Insights into the Regulatory Role of MicroRNAs in Penaeus monodon Under Moderately Low Salinity Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals Used for the Experiments
2.2. Treatment of Low Salinity Stress
2.3. Total RNA Extraction and Quality Analysis
2.4. Small RNA Library Construction and Sequencing
2.5. Bioinformatics Analysis of miRNA Transcriptome
2.6. Expressed Analysis and Target Prediction of miRNA
2.7. Enrichment Analysis of Target Genes
2.8. Validation of DEMs and Statistical Analysis
3. Results
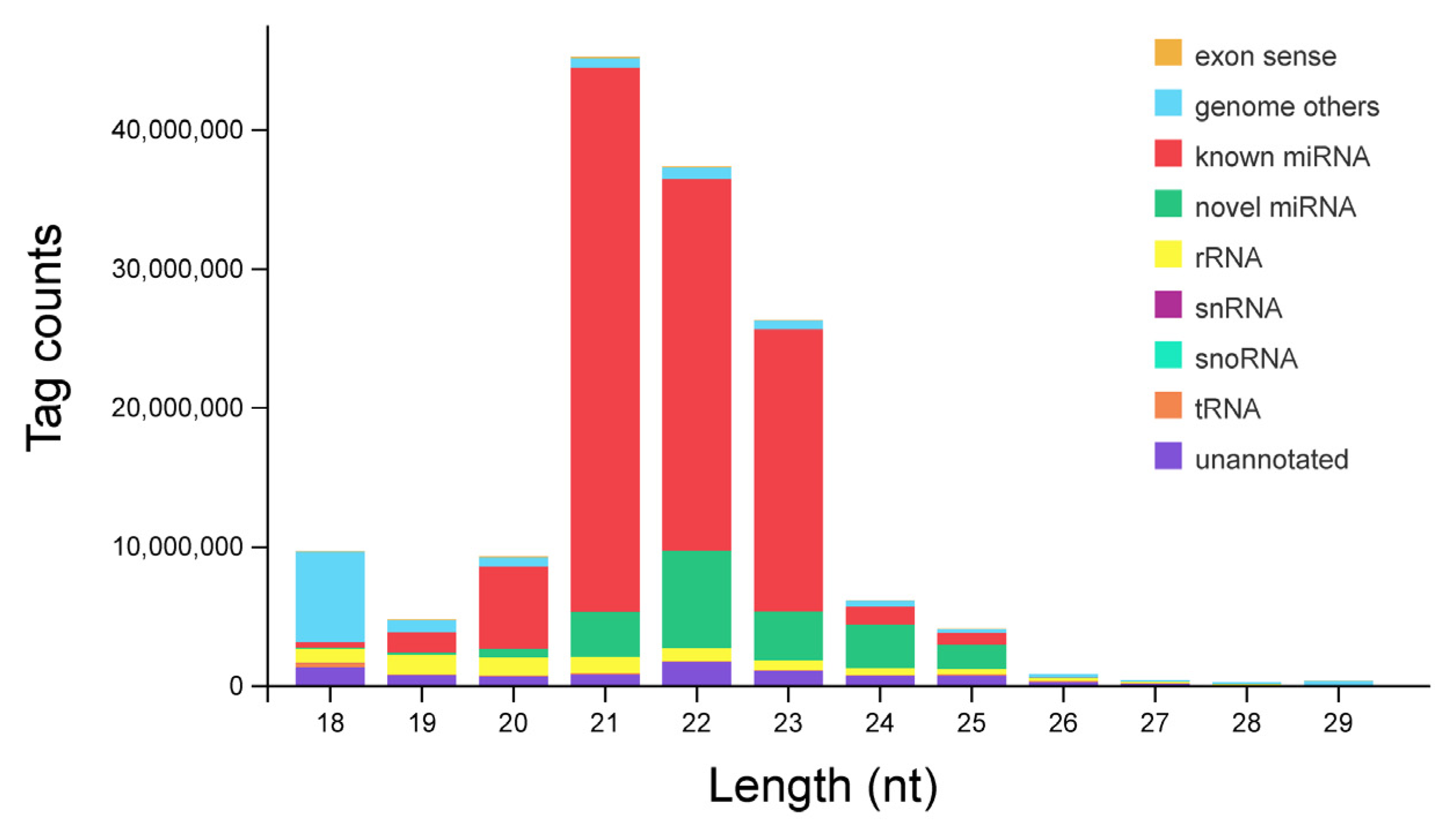
3.1. Identification and Classification of miRNAs
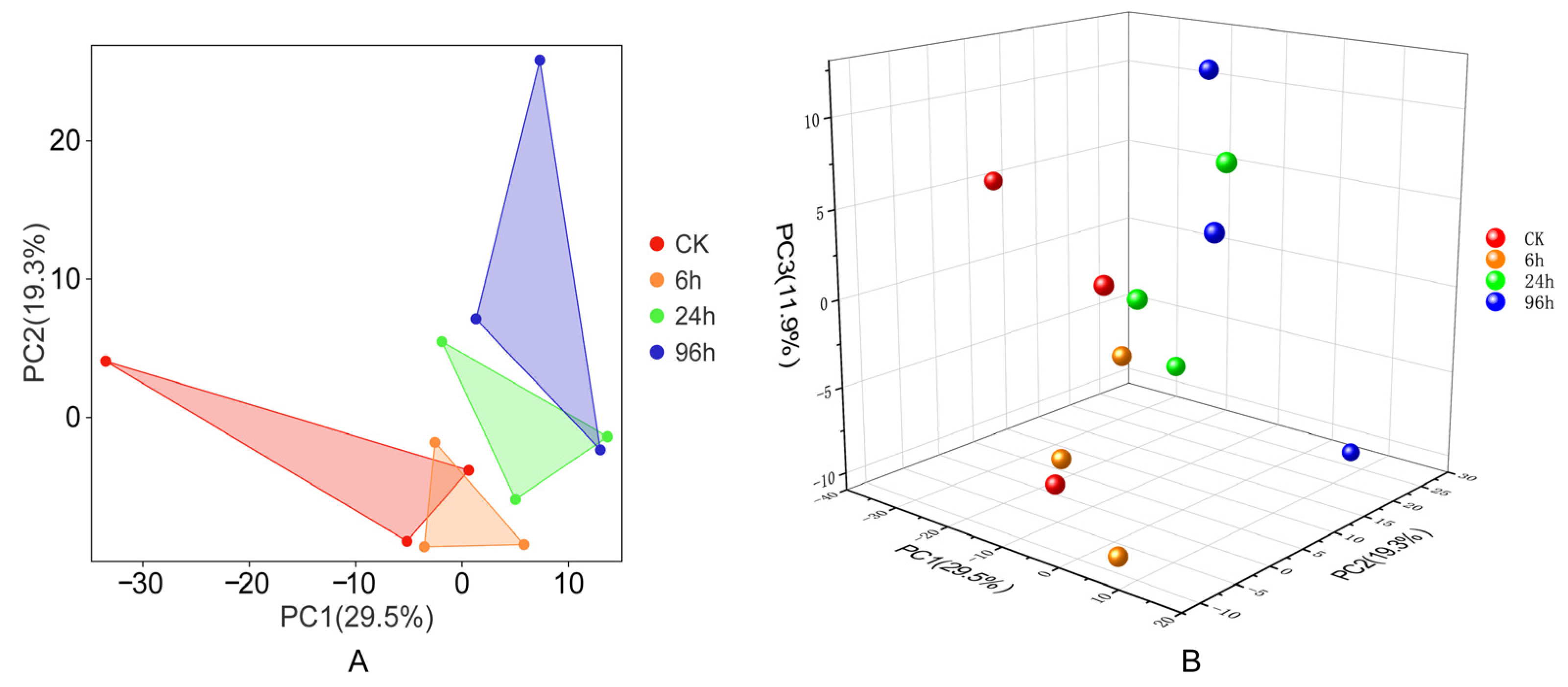
3.2. Principal Component Analysis
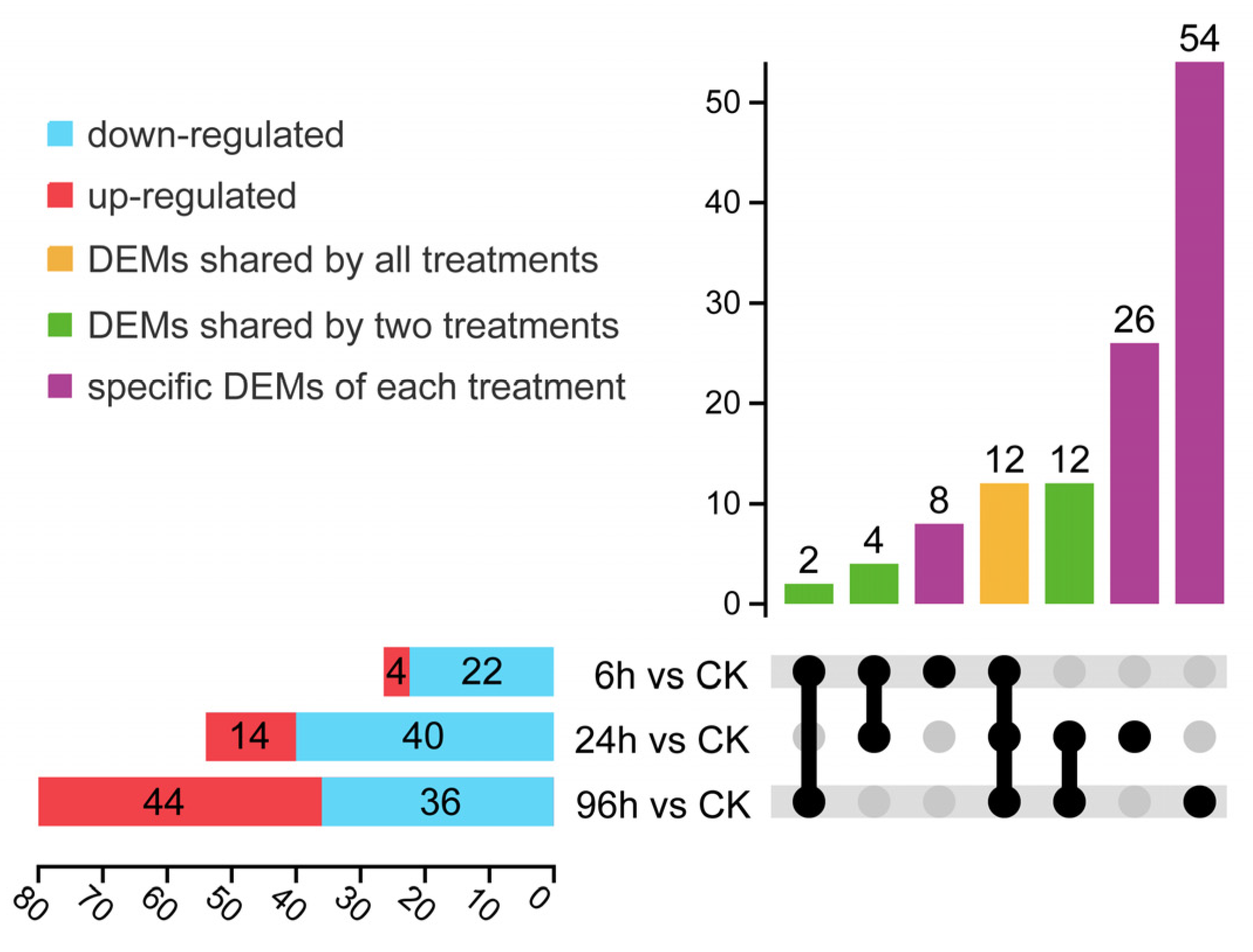
3.3. Expression Profiles of Differentially Expressed miRNA
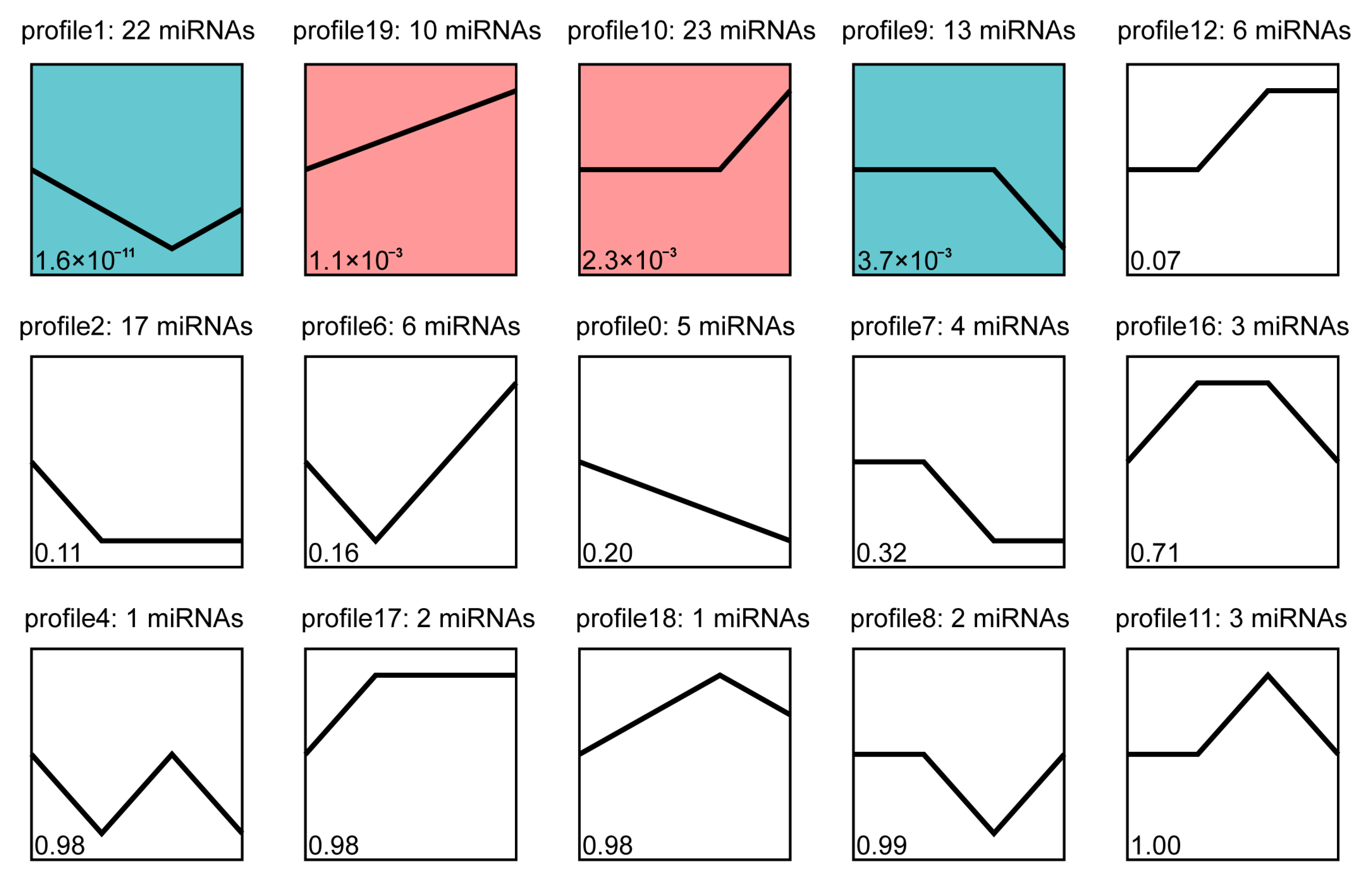
3.4. Trend Analysis of Differentially Expressed miRNA
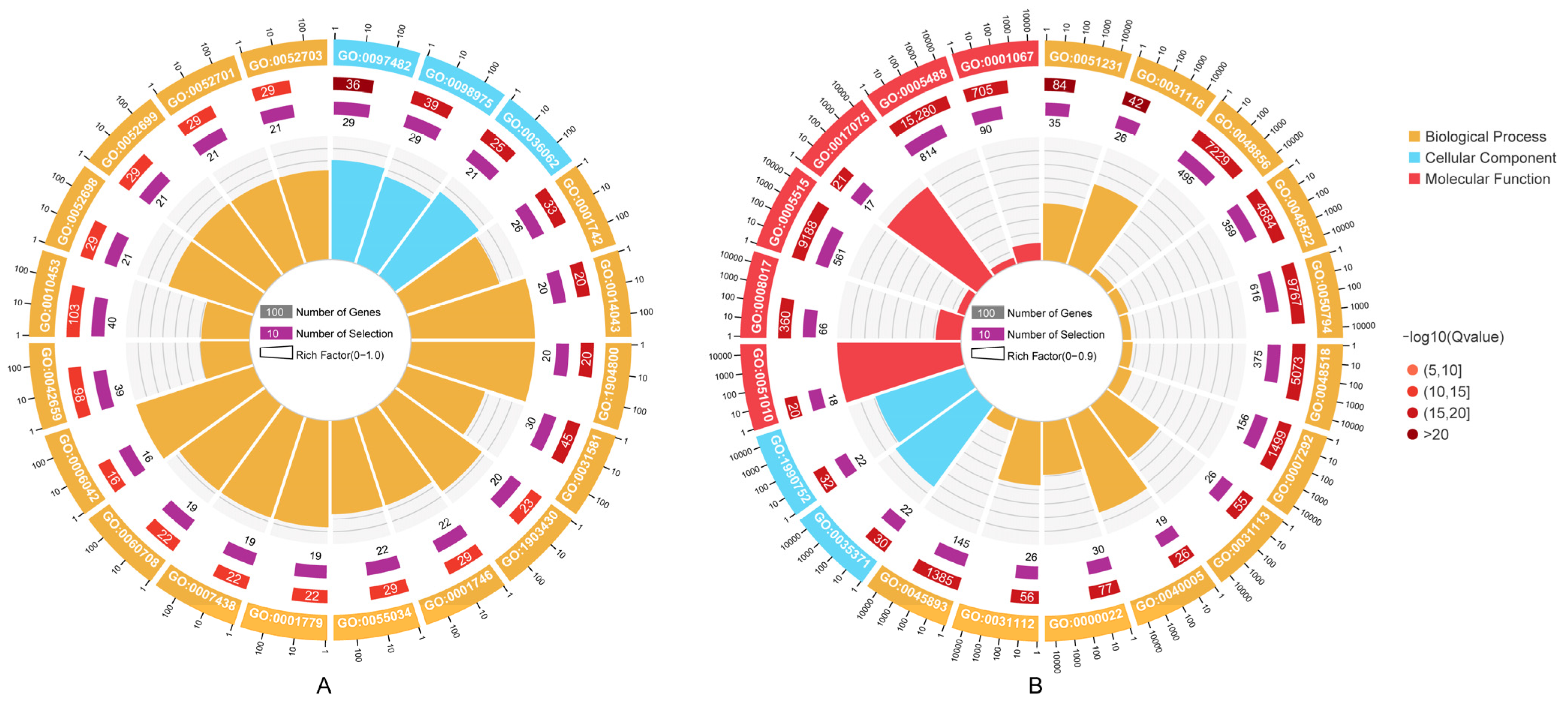
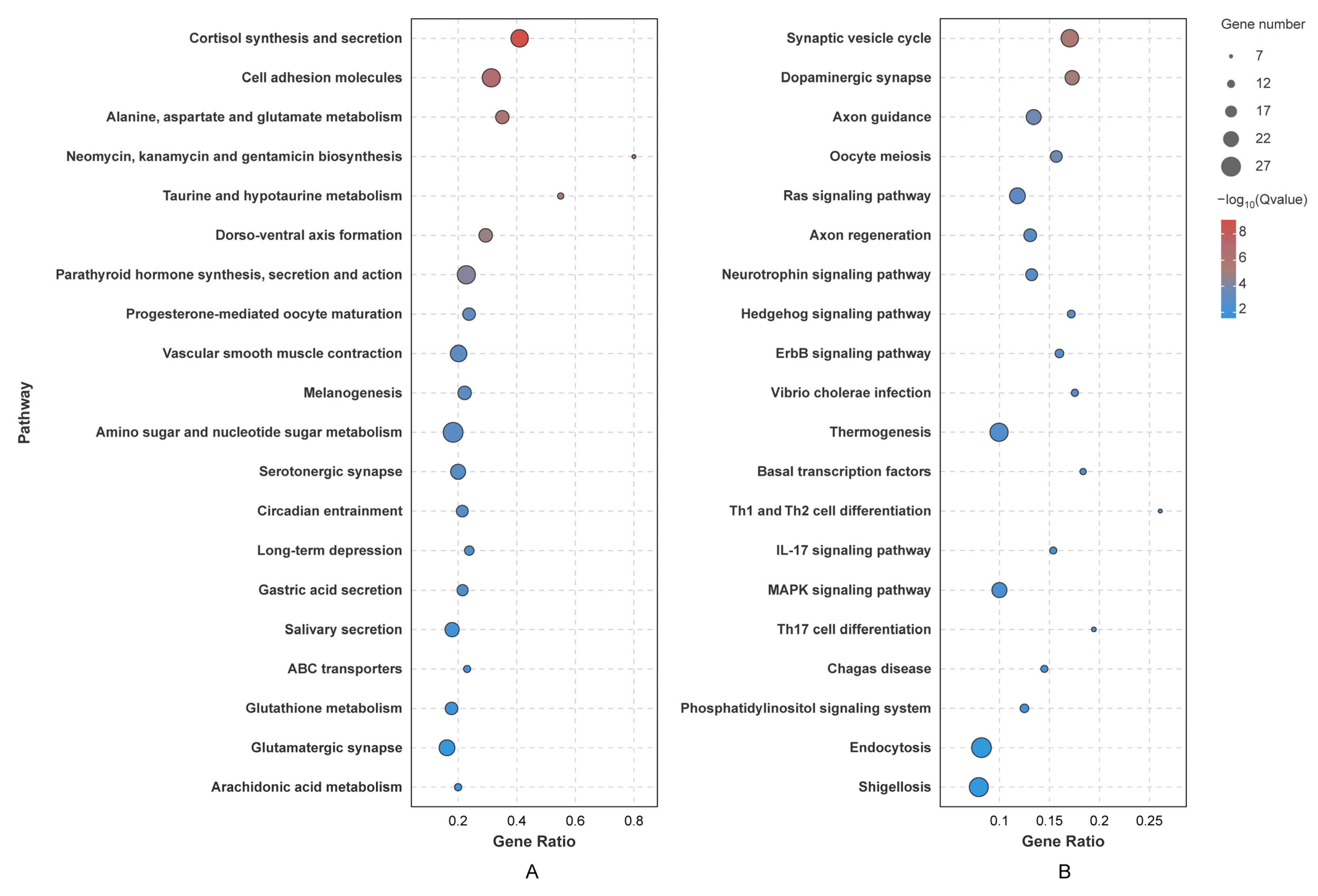
3.5. Prediction of miRNA Targets and Functional Enrichment Analysis
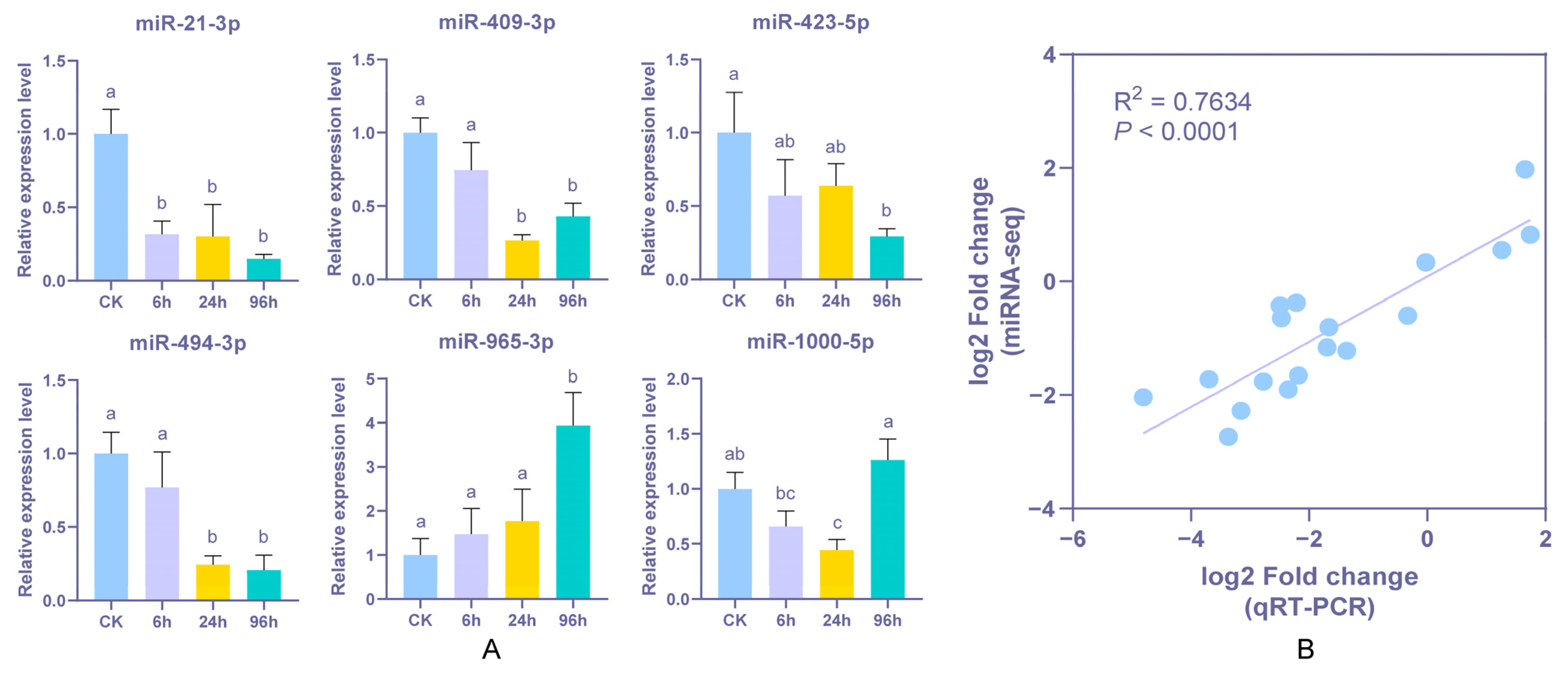
3.6. Stem-Loop qPCR Validation of Differentially Expressed miRNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Angthong, P.; Uengwetwanit, T.; Arayamethakorn, S.; Rungrassamee, W. Transcriptomic analysis of the black tiger shrimp (Penaeus monodon) reveals insights into immune development in their early life stages. Sci. Rep. 2021, 11, 13881. [Google Scholar] [CrossRef] [PubMed]
- Fisheries Administration Bureau, Ministry of Agriculture. China Fishery Statistics Year Book; China Agriculture Press: Beijing, China, 2024. [Google Scholar]
- Ahmed, N.; Diana, J.S. Threatening “white gold”: Impacts of climate change on shrimp farming in coastal Bangladesh. Ocean Coast. Manag. 2015, 114, 42–52. [Google Scholar] [CrossRef]
- Chang, Y.T.; Huang, W.T.; Wu, P.L.; Kumar, R.; Wang, H.C.; Lu, H.P. Low salinity stress increases the risk of Vibrio parahaemolyticus infection and gut microbiota dysbiosis in Pacific white shrimp. BMC Microbiol. 2024, 24, 275. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, R.P.; Parado-Estepa, E.D.; De Jesus, E.G.; Ladja, J.M. Osmotic and chloride regulation in the hemolymph of the tiger prawn Penaeus monodon during molting in various salinities. Mar. Biol. 1987, 95, 377–385. [Google Scholar] [CrossRef]
