Role of ACE1, ACE2, and CCR5-Δ32 Polymorphisms in the Transmission of SARS-CoV-2 to Intimate Contacts
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
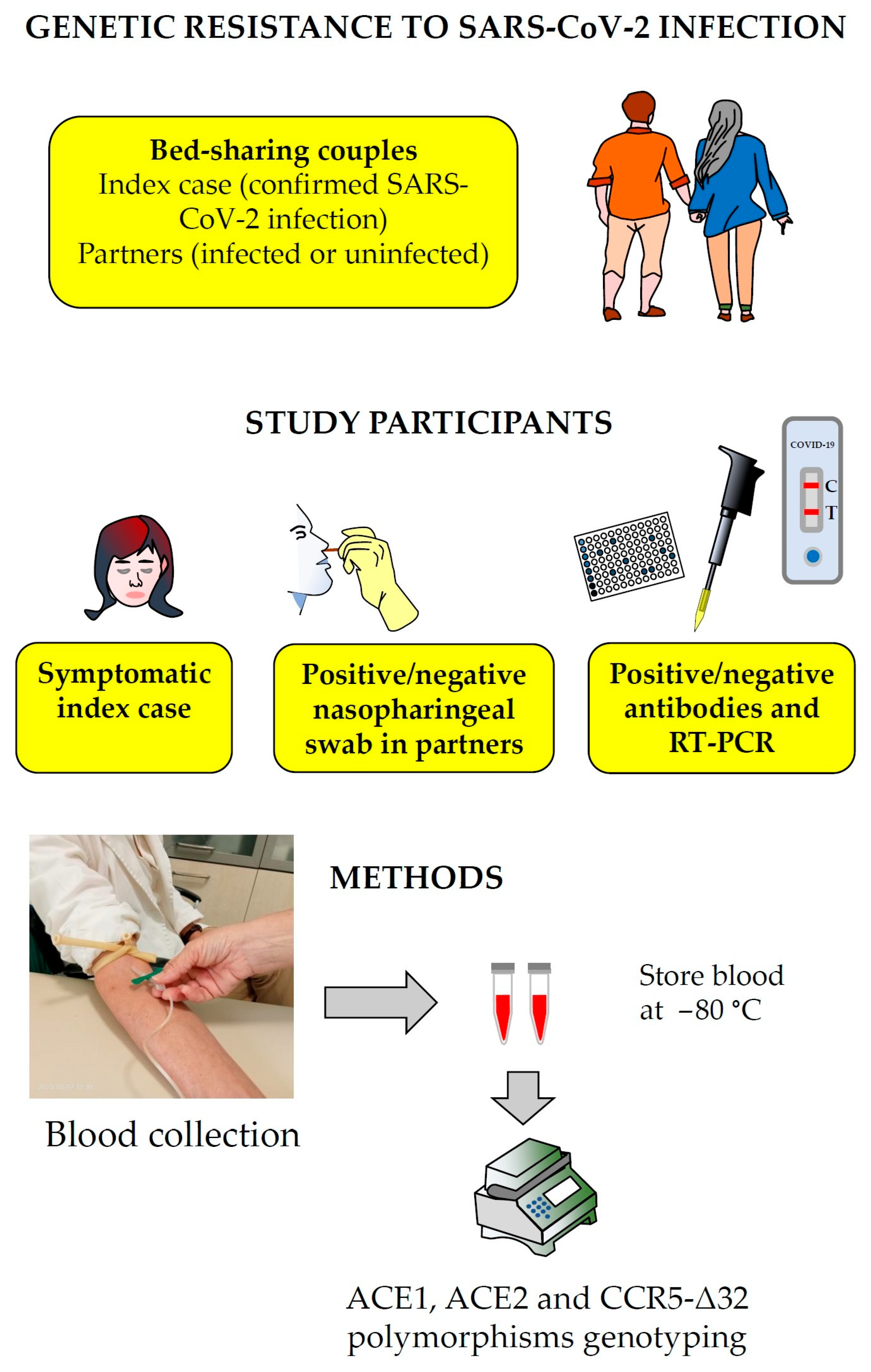
2.1. Setting
2.2. Study Population
2.3. SARS-CoV-2 Status Confirmation
2.4. Polymorphism Genotyping
2.5. Statistical Analysis
2.6. Ethical Considerations
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| COVID-19 | Coronavirus disease 2019 |
| ACE2 | Angiotensin-converting enzyme-2 |
| CCR5 | C-C Chemokine Receptor-5 |
| NPs | Nasopharyngeal swabs |
| RT-PCR | Reverse transcriptase polymerase chain reaction |
| EDTA | Ethylenediamine tetraacetic acid |
| HLA | Human leukocyte antigen |
| GWAS | Genome-wide association study |
References
- WHO. WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus (COVID-19) Dashboard with Vaccination Data n.d. Available online: https://covid19.who.int/ (accessed on 15 May 2023).
