H.; Ke, H.; Zhang, X.; Pan, S.-J.; Li, M.-S.; Yang, L.-P.; Schreckenbach, G.; Jiang, W. Molecular Recognition of Hydrophilic Molecules in Water by Combining the Hydrophobic Effect with Hydrogen Bonding. J. Am. Chem. Soc. 2018, 140, 13466–13477. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, M.; Adeniji, S.E. In-Silico Molecular Docking and ADME/Pharmacokinetic Prediction Studies of Some Novel Carboxamide Derivatives as Anti-Tubercular Agents. Chem. Afr. 2020, 3, 989–1000. [Google Scholar] [CrossRef]
- Barber, R.D. Software to Visualize Proteins and Perform Structural Alignments. Curr. Protoc. 2021, 1, e292. [Google Scholar] [CrossRef]
- Sucharitha, P.; Reddy, K.R.; Satyanarayana, S.V.; Garg, T. Absorption, Distribution, Metabolism, Excretion, and Toxicity Assessment of Drugs Using Computational Tools. In Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV-2 Infection; Elsevier: Amsterdam, The Netherlands, 2022; pp. 335–355. [Google Scholar]
- Ivanović, V.; Rančić, M.; Arsić, B.; Pavlović, A. Lipinski’s Rule of Five, Famous Extensions and Famous Exceptions. Chem. Naissensis 2020, 3, 171–177. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Smardz, P.; Anila, M.M.; Rogowski, P.; Li, M.S.; Różycki, B.; Krupa, P. A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta. Int. J. Mol. Sci. 2024, 25, 6698. [Google Scholar] [CrossRef]
- Shamsi, A.; Khan, M.S.; Yadav, D.K.; Shahwan, M.; Furkan, M.; Khan, R.H. Structure-Based Drug-Development Study against Fibroblast Growth Factor Receptor 2: Molecular Docking and Molecular Dynamics Simulation Approaches. Sci. Rep. 2024, 14, 19439. [Google Scholar] [CrossRef]
- Raad, M.T.; El Said, H.; Al Jammal, A.; Alhage, J.; Albahri, G.; Baassiry, N.; HajjHussein, H.S. Molecular Dynamics Insights into the Adsorption of the COVID-19 Antiviral Remdesivir on Silica-Functionalized Graphene Oxide: Enthalpic Prevalence and Comparative Evaluation for Targeted Antiviral Delivery. Chem. Methodol. 2025, 9, 758–770. [Google Scholar]
- Ban, X.; Xie, X.; Li, C.; Gu, Z.; Hong, Y.; Cheng, L.; Kaustubh, B.; Li, Z. The Desirable Salt Bridges in Amylases: Distribution, Configuration and Location. Food Chem. 2021, 354, 129475. [Google Scholar] [CrossRef]
- Kumar, D.; Meena, M.K.; Kumari, K.; Patel, R.; Jayaraj, A.; Singh, P. In-Silico Prediction of Novel Drug-Target Complex of Nsp3 of CHIKV through Molecular Dynamic Simulation. Heliyon 2020, 6, e04720. [Google Scholar] [CrossRef] [PubMed]
- Morris, H. Modern Triple Resonance Protein NMR Backbone Assignment, Using AlphaFold and Unlabelling to Drive Chemical Shifts Assignment in Proteins. Master’s Thesis, University of Kent, Canterbury, UK, 2025. [Google Scholar]
- Raguette, L.E.; Cuomo, A.E.; Belfon, K.A.A.; Tian, C.; Hazoglou, V.; Witek, G.; Telehany, S.M.; Wu, Q.; Simmerling, C. Phosaa14SB and Phosaa19SB: Updated Amber Force Field Parameters for Phosphorylated Amino Acids. J. Chem. Theory Comput. 2024, 20, 7199–7209. [Google Scholar] [CrossRef] [PubMed]