- Rahi, M.L.; Azad, K.N.; Tabassum, M.; Irin, H.H.; Hossain, K.S.; Aziz, D.; Moshtaghi, A.; Hurwood, D.A. Effects of Salinity on Physiological, Biochemical and Gene Expression Parameters of Black Tiger Shrimp (Penaeus monodon): Potential for Farming in Low-Salinity Environments. Biology 2021, 10, 1220. [Google Scholar] [CrossRef]
- Roy, L.A.; Davis, D.A.; Saoud, I.P.; Henry, R.P. Effects of varying levels of aqueous potassium and magnesium on survival, growth, and respiration of the Pacific white shrimp, Litopenaeus vannamei, reared in low salinity waters. Aquaculture 2007, 262, 461–469. [Google Scholar] [CrossRef]
- Guo, H.; Xian, J.A.; Zheng, P.H.; Lu, Y.P.; Wang, L.; Zhang, X.X.; Li, J.T.; Zhang, Z.L.; Wang, A.L. Dietary copper affects antioxidant status of shrimp (Penaeus monodon) reared in low salinity water. Aquac. Rep. 2022, 22, 100979. [Google Scholar] [CrossRef]
- Pan, L.; Liu, H.; Zhao, Q. Effect of salinity on the biosynthesis of amines in Litopenaeus vannamei and the expression of gill related ion transporter genes. J. Ocean Univ. China 2014, 13, 453–459. [Google Scholar] [CrossRef]
- Liu, M.; Liu, S.; Hu, Y.; Pan, L. Cloning and expression analysis of two carbonic anhydrase genes in white shrimp Litopenaeus vannamei, induced by pH and salinity stresses. Aquaculture 2015, 448, 391–400. [Google Scholar] [CrossRef]
- Gao, W.; Tian, L.; Huang, T.; Yao, M.; Hu, W.; Xu, Q. Effect of salinity on the growth performance, osmolarity and metabolism-related gene expression in white shrimp Litopenaeus vannamei. Aquac. Rep. 2016, 4, 125–129. [Google Scholar] [CrossRef]
- Li, E.; Wang, X.; Chen, K.; Xu, C.; Qin, J.G.; Chen, L. Physiological change and nutritional requirement of Pacific white shrimp Litopenaeus vannamei at low salinity. Rev. Aquac. 2017, 9, 57–75. [Google Scholar] [CrossRef]