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Ware, L.B. Physiological and biological heterogeneity in COVID-19-associated acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 1163–1165. [Google Scholar] [CrossRef] [PubMed]
- Sonnenberg, P.; Menezes, D.; Freeman, L.; Maxwell, K.J.; Reid, D.; Clifton, S.; Tanton, C.; Copas, A.; Riddell, J.; Dema, E.; et al. Intimate physical contact between people from different households during the COVID-19 pandemic: A mixed-methods study from a large, quasi-representative survey (Natsal-COVID). BMJ Open 2022, 12, e055284. [Google Scholar] [CrossRef] [PubMed]
- Adam, D.C.; Wu, P.; Wong, J.Y.; Lau, E.H.Y.; Tsang, T.K.; Cauchemez, S.; Leung, G.M.; Cowling, B.J. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020, 26, 1714–1719. [Google Scholar] [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Stralin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183, 158–168.e14. [Google Scholar] [CrossRef]
- Feng, Z.; Zou, K.; Savani, K. Cultural antecedents of virus transmission: Individualism is associated with lower compliance with social distancing rules during the COVID-19 pandemic. J. Pers. Soc. Psychol. 2023, 124, 461–482. [Google Scholar] [CrossRef]
- El-Sokkary, R.H.; Khater, W.S.; El-Kholy, A.; Mohy Eldin, S.; Gad, D.M.; Bahgat, S.; Negm, E.E.M.; El Kholy, J.A.; Mowafy, S.; Mahmoud, E.; et al. Compliance of healthcare workers to the proper use of personal protective equipment during the first wave of COVID-19 pandemic. J. Infect. Public. Health 2021, 14, 1404–1410. [Google Scholar] [CrossRef]
- Goldberg, L.; Levinsky, Y.; Marcus, N.; Hoffer, V.; Gafner, M.; Hadas, S.; Kraus, S.; Mor, M.; Scheuerman, O. SARS-CoV-2 Infection Among Health Care Workers Despite the Use of Surgical Masks and Physical Distancing-the Role of Airborne Transmission. Open Forum Infect. Dis. 2021, 8, ofab036. [Google Scholar] [CrossRef]
- Malune, P.; Piras, G.; Monne, M.; Fiamma, M.; Asproni, R.; Fancello, T.; Manai, A.; Carta, F.; Pira, G.; Fancello, P.; et al. Molecular Characterization of Severe Acute Respiratory Syndrome Coronavirus 2 Isolates From Central Inner Sardinia. Front. Microbiol. 2021, 12, 827799. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nishida, N.; Yamamoto, R.; Gojobori, T.; Shimotohno, K.; Mizokami, M.; Ariumi, Y. Angiotensin-Converting Enzyme (ACE) 1 Gene Polymorphism and Phenotypic Expression of COVID-19 Symptoms. Genes 2021, 12, 1572. [Google Scholar] [CrossRef]
- Delanghe, J.R.; Speeckaert, M.M.; De Buyzere, M.L. ACE Ins/Del genetic polymorphism and epidemiological findings in COVID-19. Clin. Chem. Lab. Med. 2020, 58, 1129–1130. [Google Scholar] [CrossRef] [PubMed]
- Gemmati, D.; Bramanti, B.; Serino, M.L.; Secchiero, P.; Zauli, G.; Tisato, V. COVID-19 and Individual Genetic Susceptibility/Receptivity: Role of ACE1/ACE2 Genes, Immunity, Inflammation and Coagulation. Might the Double X-chromosome in Females Be Protective against SARS-CoV-2 Compared to the Single X-Chromosome in Males? Int. J. Mol. Sci. 2020, 21, 3474. [Google Scholar] [CrossRef] [PubMed]
- Starčević Čizmarević, N.; Tota, M.; Ristic, S. Does the CCR5-Delta32 mutation explain the variable coronavirus-2019 pandemic statistics in Europe? Croat. Med. J. 2020, 61, 525–526. [Google Scholar] [CrossRef]
- Hartley, O.; Martins, E.; Scurci, I. Preventing HIV transmission through blockade of CCR5: Rationale, progress and perspectives. Swiss Med. Wkly. 2018, 148, w14580. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Real-Time RT-PCR Panel for Detection 2019-Novel Coronavirus. Published 2020. Available online: https://www.cdc.gov/covid/testing/index.html (accessed on 1 March 2020).