- Zaman, Z.; Khan, S.; Nouroz, F.; Farooq, U.; Urooj, A. Targeting Protein Tyrosine Phosphatase to Unravel Possible Inhibitors for Streptococcus pneumoniae Using Molecular Docking, Molecular Dynamics Simulations Coupled with Free Energy Calculations. Life Sci. 2021, 264, 118621. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.E.; Valdés-Tresanco, M.S.; Moreno, E.; Valiente, P.A. Assessment of Different Parameters on the Accuracy of Computational Alanine Scanning of Protein–Protein Complexes with the Molecular Mechanics/Generalized Born Surface Area Method. J. Phys. Chem. B 2023, 127, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, A.S.; Altwaim, S.A.; El-Daly, M.M.; Hassan, A.M.; Al-Zahrani, I.A.; Bajrai, L.H.; Alsaady, I.M.; Dwivedi, V.D.; Azhar, E.I. Marine Fungal Diversity Unlocks Potent Antivirals against Monkeypox through Methyltransferase Inhibition Revealed by Molecular Dynamics and Free Energy Landscape. BMC Chem. 2024, 18, 141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dai, X.; Gao, L.; Xu, D.; Wan, H.; Wang, Y.; Yan, L.-T. The Entropy-Controlled Strategy in Self-Assembling Systems. Chem. Soc. Rev. 2023, 52, 6806–6837. [Google Scholar] [CrossRef]
- Mohammad, T.; Mathur, Y.; Hassan, M.I. InstaDock: A Single-Click Graphical User Interface for Molecular Docking-Based Virtual High-Throughput Screening. Brief. Bioinform. 2021, 22, bbaa279. [Google Scholar] [CrossRef]
- Pantaleão, S.Q.; Fernandes, P.O.; Gonçalves, J.E.; Maltarollo, V.G.; Honorio, K.M. Recent Advances in the Prediction of Pharmacokinetics Properties in Drug Design Studies: A Review. ChemMedChem 2022, 17, e202100542. [Google Scholar] [CrossRef]
- Kumar, V.G.; Polasa, A.; Agrawal, S.; Kumar, T.K.S.; Moradi, M. Binding Affinity Estimation from Restrained Umbrella Sampling Simulations. Nat. Comput. Sci. 2023, 3, 59–70. [Google Scholar] [CrossRef]
- Samad, A.; Ajmal, A.; Mahmood, A.; Khurshid, B.; Li, P.; Jan, S.M.; Rehman, A.U.; He, P.; Abdalla, A.N.; Umair, M.; et al. Identification of Novel Inhibitors for SARS-CoV-2 as Therapeutic Options Using Machine Learning-Based Virtual Screening, Molecular Docking and MD Simulation. Front. Mol. Biosci. 2023, 10, 1060076. [Google Scholar] [CrossRef]
- Funari, R.; Bhalla, N.; Gentile, L. Measuring the Radius of Gyration and Intrinsic Flexibility of Viral Proteins in Buffer Solution Using Small-Angle x-Ray Scattering. ACS Meas. Sci. Au 2022, 2, 547–552. [Google Scholar] [CrossRef]
- Anwar, T.; Ismail, S.; Parvaiz, F.; Abbasi, S.W.; A. Al-Abbasi, F.; Alghamdi, A.M.; Al-Regaiey, K.; Ul-Haq, A.; Kaleem, I.; Bashir, S. Computational Design of Experimentally Validated Multi-Epitopes Vaccine against Hepatitis E Virus: An Immunological Approach. PLoS ONE 2023, 18, e0294663. [Google Scholar] [CrossRef]
