Transcriptome Sequencing Reveals Survival Strategies and Pathogenic Potential of Vibrio parahaemolyticus Under Gastric Acid Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Selected Experimental Strains and Culture Methods
2.2. Preparation of Simulated Gastric Fluid (SGF) and Simulated Intestinal Fluid (SIF)
2.3. Determination of the Experimental Gastric Acid Value
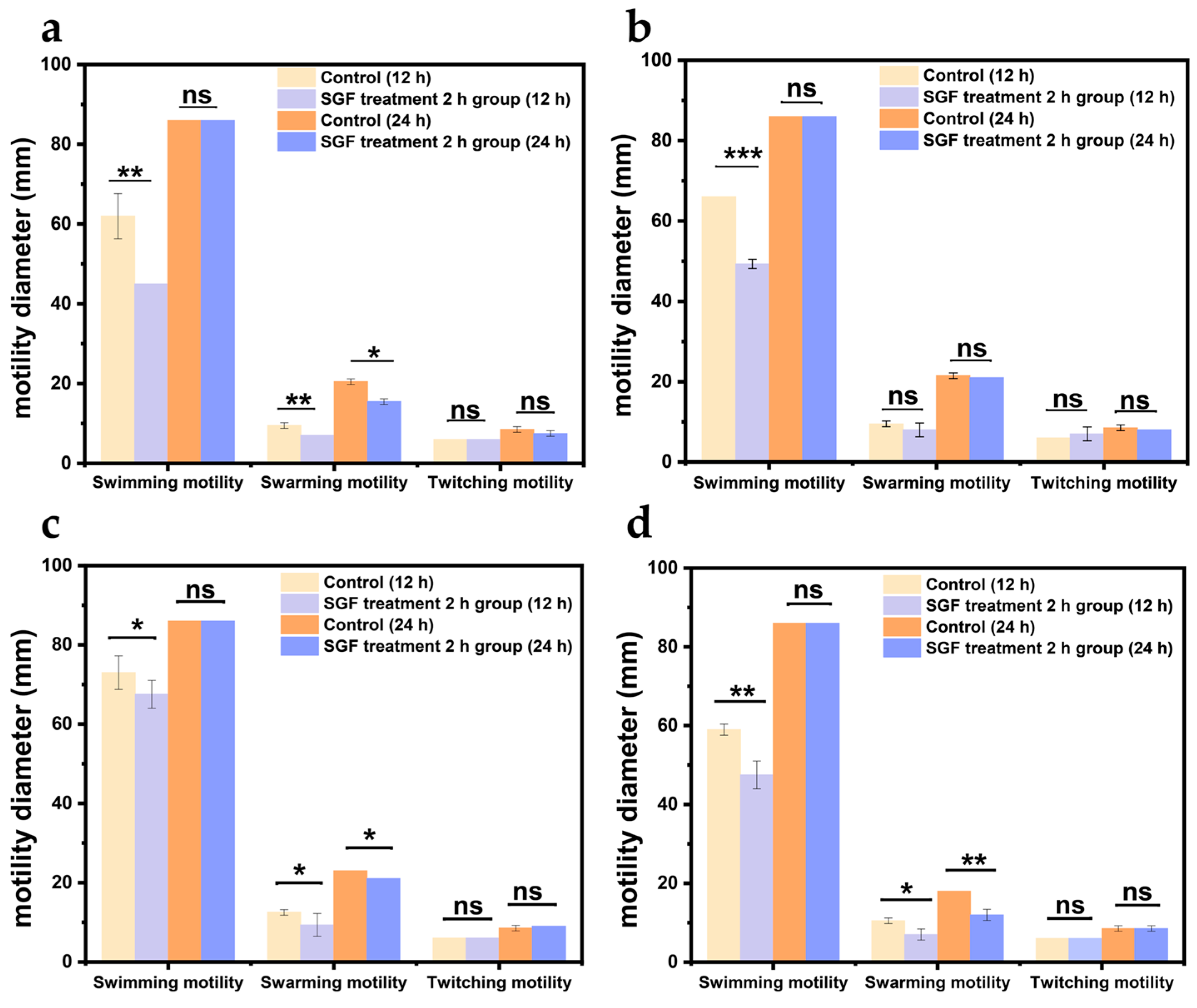
2.4. Determination of the Motility and Adhesion Capacity of V. parahaemolyticus
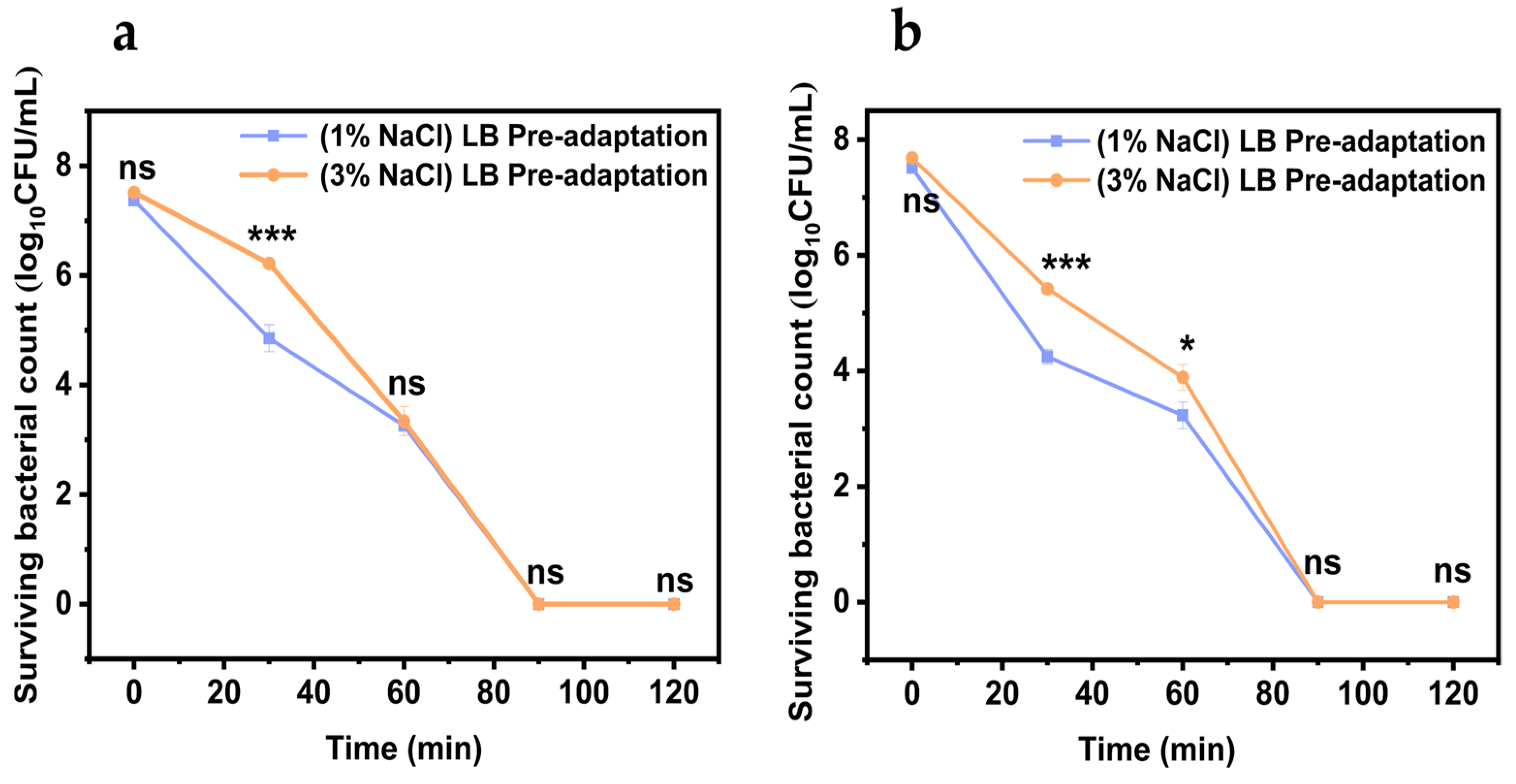
2.5. Determination of the Survival Rate
2.6. Preparation and Pretreatment of Litopenaeus vannamei
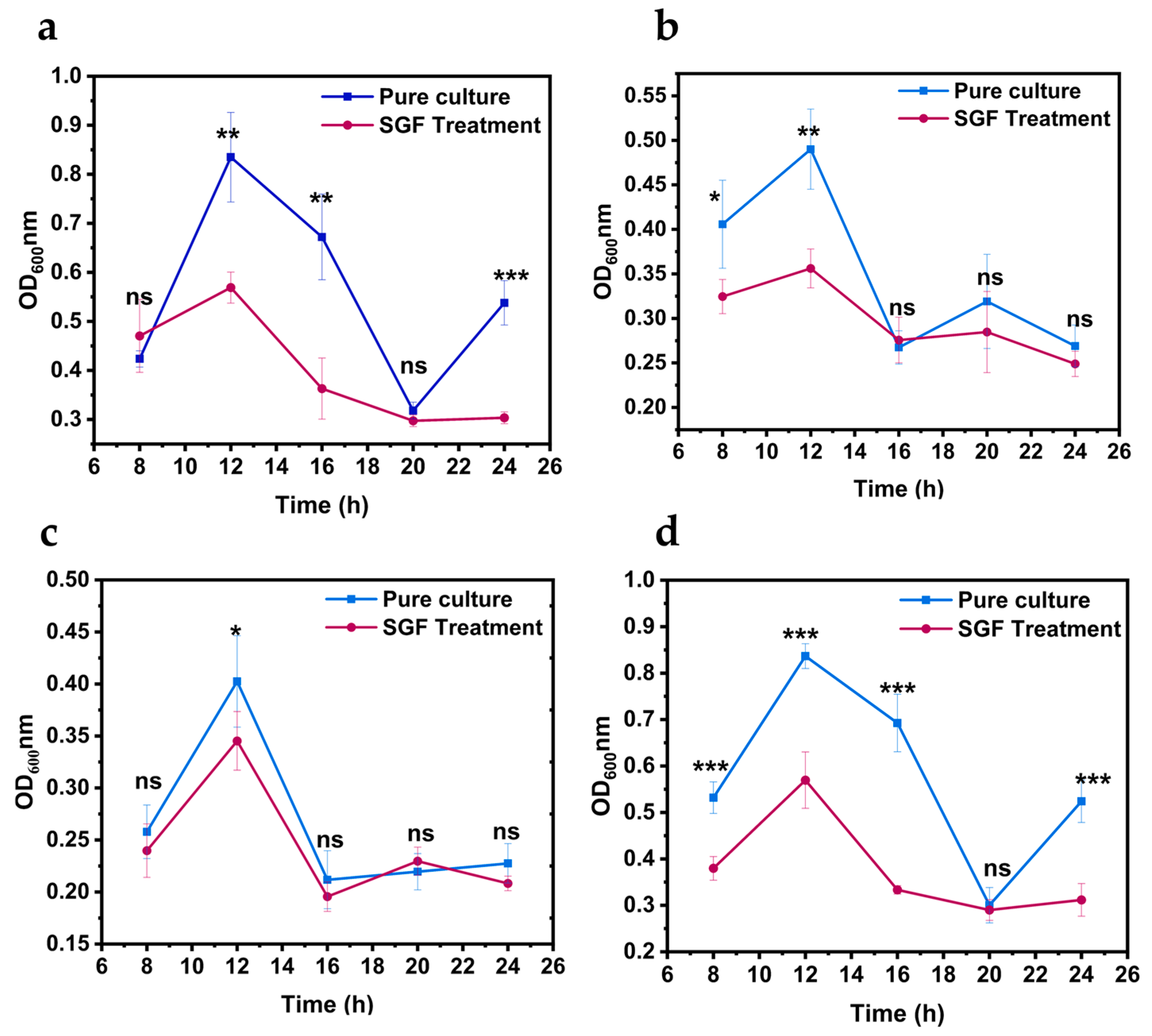