- Skvarc, M. Clinical validation of two immunochromatographic SARS-CoV-2 antigen tests in near hospital facilities. J. Infect. Dev. Ctries. 2022, 16, 418–421. [Google Scholar] [CrossRef]
- Van Elslande, J.; Houben, E.; Depypere, M.; Brackenier, A.; Desmet, S.; Andre, E.; Van Ranst, M.; Lagrou, K.; Vermeersch, P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1082–1087. [Google Scholar] [CrossRef]
- Cavalli-Sforza, L.L. Genes, Peoples and Languages; University of California Press: Berkeley, CA, USA, 2001. [Google Scholar]
- Errigo, A.; Dore, M.P.; Mocci, G.; Pes, G.M. Lack of association between common polymorphisms associated with successful aging and longevity in the population of Sardinian Blue Zone. Sci. Rep. 2024, 14, 30773. [Google Scholar] [CrossRef]
- Corbo, R.M.; Scacchi, R.; Mureddu, L.; Mulas, G.; Castrechini, S.; Rivasi, A.P. Apolipoprotein B, apolipoprotein E, and angiotensin-converting enzyme polymorphisms in 2 Italian populations at different risk for coronary artery disease and comparison of allele frequencies among European populations. Hum. Biol. 1999, 71, 933–945. [Google Scholar]
- Benetti, E.; Tita, R.; Spiga, O.; Ciolfi, A.; Birolo, G.; Bruselles, A.; Doddato, G.; Giliberti, A.; Marconi, C.; Musacchia, F.; et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020, 28, 1602–1614. [Google Scholar] [CrossRef]
- Libert, F.; Cochaux, P.; Beckman, G.; Samson, M.; Aksenova, M.; Cao, A.; Czeizel, A.; Claustres, M.; de la Rua, C.; Ferrari, M.; et al. The deltaccr5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum. Mol. Genet. 1998, 7, 399–406. [Google Scholar] [CrossRef]
- Tao, S.; You, X.; Norman, P.J.; Kichula, K.M.; Dong, L.; Chen, N.; He, J.; Zhang, W.; Zhu, F. Analysis of KIR and HLA Polymorphism in Chinese Individuals With COVID-19. HLA 2024, 104, e15715. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhu, Y.; Liu, N.; Hu, Y.; Chong, H.; He, Y. Resistance profile and mechanism of severe acute respiratory syndrome coronavirus-2 variants to LCB1 inhibitor targeting the spike receptor-binding motif. Front. Microbiol. 2022, 13, 1022006. [Google Scholar] [CrossRef]
- Bernas, S.N.; Baldauf, H.; Wendler, S.; Heidenreich, F.; Lange, V.; Hofmann, J.A.; Sauter, J.; Schmidt, A.H.; Schetelig, J. CCR5Delta32 mutations do not determine COVID-19 disease course. Int. J. Infect. Dis. 2021, 105, 653–655. [Google Scholar] [CrossRef]
- Swadling, L.; Diniz, M.O.; Schmidt, N.M.; Amin, O.E.; Chandran, A.; Shaw, E.; Pade, C.; Gibbons, J.M.; Le Bert, N.; Tan, A.T.; et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature 2022, 601, 110–117. [Google Scholar] [CrossRef]
- Augusto, D.G.; Murdolo, L.D.; Chatzileontiadou, D.S.M.; Sabatino, J.J., Jr.; Yusufali, T.; Peyser, N.D.; Butcher, X.; Kizer, K.; Guthrie, K.; Murray, V.W.; et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 2023, 620, 128–136. [Google Scholar] [CrossRef]