- da Fonseca, A.M.; Caluaco, B.J.; Madureira, J.M.C.; Cabongo, S.Q.; Gaieta, E.M.; Djata, F.; Colares, R.P.; Neto, M.M.; Fernandes, C.F.C.; Marinho, G.S.; et al. Screening of Potential Inhibitors Targeting the Main Protease Structure of SARS-CoV-2 via Molecular Docking, and Approach with Molecular Dynamics, RMSD, RMSF, H-Bond, SASA and MMGBSA. Mol. Biotechnol. 2024, 66, 1919–1933. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.M.; Marsee, J.D. Protein–Ligand Binding Thermodynamics; American Chemical Society: Washington, DC, USA, 2023; ISBN 084129979X. [Google Scholar]
- Bennett, J.L.; Nguyen, G.T.H.; Donald, W.A. Protein–Small Molecule Interactions in Native Mass Spectrometry. Chem. Rev. 2021, 122, 7327–7385. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and Methods for Circular Dichroism Spectroscopy of Proteins: A Tutorial Review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef]
- Hashem, H.E.; Ahmad, S.; Kumer, A.; Bakri, Y. El In Silico and in Vitro Prediction of New Synthesized N-Heterocyclic Compounds as Anti-SARS-CoV-2. Sci. Rep. 2024, 14, 1152. [Google Scholar] [CrossRef]
- Yin, S.; Zuo, Y.; Abu-Odeh, A.; Zheng, H.; Li, X.-G.; Ding, J.; Ong, S.P.; Asta, M.; Ritchie, R.O. Atomistic Simulations of Dislocation Mobility in Refractory High-Entropy Alloys and the Effect of Chemical Short-Range Order. Nat. Commun. 2021, 12, 4873. [Google Scholar] [CrossRef]
- Baquero, F.; Martinez, J.L.; Lanza, V.F.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clin. Microbiol. Rev. 2021, 34, e00050-19. [Google Scholar] [CrossRef]
- Martinez-Pacheco, S.; O’Driscoll, L. Pre-Clinical in Vitro Models Used in Cancer Research: Results of a Worldwide Survey. Cancers 2021, 13, 6033. [Google Scholar] [CrossRef]
- Riyaphan, J.; Pham, D.-C.; Leong, M.K.; Weng, C.-F. In Silico Approaches to Identify Polyphenol Compounds as α-Glucosidase and α-Amylase Inhibitors against Type-II Diabetes. Biomolecules 2021, 11, 1877. [Google Scholar] [CrossRef]
- Gayathiri, E.; Prakash, P.; Kumaravel, P.; Jayaprakash, J.; Ragunathan, M.G.; Sankar, S.; Pandiaraj, S.; Thirumalaivasan, N.; Thiruvengadam, M.; Govindasamy, R. Computational Approaches for Modeling and Structural Design of Biological Systems: A Comprehensive Review. Prog. Biophys. Mol. Biol. 2023, 185, 17–32. [Google Scholar] [CrossRef]
- Mahara, F.A.; Nuraida, L.; Lioe, H.N.; Nurjanah, S. The Occurrence of Folate Biosynthesis Genes in Lactic Acid Bacteria from Different Sources. Food Technol. Biotechnol. 2023, 61, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.W. The Prevalence of Trimethoprim-Resistance-Conferring Dihydrofolate Reductase Genes in Multidrug Resistant Bacteria from Clinical Isolates in Malaysia. Bachelor’s Thesis, Universiti Tunku Abdul Rahman, Kampar, Malaysia, 2024. [Google Scholar]
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Ahmad, S.; Raza, K.; Gautam, H.K. Computational Screening and MM/GBSA-Based MD Simulation Studies Reveal the High Binding Potential of FDA-Approved Drugs against Cutibacterium Acnes Sialidase. J. Biomol. Struct. Dyn. 2024, 42, 6245–6255. [Google Scholar] [CrossRef] [PubMed]