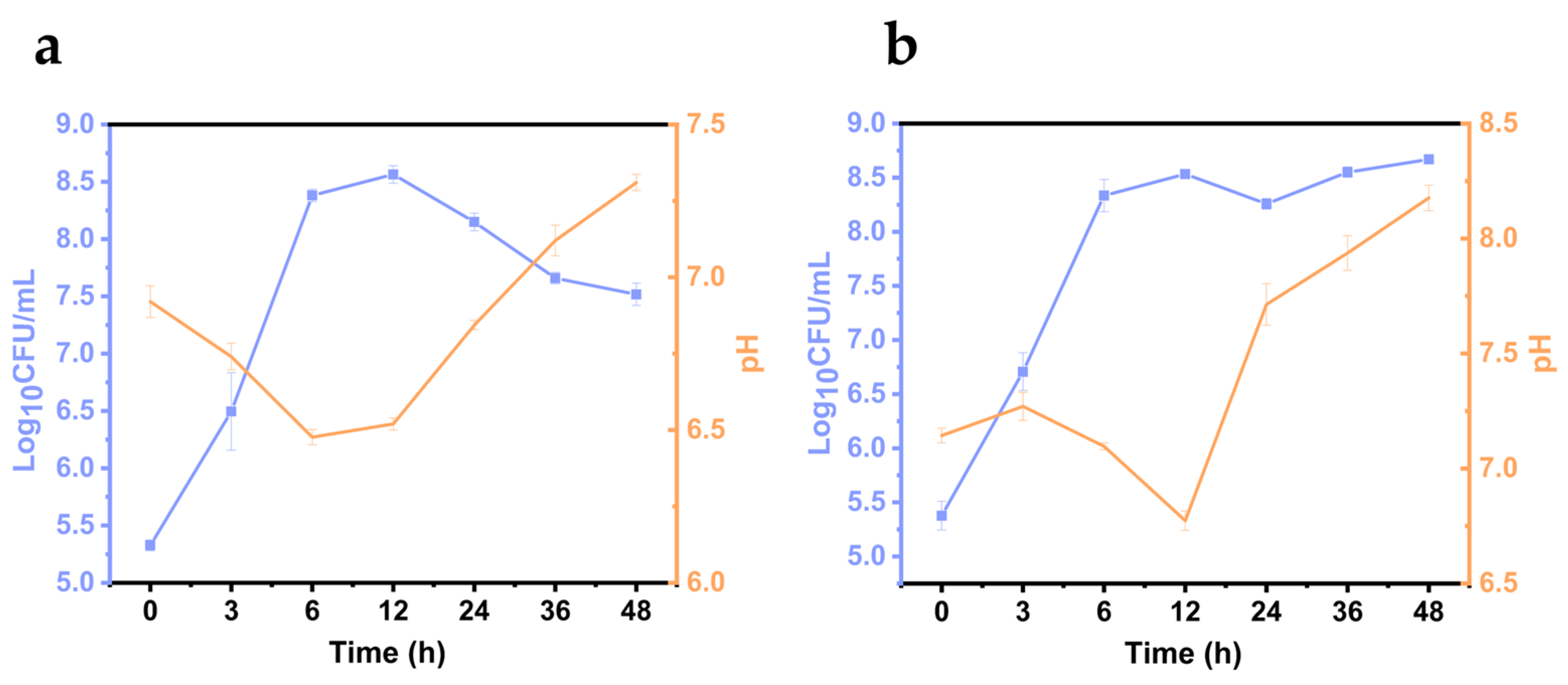
2.7. V. parahaemolyticus Growth on Shrimp Samples
2.8. Determination of the Survival Rate of V. parahaemolyticus in Different Matrices During Gastric Acid Digestion
2.9. The Recovery of V. parahaemolyticus Under SIF
2.10. Scanning Electron Microscopy (SEM) Assessment of the Morphological Changes in V. parahaemolyticus
2.11. RNA Extraction and Transcriptome Sequencing
2.12. Real-Time Quantitative Reverse Transcription PCR (RT-qPCR)
2.13. Statistical Analysis
3. Results and Discussion
3.1. The Experimental Gastric Acid Value
3.2. The Motility and Adhesion Capacity of V. parahaemolyticus
