RNA Binding to CCRRM of PABPN1 Induces Conformation Change
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
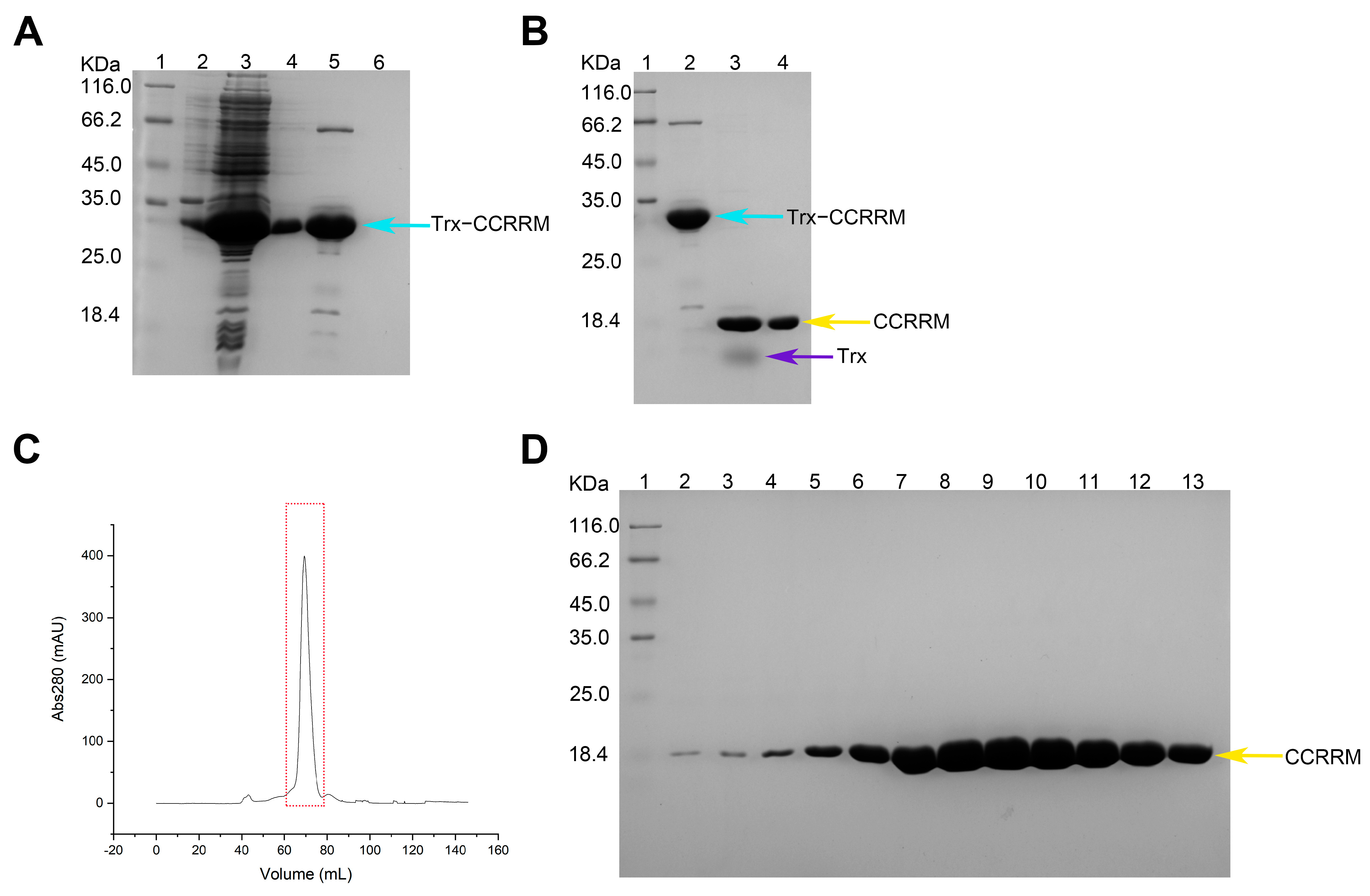
2.1.1. Protein Purification
2.1.2. RNA Preparation
2.2. SEC-FPLC
2.3. EMSA
2.4. Biolayer Interferometry [43]
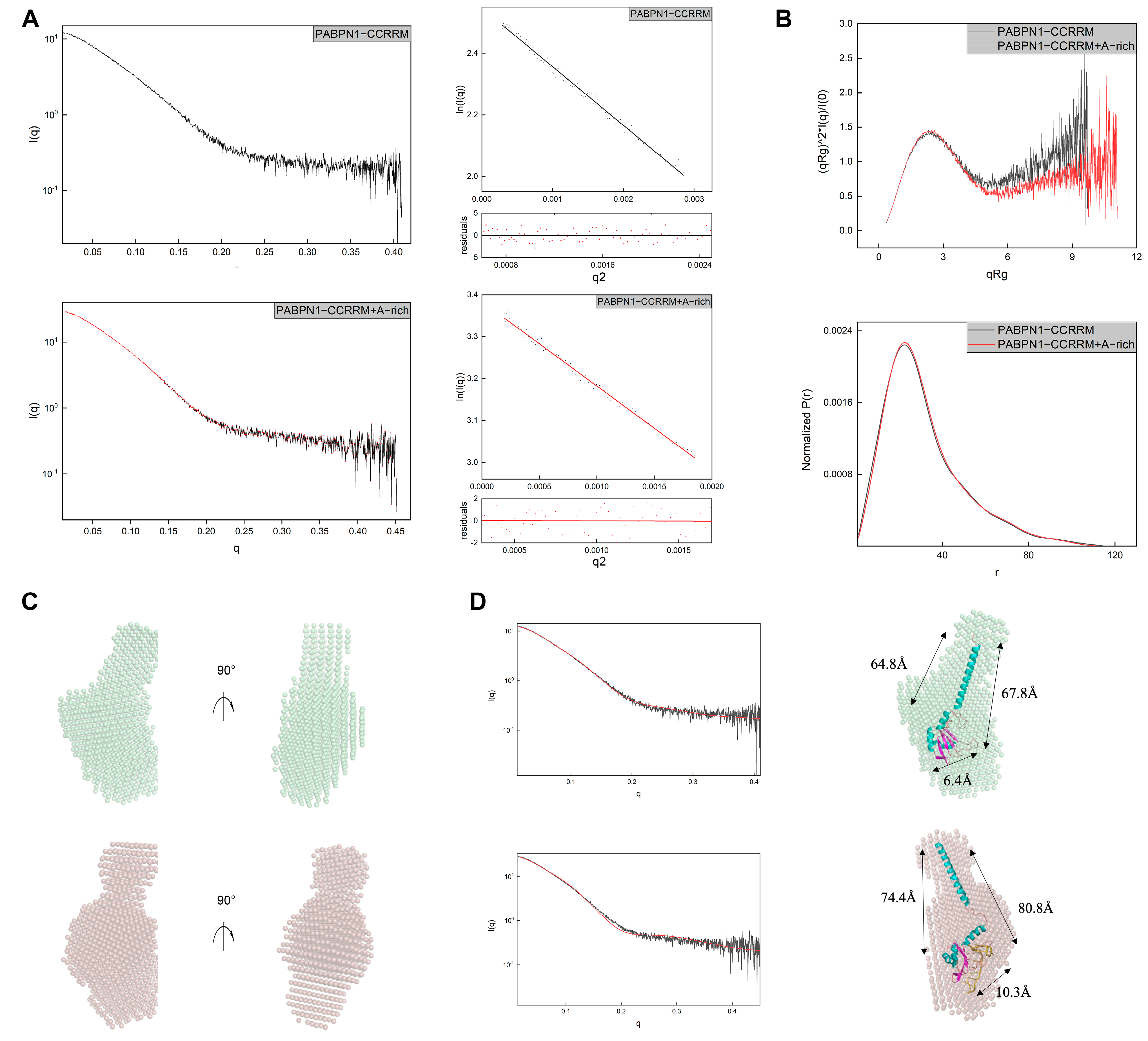
2.5. SAXS Data Collection and Analysis
2.6. SHAPE Probing Analysis
3. Results
3.1. Domain Structure of PABPN1 and Preparation of CCRRM Protein Sample
3.2. The CCRRM’s Conformation in Solution Remains Stable Irrespective of Concentration and Interact with A-Rich RNA
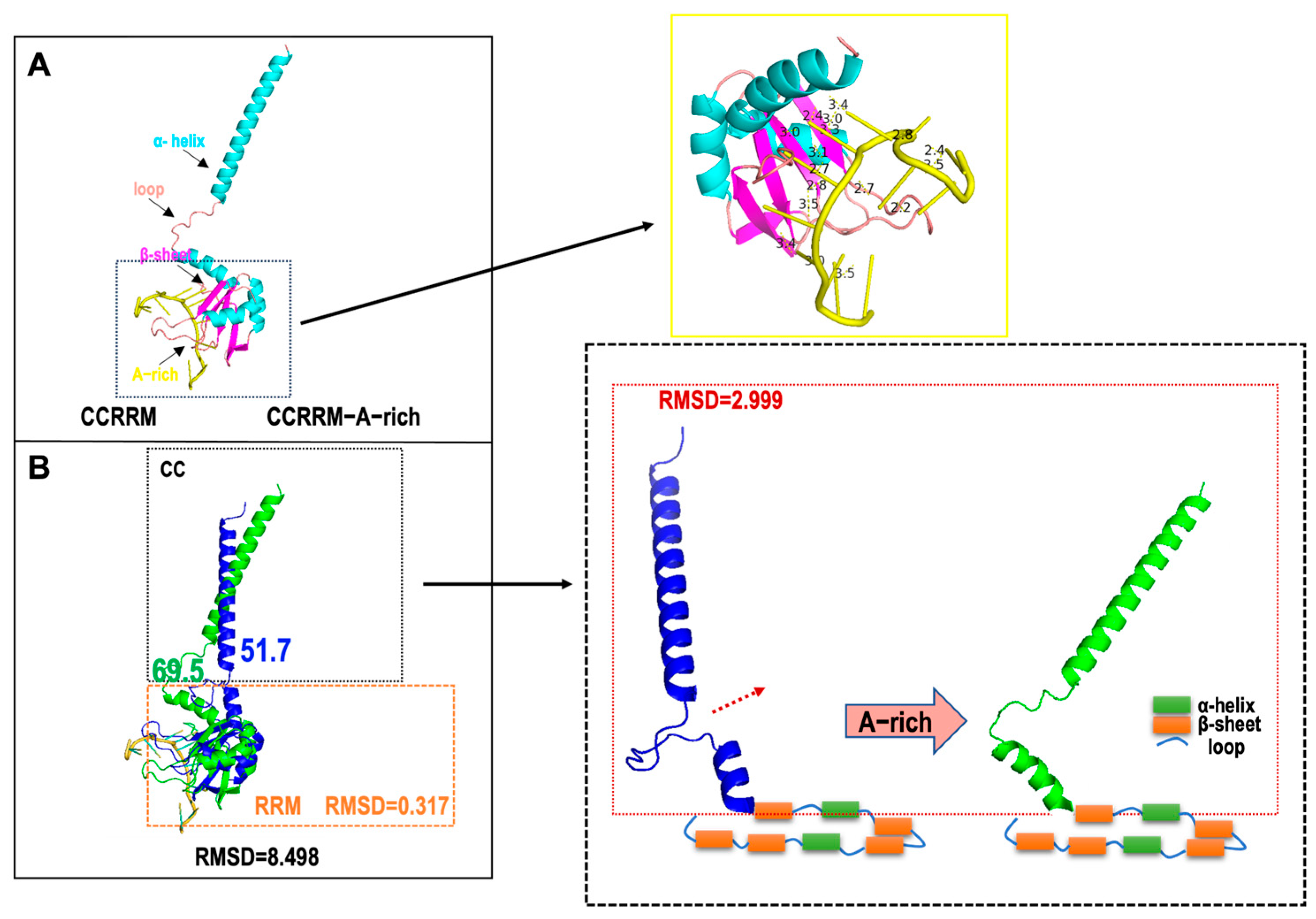
3.3. A-Rich RNA Induces Conformational Changes in CCRRM, Primarily Occurring in the CC Region
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PABPN1 | Poly(A) Binding Protein Nuclear 1 |
| CC | coiled-coil domain |
| RRM | RNA recognition motif |
| SAXS | small-angle X-ray scattering |
| SHAPE | selective 2′-hydroxyl acylation analyzed by primer extension |
| RBPs | RNA-binding proteins |
| OPMD | oculopharyngeal muscular dystrophy |
| Pabs | Poly(A) RNA binding proteins |
| APA | Alternative polyadenylation |
| ccRCC | clear cell renal cell carcinoma |
| EMSA | Electrophoretic Mobility Shift Assay |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| SEC-FPLC | Size-Exclusion Chromatography with Fast Protein Liquid Chromatography |
| BLI | Biolayer interferometry |
| SSRF | Shanghai Synchrotron Radiation Facility |
| RMSDs | root-mean-square deviations |
| 1M7 | 1-methyl-7-nitroisatoic anhydride |
| Rg | Radius of gyration |
| Dmax | maximum dimension |
| CPSF | cleavage and polyadenylation specificity factor |
References
- Wigington, C.P.; Williams, K.R.; Meers, M.P.; Bassell, G.J.; Corbett, A.H. Poly(A) RNA-binding proteins and polyadenosine RNA: New members and novel functions. Wiley Interdiscip. Rev. RNA 2014, 5, 601–622. [Google Scholar] [CrossRef] [PubMed]
- Apponi, L.H.; Corbett, A.H.; Pavlath, G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011, 32, 652–658. [Google Scholar] [CrossRef]