- Spassov, D.S.; Atanasova, M.; Doytchinova, I. A Role of Salt Bridges in Mediating Drug Potency: A Lesson from the N-Myristoyltransferase Inhibitors. Front. Mol. Biosci. 2023, 9, 1066029. [Google Scholar] [CrossRef]
- Bhati, S.K.; Anjum, F.; Shamsi, A.; Hassan, M.I.; Jain, M.; Muthukumaran, J.; Singh, R.P.; Singh, A.K. In Silico Screening and Molecular Dynamics Analysis of Natural DHPS Enzyme Inhibitors Targeting Acinetobacter Baumannii. Sci. Rep. 2025, 15, 7723. [Google Scholar] [CrossRef]
- Satuluri, S.H.; Katari, S.K.; Pasala, C.; Amineni, U. Novel and Potent Inhibitors for Dihydropteroate Synthase of Helicobacter Pylori. J. Recept. Signal Transduct. 2020, 40, 246–256. [Google Scholar] [CrossRef] [PubMed]






| Rank | Compound ID | Binding Affinity | Chemical Name and Structure |
|---|---|---|---|
| 1. | CHEMBL2322256 | −8.3 kcal/mol | 2-(4,11-dimethyl-2-oxo-6,7,8,9-tetrahydro-2H-benzofuro[3,2-g]chromen-3-yl)-N-(3-hydroxyphenyl)acetamide |
| 2. | CHEMBL2316475 | −7.8 kcal/mol | 8-((2,4-difluorophenyl)amino)-N-(2-methoxyethyl)-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulene-3-carboxamide |
| 3. | CHEMBL2334441 | −7.6 kcal/mol | 8-methoxy-5-methyl-10-(2-phenylhydrazinyl)-1,2,3,5,6,7,8,9-octahydroindeno[1,2-b]indole |
| 4. | CHEMBL2334176 | −7.5 kcal/mol | 5-(1-(2,3-dimethylbenzoyl)piperidin-3-yl)-3-phenyl-4,5-dihydro-1,2,4-oxadiazol-2-ium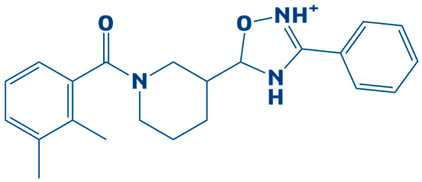 |
| 5. | CHEMBL2334557 | −7.5 kcal/mol | 4-amino-5-((4-(tert-butyl)phenyl)ethynyl)-1-(3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)pyrimidin-1,3-diium-2-olate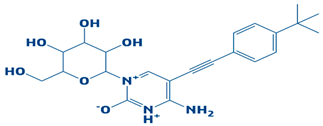 |
| 6. | CHEMBL15935 | −7.4 kcal/mol | 4-((3-(4-methylpiperazin-1-yl)propyl)carbamoyl)benzyl 2-oxo-6-propyl-4-(m-tolyl)-1,2,3,4-tetrahydropyrimidine-5-carboxylate |
| 7. | CHEMBL2260150 | −7.3 kcal/mol | 3-((3-(4,4-diphenylpiperidin-1-yl)propyl)carbamoyl)-5-(methoxycarbonyl)-4-(4-methoxyphenyl)-2,6-dimethylpyridin-1-ium |
| 8. | CHEMBL2322867 | −7.3 kcal/mol | 2-(5-(hydroxymethyl)-4-methyl-3-(trifluoromethyl)-2,3-dihydro-1H-pyrazol-1-yl)-1-(4-(3-methoxy-4-methylphenyl)piperazin-1-yl)ethanone |
| 9. | CHEMBL2334162 | −7.3 kcal/mol | 5-(1-(1-methyl-1H-indole-4-carbonyl)piperidin-3-yl)-3-phenyl-4,5-dihydro-1,2,4-oxadiazol-2-ium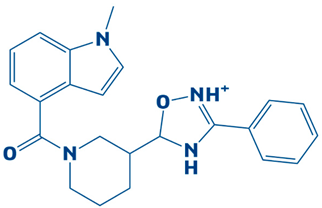 |
| 10. | CHEMBL2229522 | −7.2 kcal/mol | (E)-1-(furan-3(2H)-ylidene)-4,7a-dimethyl-3-oxo-1,3,5,6,7,7a-hexahydroisobenzofuran-5-yl 4-cyanobenzoate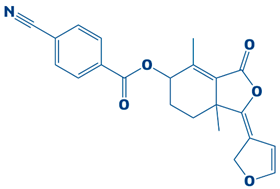 |
| 11. | Imidazole (Control) | 2,3-dihydro-1H-imidazole |
| Control | ||||||
|---|---|---|---|---|---|---|
| #Acceptor | DonorH | Donor | Frames | Frac | AvgDist | AvgAng |
| ASP_172@OD2 | LIG_266@HN | LIG_266@N1 | 854 | 0.854 | 2.8301 | 162.8601 |