- Huang, M.; Dong, Y.; Zhang, Y.; Chen, Q.; Xie, J.; Xu, C.; Zhao, Q.; Li, E. Growth and Lipidomic Responses of Juvenile Pacific White Shrimp Litopenaeus vannamei to Low Salinity. Front. Physiol. 2019, 10, 1087. [Google Scholar] [CrossRef]
- Fan, H.; Li, Y.; Yang, Q.; Jiang, S.; Yang, L.; Huang, J.; Jiang, S.; Zhou, F. Isolation and characterization of a MAPKK gene from Penaeus monodon in response to bacterial infection and low-salinity challenge. Aquac. Rep. 2021, 20, 100671. [Google Scholar] [CrossRef]
- Si, M.; Li, Y.; Jiang, S.; Yang, Q.; Jiang, S.; Yang, L.; Huang, J.; Chen, X.; Zhou, F. A CSDE1/Unr gene from Penaeus monodon: Molecular characterization, expression and association with tolerance to low salt stress. Aquaculture 2022, 561, 738660. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Hao, R.; Zhu, X.; Tian, C.; Jiang, M.; Huang, Y.; Zhu, C. Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 2022, 548, 737575. [Google Scholar] [CrossRef]
- Yue, C.; Li, Q.; Yu, H. Integrated analysis of miRNA and mRNA expression profiles identifies potential regulatory interactions during sexual development of Pacific oyster Crassostrea gigas. Aquaculture 2022, 546, 737294. [Google Scholar] [CrossRef]
- Wang, H.; Wei, H.; Tang, L.; Lu, J.; Mu, C.; Wang, C. Identification and characterization of miRNAs in the gills of the mud crab (Scylla paramamosain) in response to a sudden drop in salinity. BMC Genom. 2018, 19, 609. [Google Scholar] [CrossRef]
- Wu, W.; Dai, C.; Duan, X.; Wang, C.; Lin, X.; Ke, J.; Wang, Y.; Zhang, X.; Liu, H. miRNAs induced by white spot syndrome virus involve in immunity pathways in shrimp Litopenaeus vannamei. Fish Shellfish Immun. 2019, 93, 743–751. [Google Scholar] [CrossRef]
- Guo, H.; Lu, Z.; Zhu, X.; Zhu, C.; Wang, C.; Shen, Y.; Wang, W. Differential expression of microRNAs in hemocytes from white shrimp Litopenaeus vannamei under copper stress. Fish Shellfish Immun. 2018, 74, 152–161. [Google Scholar] [CrossRef]