3.3. Determination of Gastric Acid Resistance of V. parahaemolyticus After Pre-Adaptation with Different Salinities
3.4. Growth of Vibrio parahaemolyticus on L. vannamei Samples
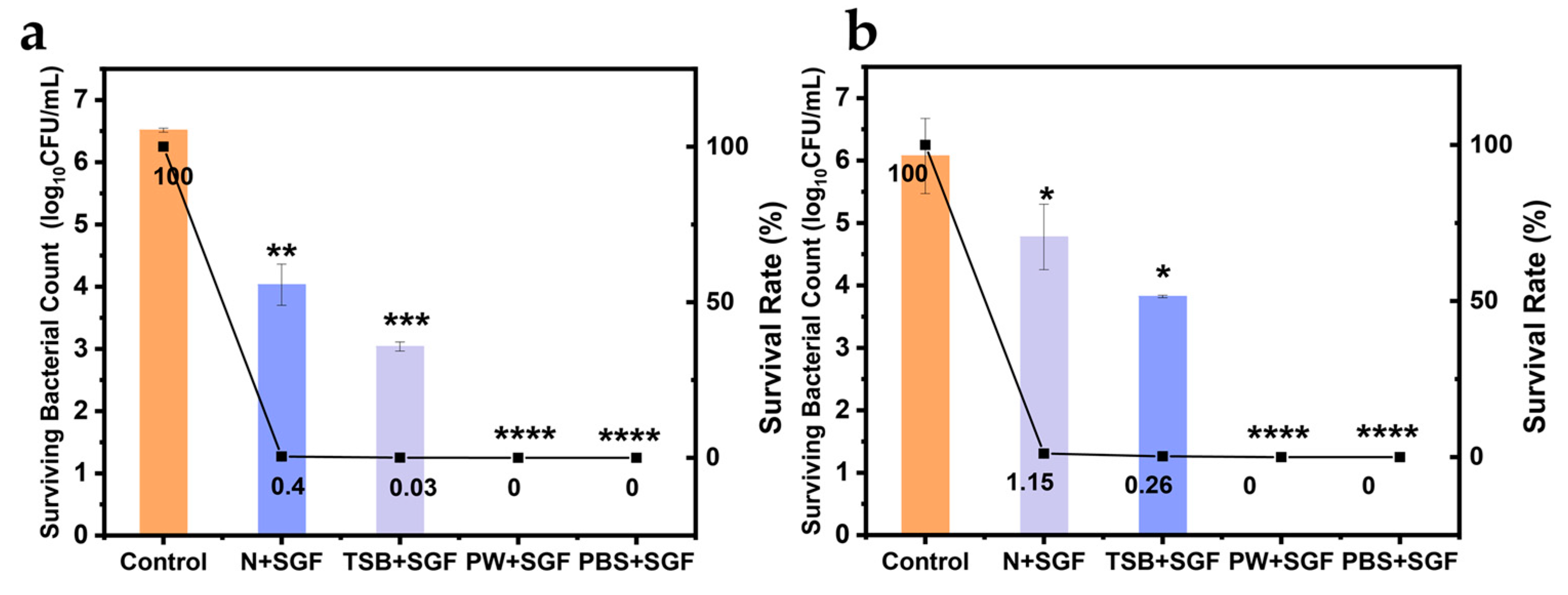
3.5. Effect of the Food Matrix on the Escape of V. parahaemolyticus from Gastric Acid Stress
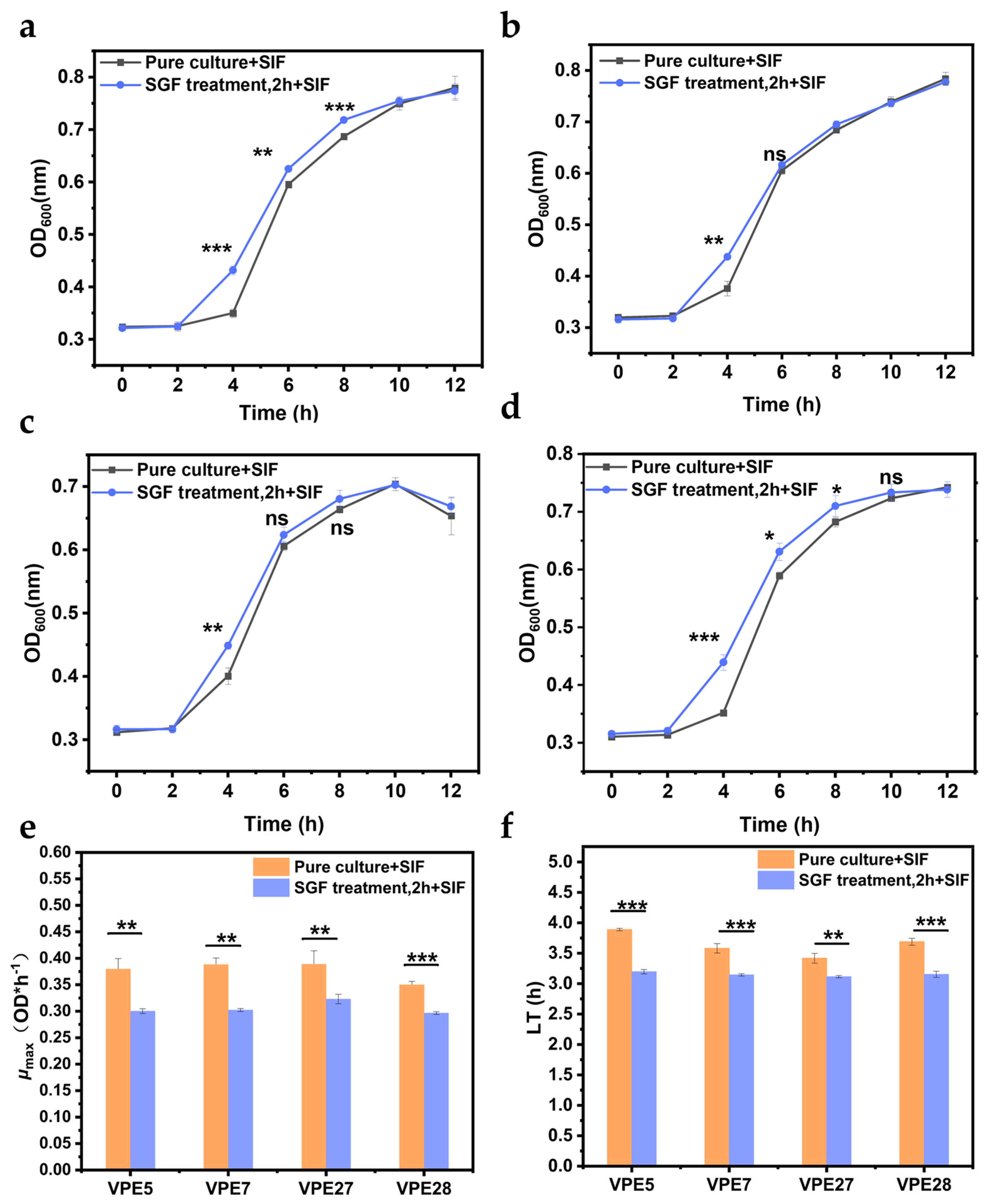
3.6. Determination of the Recovery Capacity of V. parahaemolyticus in Simulated Intestinal Fluid
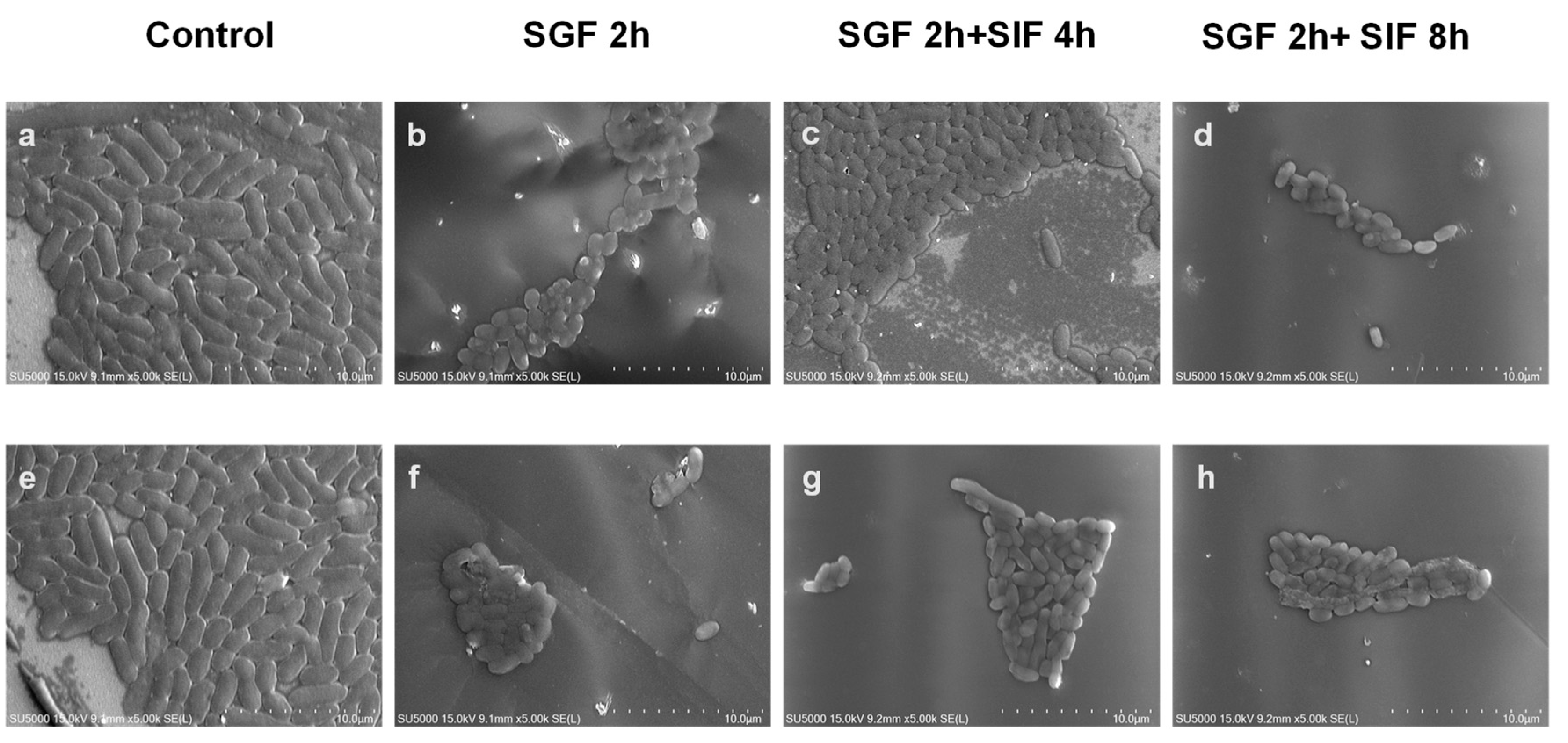
3.7. Changes in V. parahaemolyticus Morphology
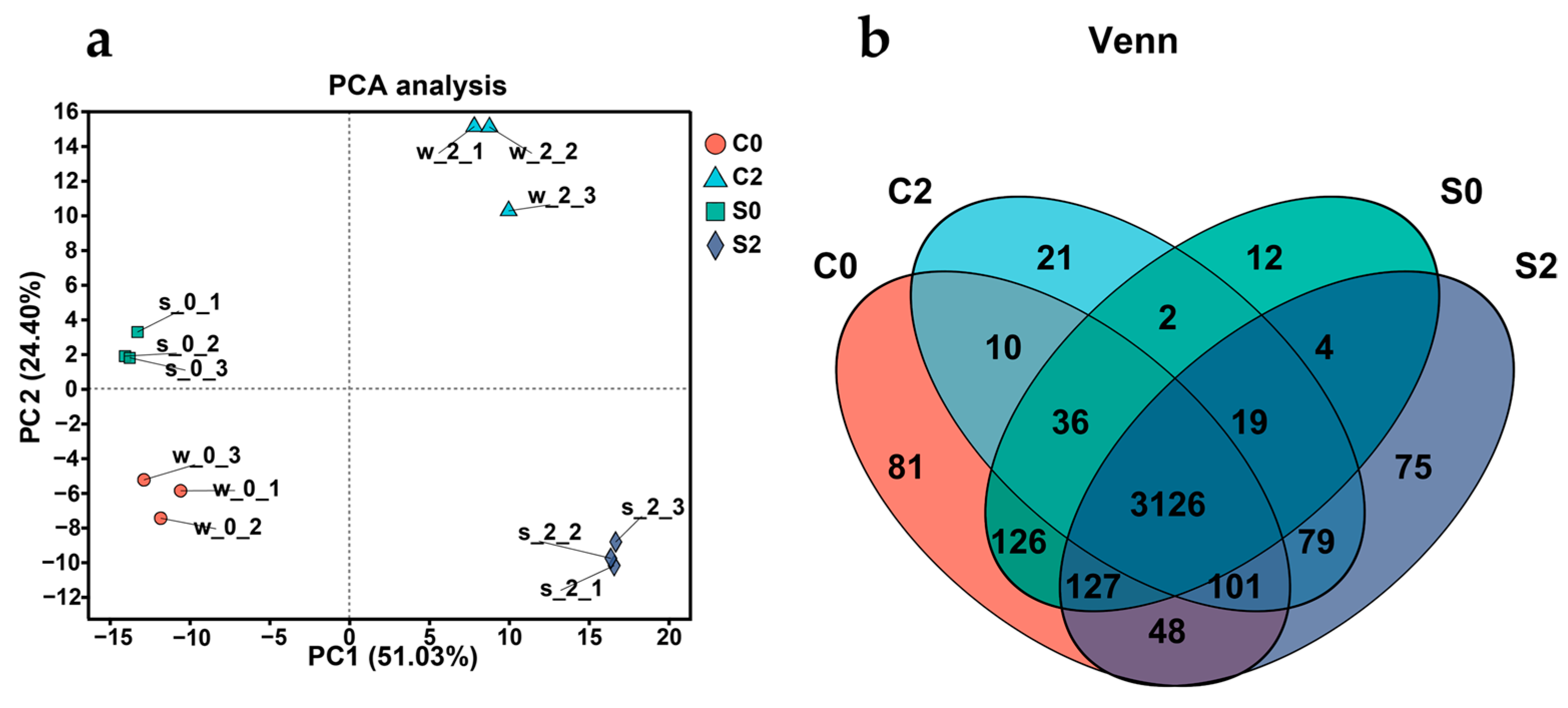
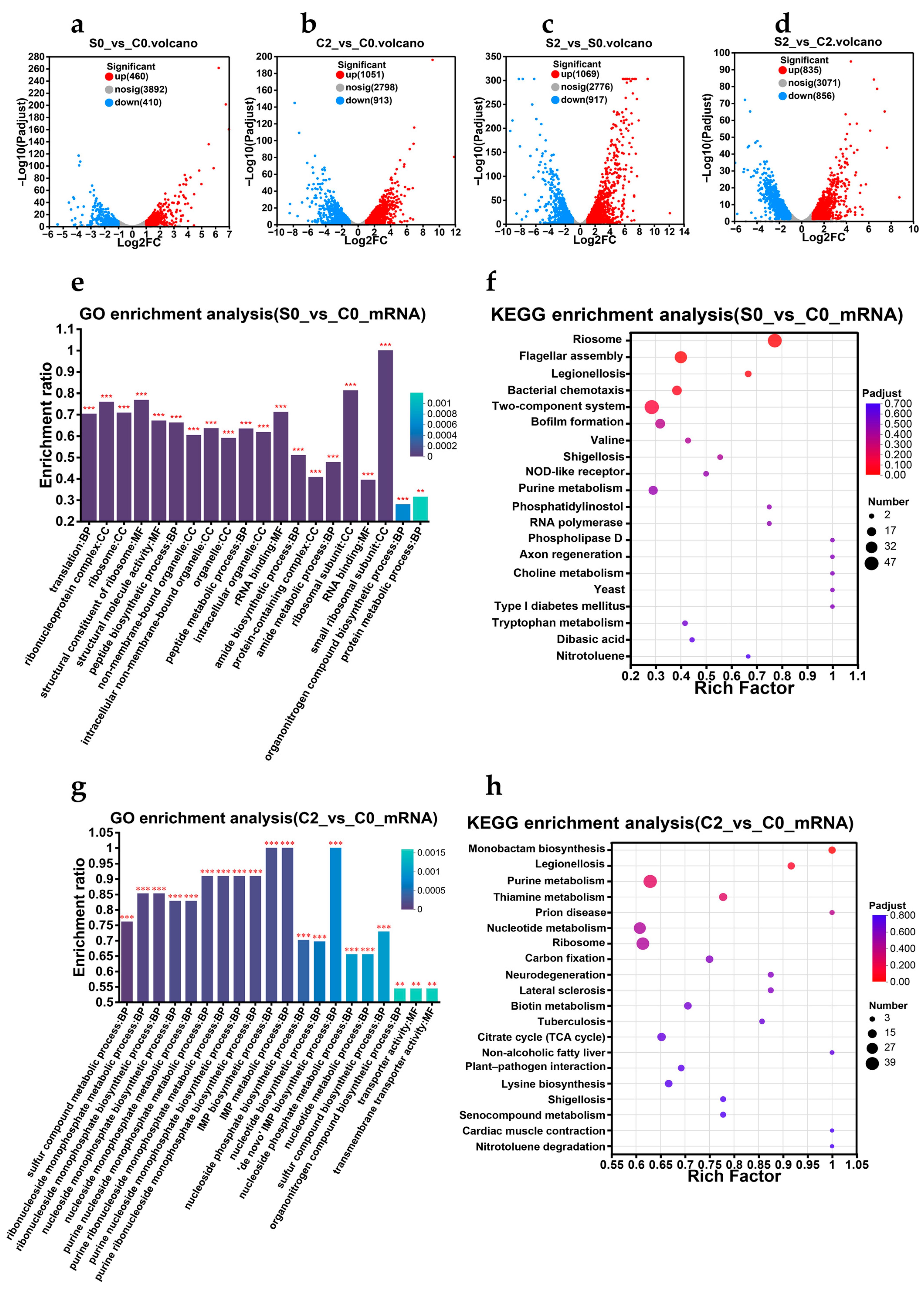
3.8. Transcriptomics Analysis
3.8.1. Exploration of Molecular Mechanisms