| ASP_83@OD2 | LIG_266@H | LIG_266@N | 274 | 0.274 | 2.8473 | 160.1394 |
| ASP_83@OD1 | LIG_266@H | LIG_266@N | 114 | 0.114 | 2.8507 | 158.6697 |
| LIG_266@N1 | ASN_102@HD22 | ASN_102@ND2 | 52 | 0.052 | 2.9458 | 155.5515 |
| ASN_102@O | LIG_266@H | LIG_266@N | 39 | 0.039 | 2.9068 | 153.4423 |
| LIG_266@N1 | ARG_243@HH11 | ARG_243@NH1 | 1 | 0.001 | 2.915 | 135.4934 |
| CHEMBL2322256 | ||||||
| #Acceptor | DonorH | Donor | Frames | Frac | AvgDist | AvgAng |
| ASP_83@OD2 | LIG_266@H1 | LIG_266@O4 | 89 | 0.089 | 2.805 | 156.8071 |
| LIG_266@O3 | HIE_245@HE2 | HIE_245@NE2 | 21 | 0.021 | 2.906 | 146.6492 |
| GLY_204@O | LIG_266@H1 | LIG_266@O4 | 15 | 0.015 | 2.8061 | 159.5714 |
| ARG_243@NH1 | LIG_266@H1 | LIG_266@O4 | 12 | 0.012 | 2.9592 | 153.7587 |
| LIG_266@O3 | ASN_23@HD21 | ASN_23@ND2 | 7 | 0.007 | 2.9273 | 153.1301 |
| ASP_172@OD1 | LIG_266@H1 | LIG_266@O4 | 1 | 0.001 | 2.8756 | 162.7102 |
| ARG_243@NH2 | LIG_266@H1 | LIG_266@O4 | 1 | 0.001 | 2.9119 | 136.3553 |
| CHEMBL2316475 | ||||||
| #Acceptor | DonorH | Donor | Frames | Frac | AvgDist | AvgAng |
| GLU_52@O | LIG_266@H | LIG_266@N | 640 | 0.64 | 2.8056 | 159.422 |
| GLY_176@O | LIG_266@H1 | LIG_266@N1 | 46 | 0.046 | 2.8677 | 148.7695 |
| GLU_52@OE1 | LIG_266@H | LIG_266@N | 29 | 0.029 | 2.8047 | 153.4161 |
| LIG_266@O2 | GLY_178@H | GLY_178@N | 5 | 0.005 | 2.8986 | 156.9123 |
| LIG_266@O1 | LYS_208@HZ2 | LYS_208@NZ | 1 | 0.001 | 2.793 | 150.6578 |
| LIG_266@O1 | LYS_208@HZ3 | LYS_208@NZ | 1 | 0.001 | 2.9694 | 138.0812 |
| LIG_266@F1 | HIE_134@HE2 | HIE_134@NE2 | 1 | 0.001 | 2.9953 | 137.6677 |
| CHEMBL2334441 | ||||||
| #Acceptor | DonorH | Donor | Frames | Frac | AvgDist | AvgAng |
| GLU_52@OE1 | LIG_266@HN | LIG_266@N1 | 156 | 0.1563 | 2.8373 | 156.3745 |
| GLU_52@OE2 | LIG_266@HN | LIG_266@N1 | 155 | 0.1553 | 2.8607 | 154.4868 |
| GLU_52@O | LIG_266@HN | LIG_266@N1 | 8 | 0.008 | 2.8815 | 148.5877 |
| GLY_133@O | LIG_266@H | LIG_266@N2 | 5 | 0.005 | 2.8763 | 151.1339 |
| S.No | DHPS–Ligand Complex | Electrostatic Interactions |
|---|---|---|
| 1. | CHEMBL2322256 | Asp252-Lys255, Asp218-Arg223, Asp252-Lys255, Glu60-Arg63, Glu261-Arg240, Glu60-Arg63, Glu160-Lys166, Asp218-Arg209, Asp218-Arg209, Asp218-Arg209, Glu222-Arg217, Glu60-Arg63, Asp143-Arg190, Glu222-Arg217, Asp143-Arg190, Glu2130arg223, Glu60-Arg63, Glu60-Arg64, Asp246-Arg207, Glu222-Arg217, Glu192-Arg237, Glu196-Arg193,Asp14-Arg12, Glu60-Arg64, Glu213-Arg209, Glu213-Arg209, Glu213-Arg209, Asp5-Lys167, Asp218-Arg223, Glu42-Lys38. |
| 2. | CHEMBL2316475 | Asp252-Lys255, Asp45-Arg17, Asp83-Arg243, Glu192-Arg193, Glu61-Lys86, Glu60-Arg63, Glu59-Arg63, Glu192-Arg193, Glu61-Lys86, Asp48-Arg243, Glu88-Arg91, Asp143-Arg190, Asp218-Arg209, Asp143-Arg190, Glu88-Arg91, Glu52-Lys86, Glu60-Arg64, Glu59-Arg63, Asp246-Arg207, Glu60-Arg64, Asp45-Arg15, Glu192-Arg190, Glu196-Arg193, Glu192-Arg190, Glu196-Arg193, Glu192-Arg237, Glu192-Arg237, Glu192-Arg190,Glu192-Arg190, Asp14-Arg17, Glu213-Arg209, Asp14-Arg17, Asp45-Arg15, Asp14-Arg12, Glu42-Lys38. |