- Banerjee, A.; Vest, K.E.; Pavlath, G.K.; Corbett, A.H. Nuclear poly(A) binding protein 1 (PABPN1) and Matrin3 interact in muscle cells and regulate RNA processing. Nucleic Acids Res. 2017, 45, 10706–10725. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Apponi, L.H.; Pavlath, G.K.; Corbett, A.H. PABPN1: Molecular function and muscle disease. FEBS J. 2013, 280, 4230–4250. [Google Scholar] [CrossRef] [PubMed]
- Apponi, L.H.; Leung, S.W.; Williams, K.R.; Valentini, S.R.; Corbett, A.H.; Pavlath, G.K. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum. Mol. Genet. 2010, 19, 1058–1065. [Google Scholar] [CrossRef]
- Malerba, A.; Klein, P.; Bachtarzi, H.; Jarmin, S.A.; Cordova, G.; Ferry, A.; Strings, V.; Espinoza, M.P.; Mamchaoui, K.; Blumen, S.C.; et al. PABPN1 gene therapy for oculopharyngeal muscular dystrophy. Nat. Commun. 2017, 8, 14848. [Google Scholar] [CrossRef]
- Brais, B. Oculopharyngeal muscular dystrophy: A polyalanine myopathy. Curr. Neurol. Neurosci. Rep. 2009, 9, 76–82. [Google Scholar] [CrossRef]
- Raz, Y.; Raz, V. Oculopharyngeal muscular dystrophy as a paradigm for muscle aging. Front. Aging Neurosci. 2014, 6, 317. [Google Scholar] [CrossRef]
- Harish, P.; Malerba, A.; Dickson, G.; Bachtarzi, H. Progress on gene therapy, cell therapy, and pharmacological strategies toward the treatment of oculopharyngeal muscular dystrophy. Hum. Gene Ther. 2015, 26, 286–292. [Google Scholar] [CrossRef]
- Albrecht, A.; Mundlos, S. The other trinucleotide repeat: Polyalanine expansion disorders. Curr. Opin. Genet. Dev. 2005, 15, 285–293. [Google Scholar] [CrossRef]
- Fan, X.; Rouleau, G.A. Progress in understanding the pathogenesis of oculopharyngeal muscular dystrophy. Can. J. Neurol. Sci. 2003, 30, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Phillips, B.L.; Banerjee, A.; Sanchez, B.J.; Di Marco, S.; Gallouzi, I.-E.; Pavlath, G.K.; Corbett, A.H. Post-transcriptional regulation of Pabpn1 by the RNA binding protein HuR. Nucleic Acids Res. 2018, 46, 7643–7661. [Google Scholar] [CrossRef] [PubMed]