- He, Y.; Li, Z.; Zhang, H.; Hu, S.; Wang, Q.; Li, J. Genome-wide identification of Chinese shrimp (Fenneropenaeus chinensis) microRNA responsive to low pH stress by deep sequencing. Cell Stress Chaperon. 2019, 24, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhong, P.; Yi, J.; Xu, A.; Lin, W.; Guo, Z.; Wang, C.; Sun, C.; Chan, S. Potential role for microRNA in facilitating physiological adaptation to hypoxia in the Pacific whiteleg shrimp Litopenaeus vannamei. Fish Shellfish Immun. 2019, 84, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Jiang, S.; Yang, Q.; Li, Y.; Yang, L.; Huang, J.; Jiang, S.; Zhou, F. Identification of microRNAs in black tiger shrimp (Penaeus monodon) under acute low-salinity stress. Front. Mar. Sci. 2024, 11, 1403559. [Google Scholar] [CrossRef]
- Chaiyapechara, S.; Uengwetwanit, T.; Arayamethakorn, S.; Bunphimpapha, P.; Phromson, M.; Jangsutthivorawat, W.; Tala, S.; Karoonuthaisiri, N.; Rungrassamee, W. Understanding the host-microbe-environment interactions: Intestinal microbiota and transcriptomes of black tiger shrimp Penaeus monodon at different salinity levels. Aquaculture 2022, 546, 737371. [Google Scholar] [CrossRef]
- Vogt, G. Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 2019, 280, 1405–1444. [Google Scholar] [CrossRef]
- Chen, K.; Li, E.; Li, T.; Xu, C.; Wang, X.; Lin, H.; Qin, J.; Chen, L. Transcriptome and molecular pathway analysis of the hepatopancreas in the Pacific white shrimp Litopenaeus vannamei under chronic low-salinity stress. PLoS ONE 2015, 10, e0131503. [Google Scholar] [CrossRef]
- Shekhar, M.S.; Kiruthika, J.; Ponniah, A.G. Identification and expression analysis of differentially expressed genes from shrimp (Penaeus monodon) in response to low salinity stress. Fish Shellfish Immun. 2013, 35, 1957–1968. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Argasinska, J.; Quinones-Olvera, N.; Finn, R.D.; Bateman, A.; Petrov, A.I. Non-coding RNA analysis using the rfam database. Curr. Protoc. Bioinf. 2018, 62, e51. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Mackowiak, S.D. Identification of Novel and Known miRNAs in Deep-Sequencing Data with miRDeep2. Curr. Protoc. Bioinf. 2011, 36, 12.10.1–12.10.15. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome. Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Nantapojd, T.; Panyim, S.; Ongvarrasopone, C. Identification and expression of white spot syndrome virus-encoded microRNAs in infected Penaeus monodon. Aquaculture 2019, 503, 436–445. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Q.; Liu, H.; Dai, X. Differential expression of microRNAs in giant freshwater prawn (Macrobrachium rosenbergii) during the infection of Vibrio parahaemolyticus. Fish Shellfish Immun. 2024, 153, 109827. [Google Scholar] [CrossRef]
- Li, Y.; Si, M.; Jiang, S.; Yang, Q.; Jiang, S.; Yang, L.; Huang, J.; Chen, X.; Zhou, F.; Li, E. Transcriptome and molecular regulatory mechanisms analysis of gills in the black tiger shrimp Penaeus monodon under chronic low-salinity stress. Front. Physiol. 2023, 14, 1118341. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Shang, Y.; Guo, R.; Chang, Y.; Jiang, Y. Salinity stress-induced differentially expressed miRNAs and target genes in sea cucumbers Apostichopus japonicus. Cell Stress Chaperon. 2019, 24, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Aweya, J.; Zhu, C.; Tran, N.; Hong, Y.; Li, S.; Yao, D.; Zhang, Y. Modulation of Crustacean Innate Immune Response by Amino Acids and Their Metabolites: Inferences from Other Species. Front. Immunol. 2020, 11, 574721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shi, X.; Guo, J.; Mao, X.; Fan, B. Acute stress response in gill of Pacific white shrimp Litopenaeus vannamei to high alkalinity. Aquaculture 2024, 586, 740766. [Google Scholar] [CrossRef]
- Niu, M.; Gao, G.; Qin, K.; Chen, Y.; Wang, H.; Li, X.; Liang, G.; Wang, C.; Mu, C.; Su, Q. Multiple low salinity stress modes provided novel insight into the metabolic response of Scylla paramamosain adapting to inland saline-alkaline water. Front. Mar. Sci. 2022, 9, 977599. [Google Scholar] [CrossRef]