| 3. | CHEMBL2334441 | Asp252-Lys255, Asp14-Arg17, Asp252-Lys255, Glu60-Arg63, Glu42-Lys38, Asp83-Arg243, Glu160-Lys166, Asp218-Arg209, Asp14-Arg15, Glu88-Arg91, Glu160-Lys166, Asp143-Arg190, Asp246-Arg207, Asp48-Arg243, Glu41-Lys38, Asp14-Arg15, Asp246-Arg207, Asp246-Arg207, Asp138-Arg217, Asp138-Arg209, Glu192-Arg237, Glu213-Arg209, Glu213-Arg209, Asp14-Arg17, Glu213-Arg219, Glu213-Arg209, Asp5-Lys167, Asp218-Arg223, Asp14-Arg17. |
| 4. | Control | Asp252-Lys255, Asp45-Arg17, Glu261-Arg240, Glu131-Arg108, Glu61-Lys86, Glu60-Arg63, Glu41-Lys38, Asp218-Arg209, Asp218-Arg209, Asp218-Arg209, Glu222-Arg217, Asp143-Arg190, Glu222-Arg217, Asp218-Arg209, Asp48-Arg243, Glu192-Arg237,3 Glu60-Arg64, Glu88-Arg91, Asp48-Arg243, Glu88-Arg91, Glu196-Arg193, Asp246-Arg207, Asp45-Arg15, Asp14-Arg12, Glu60-Arg64, Glu60-Arg64, Glu213-Arg209, Asp14-Arg12, Asp218-Arg223, Glu30-Arg75. |
| Energy Parameter | CHEMBL2322256 | CHEMBL2316475 | CHEMBL2334441 | Control |
|---|---|---|---|---|
| MM/GBSA | ||||
| Energy van der Waals | −114.20 | −98.62 | −111.08 | −84.08 |
| Energy electrostatic | −26.38 | −22.01 | −25.67 | −16.34 |
| Total gas phase energy | −140.58 | −120.63 | −85.41 | −100.42 |
| Total salvation energy | 14.09 | 15.00 | 19.84 | 15.38 |
| Net energy | −126.49 | −105.63 | −65.57 | −85.04 |
| MMPB/SA | ||||
| Energy van der Waals | −114.20 | −98.62 | −111.08 | −84.08 |
| Energy electrostatics | −26.38 | −22.01 | −25.67 | −16.34 |
| Total gas phase energy | −140.58 | −120.63 | −85.41 | −100.42 |
| Total energy salvation | 16.09 | 16.88 | 20.13 | 16.82 |
| Net energy | −124.49 | −103.75 | −65.28 | −83.6 |
| Complex | Translational | Vibrational | Rotational | DELTA S Total |
|---|---|---|---|---|
| CHEMBL2322256 | 16.35 | 1023.01 | 18.64 | 8.63 |
| CHEMBL2316475 | 15.14 | 984.05 | 19.37 | 15.03 |
| CHEMBL2334441 | 18.06 | 1102.36 | 21.01 | 18.01 |
| Control | 20.59 | 975.05 | 19.80 | 16.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alabbas, A. Integrating Multi-Domain Approach for Identification of Neo Anti-DHPS Inhibitors Against Pathogenic Stenotrophomonas maltophilia. Biology 2025, 14, 1030. https://doi.org/10.3390/biology14081030
Alabbas A. Integrating Multi-Domain Approach for Identification of Neo Anti-DHPS Inhibitors Against Pathogenic Stenotrophomonas maltophilia. Biology. 2025; 14(8):1030. https://doi.org/10.3390/biology14081030
Chicago/Turabian StyleAlabbas, Alhumaidi. 2025. "Integrating Multi-Domain Approach for Identification of Neo Anti-DHPS Inhibitors Against Pathogenic Stenotrophomonas maltophilia" Biology 14, no. 8: 1030. https://doi.org/10.3390/biology14081030
APA StyleAlabbas, A. (2025). Integrating Multi-Domain Approach for Identification of Neo Anti-DHPS Inhibitors Against Pathogenic Stenotrophomonas maltophilia. Biology, 14(8), 1030. https://doi.org/10.3390/biology14081030