- Brais, B.; Bouchard, J.P.; Xie, Y.G.; Rochefort, D.L.; Chrétien, N.; Tomé, F.M.; Lafrenière, R.G.; Rommens, J.M.; Uyama, E.; Nohira, O. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998, 18, 164–167. [Google Scholar] [CrossRef]
- Wigington, C.P.; Morris, K.J.; Newman, L.E.; Corbett, A.H. The Polyadenosine RNA-binding Protein, Zinc Finger Cys3His Protein 14 (ZC3H14), Regulates the Pre-mRNA Processing of a Key ATP Synthase Subunit mRNA. J. Biol. Chem. 2016, 291, 22442–22459. [Google Scholar] [CrossRef]
- Curinha, A.; Braz, S.O.; Pereira-Castro, I.; Cruz, A.; Moreira, A. Implications of polyadenylation in health and disease. Nucleus 2014, 5, 508–519. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Lemay, J.F.; Lemieux, C.; St-André, O.; Bachand, F. Crossing the borders: Poly(A)-binding proteins working on both sides of the fence. RNA Biol. 2010, 7, 291–295. [Google Scholar] [CrossRef]
- Kühn, U.; Wahle, E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 2004, 1678, 67–84. [Google Scholar] [CrossRef]
- Brais, B.; Xie, Y.G.; Sanson, M.; Morgan, K.; Weissenbach, J.; Korczyn, A.D.; Blumen, S.C.; Fardeau, M.; Tomé, F.M.; Bouchard, J.P.; et al. The oculopharyngeal muscular dystrophy locus maps to the region of the cardiac alpha and beta myosin heavy chain genes on chromosome 14q11.2-q13. Hum. Mol. Genet. 1995, 4, 429–434. [Google Scholar] [CrossRef]
- Brais, B. Oculopharyngeal muscular dystrophy: A late-onset polyalanine disease. Cytogenet. Genome Res. 2003, 100, 252–260. [Google Scholar] [CrossRef]
- Harvey, R.; Dezi, V.; Pizzinga, M.; Willis, A.E. Post-transcriptional control of gene expression following stress: The role of RNA-binding proteins. Biochem. Soc. Trans. 2017, 45, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Winter, R.; Liebold, J.; Schwarz, E. The unresolved puzzle why alanine extensions cause disease. Biol. Chem. 2013, 394, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Tudek, A.; Schmid, M.; Jensen, T.H. Escaping nuclear decay: The significance of mRNA export for gene expression. Curr. Genet. 