- Nayak, S.; Koven, W.; Meiri, I.; Khozin-Goldberg, I.; Isakov, N.; Zibdeh, M.; Zilberg, D. Dietary arachidonic acid affects immune function and fatty acid composition in cultured rabbitfish Siganus rivulatus. Fish Shellfish Immun. 2017, 68, 46–53. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.; Liu, C.; Liu, R.; Yang, C.; Wang, L.; Song, L. Cortisol modulates glucose metabolism and oxidative response after acute high temperature stress in Pacific oyster Crassostrea gigas. Fish Shellfish Immun. 2022, 126, 141–149. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Dou, Y.; Shen, H.; Qiao, Y.; Cao, X.; Fan, X.; Hu, Y.; Qian, J. Hormonal regulation changes in shrimp infected with Enterocytozoon hepatopenaei: A transcriptomic analysis. Aquac. Int. 2024, 32, 841–2863. [Google Scholar] [CrossRef]
- Xu, C.; Li, E.; Liu, Y.; Wang, X.; Qin, J.; Chen, L. Comparative proteome analysis of the hepatopancreas from the Pacific white shrimp Litopenaeus vannamei under long-term low salinity stress. J. Proteom. 2017, 162, 1–10. [Google Scholar] [CrossRef]
- Medina Gomez, H.; Adame Rivas, G.; Hernández-Quintero, A.; González Hernández, A.; Torres Guzmán, J.C.; Mendoza, H.L.; Contreras-Garduño, J. The occurrence of immune priming can be species-specific in entomopathogens. Microb. Pathog. 2018, 118, 361–364. [Google Scholar] [CrossRef]
- Zhai, Y.; Xu, R.; He, P.; Jia, R. A proteomics investigation of ‘immune priming’ in Penaeus vannamei as shown by isobaric tags for relative and absolute quantification. Fish Shellfish Immun. 2021, 117, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chu, K.H.; Ma, K. Transcriptome Analysis of Multiple Tissues in the Shrimp Penaeus vannamei Reveals the Typical Physiological Response to Three Pathogens. J. Mar. Sci. Eng. 2023, 11, 389. [Google Scholar] [CrossRef]
- Ren, X.; Wang, Q.; Shao, H.; Xu, Y.; Liu, P.; Li, J. Effects of Low Temperature on Shrimp and Crab Physiology, Behavior, and Growth: A Review. Front. Mar. Sci. 2021, 8, 746177. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Kumai, Y.; Bernier, N.J.; Perry, S.F. Angiotensin-II promotes Na+ uptake in larval zebrafish, Danio rerio, in acidic and ion-poor water. J. Endocrinol. 2014, 220, 195–205. [Google Scholar] [CrossRef]