- Horowitz, J.E.; Kosmicki, J.A.; Damask, A.; Sharma, D.; Roberts, G.H.L.; Justice, A.E.; Banerjee, N.; Coignet, M.V.; Yadav, A.; Leader, J.B.; et al. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat. Genet. 2022, 54, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef] [PubMed]
- Andreakos, E.; Abel, L.; Vinh, D.C.; Kaja, E.; Drolet, B.A.; Zhang, Q.; O’Farrelly, C.; Novelli, G.; Rodriguez-Gallego, C.; Haerynck, F.; et al. A global effort to dissect the human genetic basis of resistance to SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 159–164. [Google Scholar] [CrossRef]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef]
| Symptomatic Spouses Infected with SARS-CoV-2 | |||
|---|---|---|---|
| Features | Resistant Partners (n = 33) | Infected Partners (n = 30) | p |
| Sex (F/M) | 19 F/14 M | 15 F/15 M | 0.547 |
| Mean age | 48 ± 11 years | 48 ± 11 years | |
| Exposure duration (days) | 3–15 (mean 8 ± 4) | 3–12 (mean 7 ± 3) | |
| Comorbidities | 5/33 (15%) * | 6/30 (20%) * | |
| IgG/IgM seroconversion | 0/33 (0%) | 30/30 (100%) | |
| Polymorphism | Genotype | Genotype | p-Value | Allele | ||
|---|---|---|---|---|---|---|
| Resistant (n = 33) | Infected (n = 30) | Resistant (n = 33) | Infected (n = 30) | |||
| ACE1 | II | 2 (6%) | 1 (3%) | I allele: 26% D allele: 74% | I allele: 27% D allele: 73% | |
| ID | 13 (39%) | 14 (47%) | p = 0.78 | |||
| DD | 18 (55%) | 15 (50%) | ||||
| ACE2 | GG | 7 (21%) | 11 (37%) | G allele: 46% A allele: 54% | G allele: 53% A allele: 47% | |
| GA | 16 (49%) | 7 (23%) | p = 0.11 | |||
| AA | 10 (30%) | 12 (40%) | ||||
| CCR5-Δ32 * | WT/WT | 33 (100%) | 30 (100%) | Δ32 allele: 0% | Δ32 allele: 0% | |
| WT/Δ32 | 0 | 0 | ||||
| Δ32/Δ32 | 0 | 0 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dore, M.P.; Errigo, A.; Merola, E.; Pes, G.M. Role of ACE1, ACE2, and CCR5-Δ32 Polymorphisms in the Transmission of SARS-CoV-2 to Intimate Contacts. Biology 2025, 14, 587. https://doi.org/10.3390/biology14060587
Dore MP, Errigo A, Merola E, Pes GM. Role of ACE1, ACE2, and CCR5-Δ32 Polymorphisms in the Transmission of SARS-CoV-2 to Intimate Contacts. Biology. 2025; 14(6):587. https://doi.org/10.3390/biology14060587
Chicago/Turabian StyleDore, Maria Pina, Alessandra Errigo, Elettra Merola, and Giovanni Mario Pes. 2025. "Role of ACE1, ACE2, and CCR5-Δ32 Polymorphisms in the Transmission of SARS-CoV-2 to Intimate Contacts" Biology 14, no. 6: 587. https://doi.org/10.3390/biology14060587
APA StyleDore, M. P., Errigo, A., Merola, E., & Pes, G. M. (2025). Role of ACE1, ACE2, and CCR5-Δ32 Polymorphisms in the Transmission of SARS-CoV-2 to Intimate Contacts. Biology, 14(6), 587. https://doi.org/10.3390/biology14060587