2019, 65, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Olie, C.S.; Riaz, M.; Konietzny, R.; Charles, P.D.; Pinto-Fernandez, A.; Kiełbasa, S.M.; Aartsma-Rus, A.; Goeman, J.J.; Kessler, B.M.; Raz, V. Deacetylation Inhibition Reverses PABPN1-Dependent Muscle Wasting. iScience 2019, 12, 318–332. [Google Scholar] [CrossRef]
- Liu, X.; Xie, H.; Liu, W.; Zuo, J.; Li, S.; Tian, Y.; Zhao, J.; Bai, M.; Li, J.; Bao, L.; et al. Dynamic regulation of alternative polyadenylation by PQBP1 during neurogenesis. Cell Rep. 2024, 43, 114525. [Google Scholar] [CrossRef]
- Ichinose, J.; Watanabe, K.; Sano, A.; Nagase, T.; Nakajima, J.; Fukayama, M.; Yatomi, Y.; Ohishi, N.; Takai, D. Alternative polyadenylation is associated with lower expression of PABPN1 and poor prognosis in non-small cell lung cancer. Cancer Sci. 2014, 105, 1135–1141. [Google Scholar] [CrossRef]
- Jenal, M.; Elkon, R.; Loayza-Puch, F.; van Haaften, G.; Kühn, U.; Menzies, F.M.; Vrielink, J.A.F.O.; Bos, A.J.; Drost, J.; Rooijers, K.; et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 2012, 149, 538–553. [Google Scholar] [CrossRef]
- Simonelig, M. PABPN1 shuts down alternative poly(A) sites. Cell Res. 2012, 22, 1419–1421. [Google Scholar] [CrossRef]
- de Klerk, E.; Venema, A.; Anvar, S.Y.; Goeman, J.J.; Hu, O.; Trollet, C.; Dickson, G.; den Dunnen, J.T.; van der Maarel, S.M.; Raz, V.; et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012, 40, 9089–9101. [Google Scholar] [CrossRef]
- Xiong, M.; Liu, C.; Li, W.; Jiang, H.; Long, W.; Zhou, M.; Yang, C.; Kazobinka, G.; Sun, Y.; Zhao, J.; et al. PABPN1 promotes clear cell renal cell carcinoma progression by suppressing the alternative polyadenylation of SGPL1 and CREG1. Carcinogenesis 2023, 44, 576–586. [Google Scholar] [CrossRef]