- Farhadi, A.; Tang, S.; Huang, M.; Yu, Q.; Xu, C.; Li, E. Identification of key immune and stress related genes and pathways by comparative analysis of the gene expression profile under multiple environmental stressors in pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immun. 2023, 135, 108695. [Google Scholar] [CrossRef]
- Chen, D.; Xin, Y.; Teng, J.; Zhao, X.; Lu, J.; Li, Y.; Wang, H. Comparative transcriptomic and molecular biology analyses to explore potential immune responses to Vibrio parahaemolyticus challenge in Eriocheir sinensis. Front. Cell Infect. Microbiol. 2024, 14, 1456130. [Google Scholar] [CrossRef]
- Lai, X.; Zhang, C.; Wu, Y.; Yang, Y.; Zhang, M.; Qin, W.; Wang, R.; Shu, H. Comparative transcriptome analysis reveals physiological responses in liver tissues of Epinephelus coioides under acute hypoxia stress. Comp. Biochem. Phys. D 2022, 43, 101005. [Google Scholar] [CrossRef]
- Li, Z.; Chang, T.; Han, F.; Fan, X.; Liu, W.; Wu, P.; Xu, C.; Li, E. Effects of myo-inositol on growth and biomarkers of environmental stress and metabolic regulation in Pacific white shrimp (Litopenaeus vannamei) reared at low salinity. Comp. Biochem. Phys. D 2024, 50, 101216. [Google Scholar] [CrossRef]
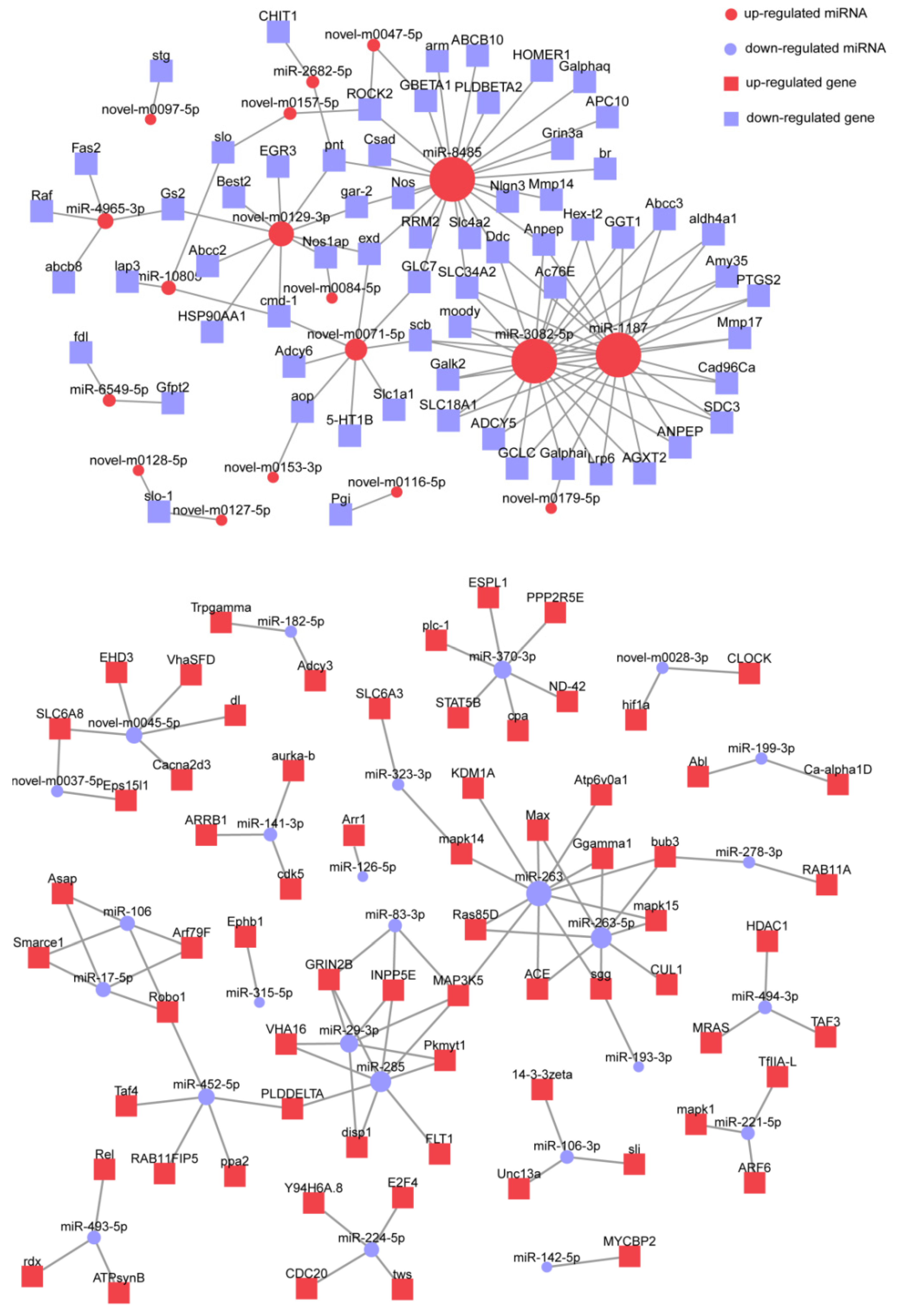
| Gene Symbol | Description | Related miRNA | Gene Function |
|---|---|---|---|
| Anpep | aminopeptidase N-like | miR-1187, miR-3082-5p, miR-8485 | Amino acid metabolism |
| cmd-1 | calmodulin isoform X1 | miR-10805, novel-m0071-5p, novel-m0129-3p | Signal transduction |
| Ddc | aromatic-L-amino-acid decarboxylase-like | miR-1187, miR-3082-5p, miR-8485 | Amino acid metabolism |
| exd | homeobox protein extradenticle-like isoform X1 | miR-8485, novel-m0071-5p, novel-m0129-3p | Development and differentiation |
| Galphai | guanine nucleotide-binding protein G(i) subunit alpha isoform X1 | miR-1187, miR-3082-5p, novel-m0179-5p | Lipid metabolism |
| pnt | ETS-like protein pointed isoform X1 | miR-2682-5p, miR-8485, novel-m0129-3p | Development and differentiation |
| ROCK | rho-associated protein kinase 1-like | miR-8485, novel-m0047-5p, novel-m0157-5p | Regulation of cytoskeleton |