- Nguyen, D.; St-Sauveur, V.G.; Bergeron, D.; Dupuis-Sandoval, F.; Scott, M.S.; Bachand, F. A Polyadenylation-Dependent 3' End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA. Cell Rep. 2015, 13, 2244–2257. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. SDS-Polyacrylamide Gel Electrophoresis of Proteins. CSH Protoc 2006, 2006, pdb-prot4540. [Google Scholar] [CrossRef]
- Domingues, M.N.; Sforça, M.L.; Soprano, A.S.; Lee, J.; de Arruda Campos Brasil de Souza, T.; Cassago, A.; Portugal, R.V.; de Mattos Zeri, A.C.; Murakami, M.T.; Sadanandom, A.; et al. Structure and Mechanism of Dimer-Monomer Transition of a Plant Poly(A)-Binding Protein upon RNA Interaction: Insights into Its Poly(A) Tail Assembly. J. Mol. Biol. 2015, 427, 2491–2506. [Google Scholar] [CrossRef]
- Kühn, U.; Nemeth, A.; Meyer, S.; Wahle, E. The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 2003, 278, 16916–16925. [Google Scholar] [CrossRef]
- Meyer, S.; Urbanke, C.; Wahle, E. Equilibrium studies on the association of the nuclear poly(A) binding protein with poly(A) of different lengths. Biochemistry 2002, 41, 6082–6089. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Zubiaga, A.M.; Belasco, J.G.; Greenberg, M.E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell Biol. 1995, 15, 2219–2230. [Google Scholar] [CrossRef]
- Hui, J.; Reither, G.; Bindereif, A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA 2003, 9, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, I.A.; Tahoe, N.M.; Fan, D.; Larsson, O.; Rattenbacher, B.; Sternjohn, J.R.; Vasdewani, J.; Karypis, G.; Reilly, C.S.; Bitterman, P.B.; et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 2008, 29, 263–270. [Google Scholar] [CrossRef]
- Ke, A.; Doudna, J.A. Crystallization of RNA and RNA-protein complexes. Methods 2004, 34, 408–414. [Google Scholar] [CrossRef]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Mansouri-Noori, F.; Bayfield, M.A. Electrophoretic mobility shift assays (EMSAs) for in vitro detection of protein-nucleic acid interactions. STAR Protoc. 2024, 5, 103128. [Google Scholar] [CrossRef] [PubMed]
- Wallner, J.; Lhota, G.; Jeschek, D.; Mader, A.; Vorauer-Uhl, K. Application of Bio-Layer Interferometry for the analysis of protein/liposome interactions. J. Pharm. Biomed. Anal. 2013, 72, 150–154. [Google Scholar] [CrossRef]
- Dmitri, I.S.; Michel, H.J.K. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003, 66, 1735. [Google Scholar]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.B.; Gillilan, R.E.; Skou, S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 2017, 50, 1545–1553. [Google Scholar] [CrossRef]
- Putnam, C.D.; Hammel, M.; Hura, G.L.; Tainer, J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007, 40, 191–285. [Google Scholar] [CrossRef]
- Burrill, C.P.; Andino, R. RNA structure analysis of viruses using SHAPE. Curr. Protoc. Microbiol. 2013, 30, 15h.3.1–15h.3.12. [Google Scholar] [CrossRef]
- Mortimer, S.A.; Weeks, K.M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007, 129, 4144–4145. [Google Scholar] [CrossRef]
- Vasa, S.M.; Guex, N.; Wilkinson, K.A.; Weeks, K.M.; Giddings, M.C. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 2008, 14, 1979–1990. [Google Scholar] [CrossRef]
- Bear, D.G.; Fomproix, N.; Soop, T.; Björkroth, B.; Masich, S.; Daneholt, B. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 2003, 286, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Wahle, E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995, 270, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Ishigaki, Y.; Li, X.; Serin, G.; Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 2001, 106, 607–617. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Krug, R.M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. Embo J. 1999, 18, 2273–2283. [Google Scholar] [CrossRef] [PubMed]
- Calado, A.; Kutay, U.; Kühn, U.; Wahle, E.; Carmo-Fonseca, M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 2000, 6, 245–256. [Google Scholar] [CrossRef]