| scb | integrin alpha-7-like | miR-1187, miR-3082-5p, novel-m0071-5p | Regulation of cytoskeleton |
| SLC34A2 | sodium-dependent phosphate transport protein 2B-like | miR-1187, miR-3082-5p, miR-8485 | Phosphate digestive absorption |
| MAP3K5 | mitogen-activated protein kinase kinase kinase 15-like isoform X1 | miR-263, miR-285, miR-29-3p, miR-83-3p | Signal transduction |
| bub3 | mitotic checkpoint protein BUB3-like isoform X1 | miR-263-5p, miR-263, miR-278-3p | Pathogen infection |
| GRIN2B | glutamate receptor ionotropic, NMDA 2B-like isoform X1 | miR-285, miR-29-3p, miR-83-3p | Synapse maturation and plasticity |
| INPP5E | phosphatidylinositol polyphosphate 5-phosphatase type IV-like isoform X1 | miR-285, miR-29-3p, miR-83-3p | Signal transduction |
| Robo1 | protein sax-3-like isoform X1 | miR-106, miR-17-5p, miR-452-5p | Axon guidance and regeneration |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Jiang, S.; Ding, Y.; Diao, H.; Li, W.; Li, Y.; Huang, J.; Yang, L.; Yang, Q.; Zhou, F. Insights into the Regulatory Role of MicroRNAs in Penaeus monodon Under Moderately Low Salinity Stress. Biology 2025, 14, 440. https://doi.org/10.3390/biology14040440
Shi J, Jiang S, Ding Y, Diao H, Li W, Li Y, Huang J, Yang L, Yang Q, Zhou F. Insights into the Regulatory Role of MicroRNAs in Penaeus monodon Under Moderately Low Salinity Stress. Biology. 2025; 14(4):440. https://doi.org/10.3390/biology14040440
Chicago/Turabian StyleShi, Jianzhi, Song Jiang, Yangyang Ding, Hongshan Diao, Wenzhe Li, Yundong Li, Jianhua Huang, Lishi Yang, Qibin Yang, and Falin Zhou. 2025. "Insights into the Regulatory Role of MicroRNAs in Penaeus monodon Under Moderately Low Salinity Stress" Biology 14, no. 4: 440. https://doi.org/10.3390/biology14040440
APA StyleShi, J., Jiang, S., Ding, Y., Diao, H., Li, W., Li, Y., Huang, J., Yang, L., Yang, Q., & Zhou, F. (2025). Insights into the Regulatory Role of MicroRNAs in Penaeus monodon Under Moderately Low Salinity Stress. Biology, 14(4), 440. https://doi.org/10.3390/biology14040440