- Kwiatek, L.; Landry-Voyer, A.-M.; Latour, M.; Yague-Sanz, C.; Bachand, F. PABPN1 prevents the nuclear export of an unspliced RNA with a constitutive transport element and controls human gene expression via intron retention. RNA 2023, 29, 644–662. [Google Scholar] [CrossRef]
- Ge, H.; Zhou, D.; Tong, S.; Gao, Y.; Teng, M.; Niu, L. Crystal structure and possible dimerization of the single RRM of human PABPN1. Proteins 2008, 71, 1539–1545. [Google Scholar] [CrossRef]
- Gordon, J.M.; Phizicky, D.V.; Schärfen, L.; Brown, C.L.; Escayola, D.A.; Kanyo, J.; Lam, T.T.; Simon, M.; Neugebauer, K.M. Phosphorylation of the nuclear poly(A) binding protein (PABPN1) during mitosis protects mRNA from hyperadenylation and maintains transcriptome dynamics. Nucleic Acids Res. 2024, 52, 9886–9903. [Google Scholar] [CrossRef]
- Huang, L.; Li, G.; Du, C.; Jia, Y.; Yang, J.; Fan, W.; Xu, Y.-Z.; Cheng, H.; Zhou, Y. The polyA tail facilitates splicing of last introns with weak 3’ splice sites via PABPN1. EMBO Rep. 2023, 24, e57128. [Google Scholar] [CrossRef]
- Li, W.; You, B.; Hoque, M.; Zheng, D.; Luo, W.; Ji, Z.; Park, J.Y.; Gunderson, S.I.; Kalsotra, A.; Manley, J.L.; et al. Systematic profiling of poly(A)+ transcripts modulated by core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015, 11, e1005166. [Google Scholar] [CrossRef]
- Liu, X.; Hoque, M.; Larochelle, M.; Lemay, J.-F.; Yurko, N.; Manley, J.L.; Bachand, F.; Tian, B. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res. 2017, 27, 1685–1695. [Google Scholar] [CrossRef]
- Kerwitz, Y.; Kühn, U.; Lilie, H.; Knoth, A.; Scheuermann, T.; Friedrich, H.; Schwarz, E.; Wahle, E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. Embo J. 2003, 22, 3705–3714. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.-X.; Pi, S.-B.; Zhao, L.-W.; Wu, Y.-W.; Shen, J.-L.; Zhang, S.-Y.; Sha, Q.-Q.; Fan, H.-Y. PABPN1 functions as a hub in the assembly of nuclear poly(A) domains that are essential for mouse oocyte development. Sci. Adv. 2022, 8, eabn9016. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dong, W.; Zhou, M.; Yang, C.; Xiong, M.; Kazobinka, G.; Chen, Z.; Xing, Y.; Hou, T. PABPN1 regulates mRNA alternative polyadenylation to inhibit bladder cancer progression. Cell Biosci. 2023, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.-W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486–490. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef]
- García-Mauriño, S.M.; Rivero-Rodríguez, F.; Velázquez-Cruz, A.; Hernández-Vellisca, M.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. RNA Binding Protein Regulation and Cross-Talk in the Control of AU-rich mRNA Fate. Front. Mol. Biosci. 2017, 4, 71. [Google Scholar] [CrossRef]

| RNA Name | Sequence |
|---|---|
| A-rich | UAAUACGACUCACUAUAGGGUGGUCAGUCGAGUGGAAAAAA AAAAAAAAAAAAGGGCGGCAUGGUCCCAGCCUCCU |
| RE_A-rich | CCUUCGGGCCAAAUUUAUUUAUUUAUUUAUUUAGCUGACGA UAAAAAAAAAAAAAAAAAACCACACACACAGGAAUCGACUC UGUUUGUUUGUGUUUGUUUGUACUGAAUUGGCACUUUUCCC CUUUUCCCUUUCUGGACUGGCAUCGAUCCGGUUCGCCGGAUC CAAAUCGGGCUUCGGUCCGGUUC |
| Structural Parameters | RNA-Free | RNA-Bound |
| I (0) (cm−1) from Guinier fit | 12.69 ± 0.03 | 29.48 ± 0.06 |
| Rg (Å) from Guinier fit | 23.75 ± 0.1 | 24.61 ± 0.11 |
| Rg (Å) from P(r) | 26.31 ± 0.34 | 26.1 ± 0.18 |
| Dmax (Å) from P(r) | 119.0 | 114.0 |
| I (0) (cm−1) from P(r) | 13.04 ± 0.06 | 29.82 ± 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Chen, T.; Zhang, Y.; Lu, C. RNA Binding to CCRRM of PABPN1 Induces Conformation Change. Biology 2025, 14, 432. https://doi.org/10.3390/biology14040432
Zhang S, Chen T, Zhang Y, Lu C. RNA Binding to CCRRM of PABPN1 Induces Conformation Change. Biology. 2025; 14(4):432. https://doi.org/10.3390/biology14040432
Chicago/Turabian StyleZhang, Shengping, Ting Chen, Yunlong Zhang, and Changrui Lu. 2025. "RNA Binding to CCRRM of PABPN1 Induces Conformation Change" Biology 14, no. 4: 432. https://doi.org/10.3390/biology14040432
APA StyleZhang, S., Chen, T., Zhang, Y., & Lu, C. (2025). RNA Binding to CCRRM of PABPN1 Induces Conformation Change. Biology, 14(4), 432. https://doi.org/10.3390/biology14040432