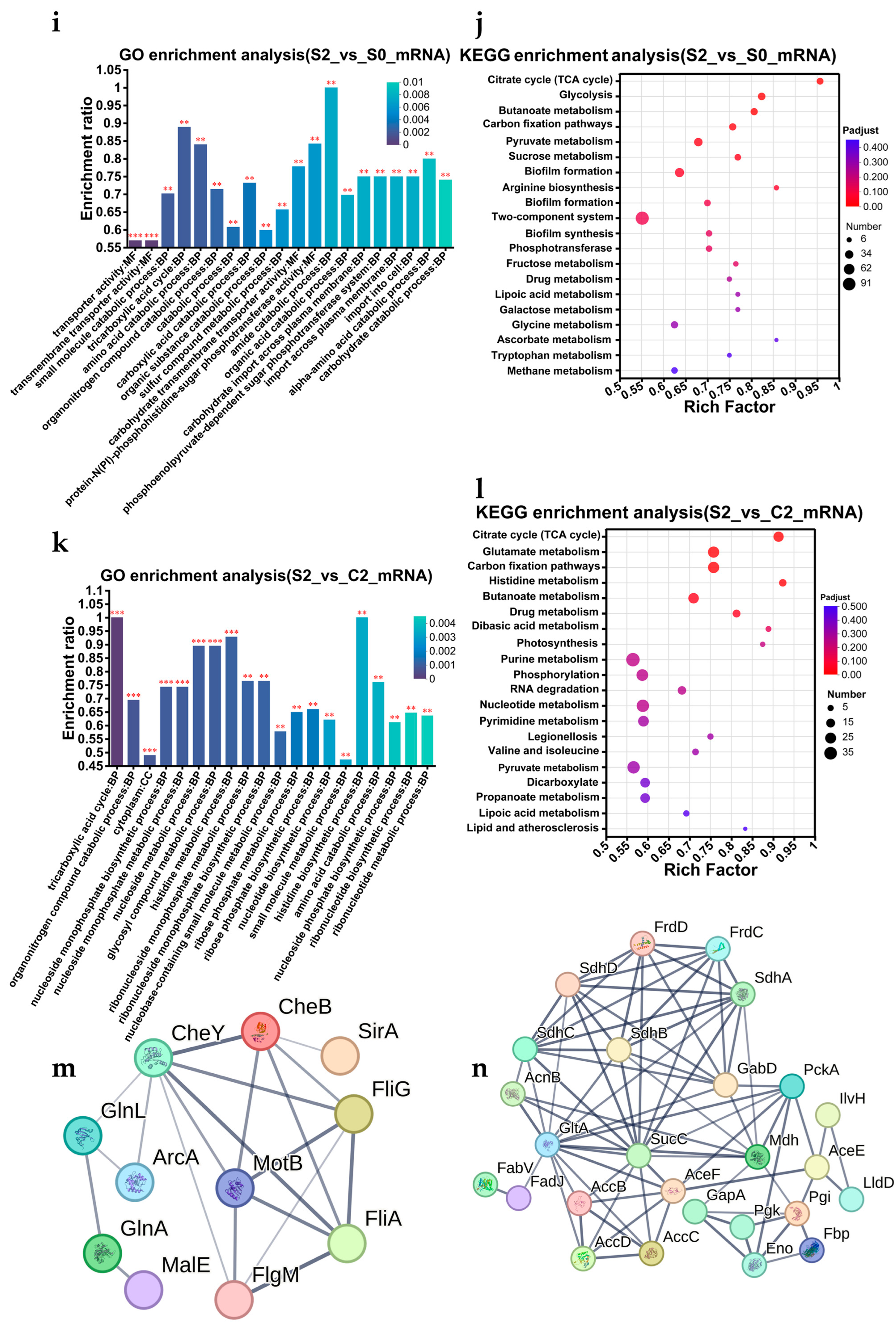
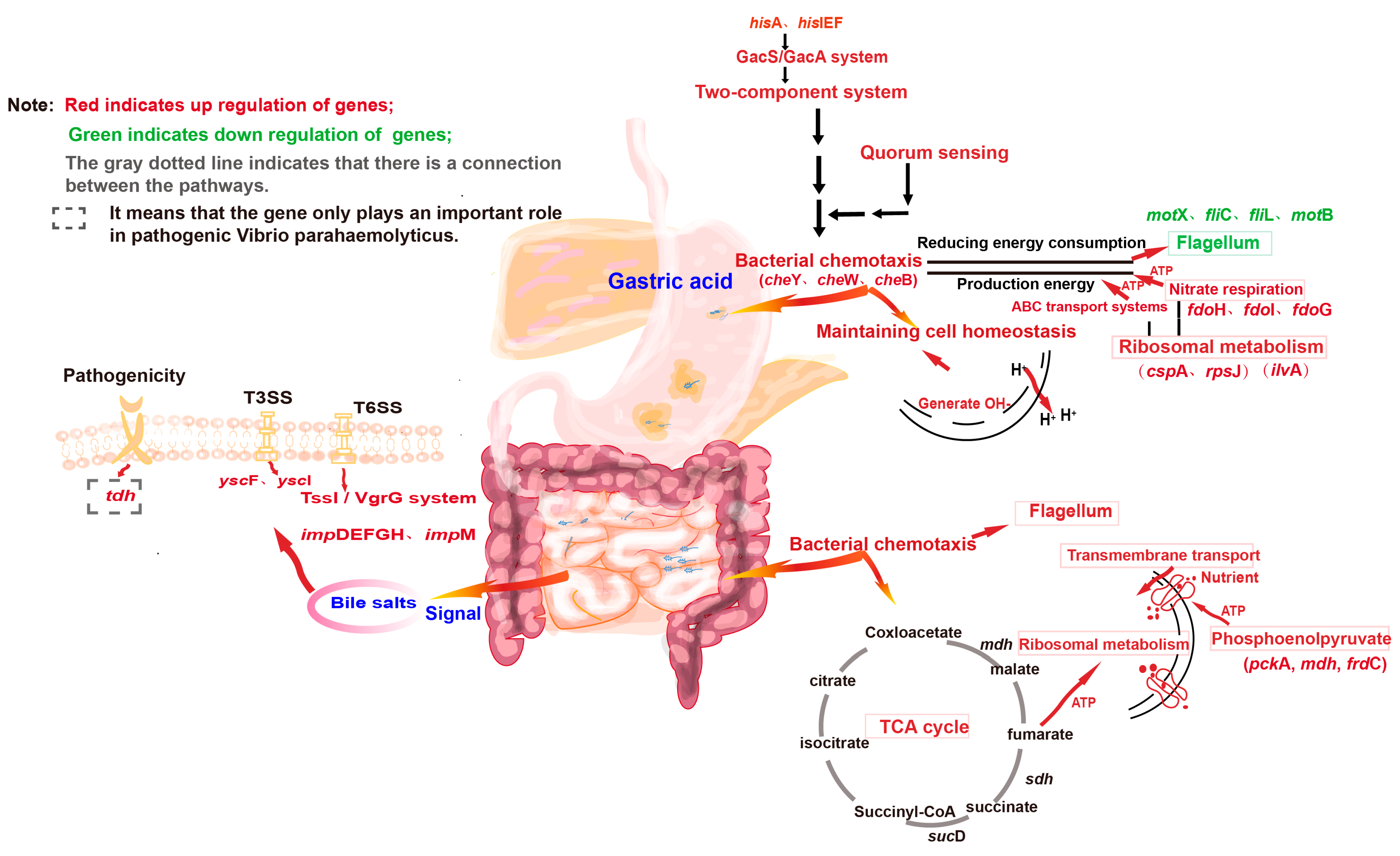
3.8.2. V. parahaemolyticus Actively Responds to Gastric Acid Stress Through Various Mechanisms and Prepares for Escaping from Stress at Any Time
3.8.3. V. parahaemolyticus That Pass the Gastric Acid Barrier Can Recover and Induce Pathogenicity
3.8.4. Regulatory Pathway Prediction
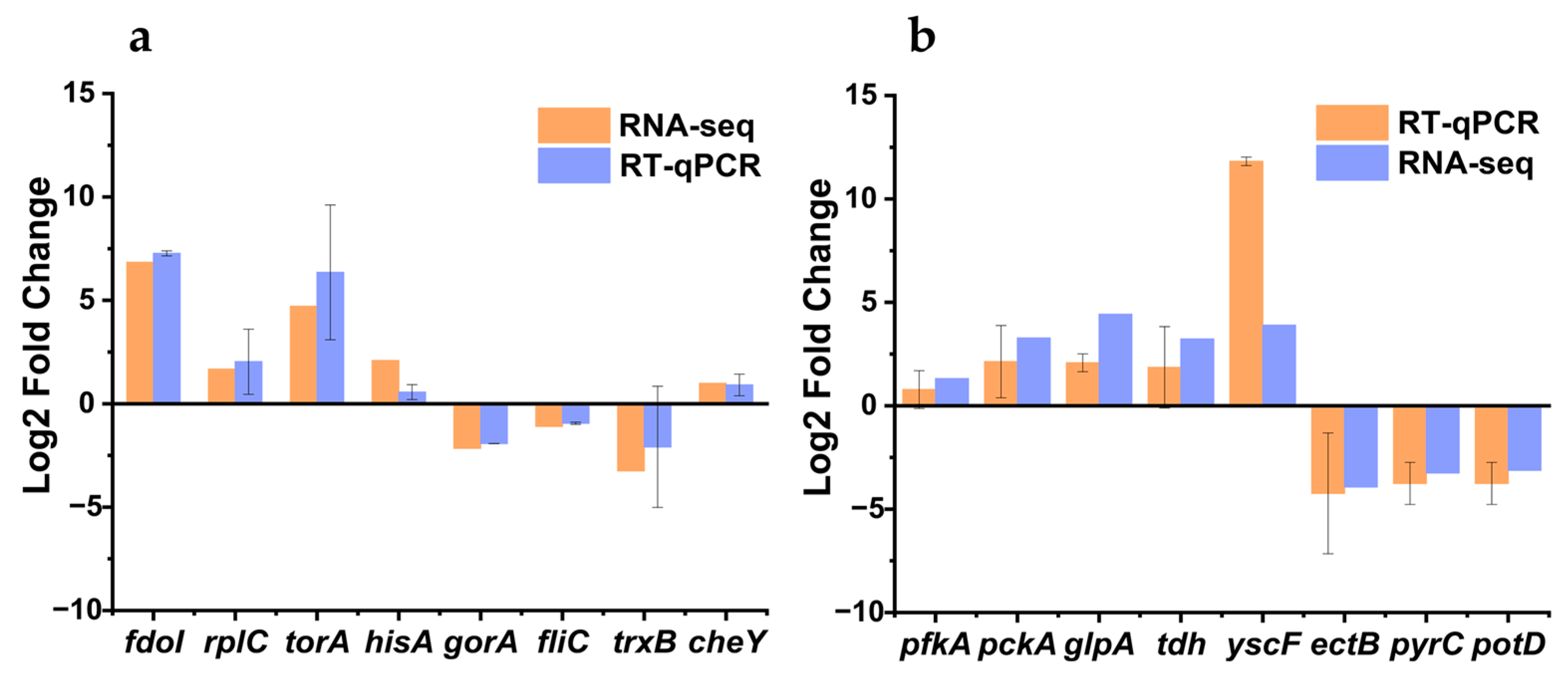
3.9. Validation of Transcriptome Data
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fung, F.; Wang, H.-S.; Menon, S. Food safety in the 21st century. Biomed. J. 2018, 41, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, Z.; Tong, W.H.; Ding, Y.M.; Liu, B.; Shi, Y.X.; Wang, J.C.; Sun, S.M.; Liu, M.; Wang, Y.H.; et al. An acid-tolerance response system protecting exponentially growing Escherichia coli. Nat. Commun. 2020, 11, 1496. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.; Cheng, J.; Sharma, D.; Bitzikos, O.; Gustafson, R.; Fyfe, M.; Greve, R.; Murti, M.; Stone, J.; Honish, L.; et al. Outbreak of Vibrio parahaemolyticus Associated with Consumption of Raw Oysters in Canada, 2015. Foodborne Pathog. Dis. 2018, 15, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Kanjee, U.; Houry, W.A. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 2013, 67, 65–81. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, D.-Y.; Li, D.; Lv, Y.; Chen, S.; Bai, F. Human gastric microbiota analysis of refractory H. pylori infection. Sci. Rep. 2024, 14, 15619. [Google Scholar] [CrossRef]
- Pienaar, J.A.; Singh, A.; Barnard, T.G. Membrane modification as a survival mechanism through gastric fluid in non-acid adapted enteropathogenic Escherichia coli (EPEC). Microb. Pathog. 2020, 144, 104180. [Google Scholar] [CrossRef]
- Wong, H.C.; Peng, P.Y.; Han, J.M.; Chang, C.Y.; Lan, S.L. Effect of mild acid treatment on the survival, enteropathogenicity, and protein production in Vibrio parahaemolyticus. Infect. Immun. 1998, 66, 3066–3071. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Z.; Malakar, P.K.; Pan, Y.; Zhao, Y. The Fate of Bacteria in Human Digestive Fluids: A New Perspective Into the Pathogenesis of Vibrio parahaemolyticus. Front. Microbiol. 2019, 10, 1614. [Google Scholar] [CrossRef]
- Kitamoto, S.; Nagao-Kitamoto, H.; Kuffa, P.; Kamada, N. Regulation of virulence: The rise and fall of gastrointestinal pathogens. J. Gastroenterol. 2016, 51, 195–205. [Google Scholar] [CrossRef]
- Trowers, E.; Tischler, M. Gastrointestinal Secretion: Aids in Digestion and Absorption. In Gastrointestinal Physiology: A Clinical Approach; Trowers, E., Tischler, M., Eds.; Springer: Cham, Switzerland, 2014; pp. 53–80. [Google Scholar]
- Wang, Y.; Zhao, Y.; Pan, Y.; Liu, H. Comparison on the Growth Variability of Vibrio parahaemolyticus Coupled With Strain Sources and Genotypes Analyses in Simulated Gastric Digestion Fluids. Front. Microbiol. 2020, 11, 212. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. Infogest static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.B.; Rybtke, M.; Tolker-Nielsen, T. The dynamics of biofilm development and dispersal should be taken into account when quantifying biofilm via the crystal violet microtiter plate assay. Biofilm 2024, 8, 100207. [Google Scholar] [CrossRef]
- Gao, R.; Liu, X.; Xiong, Z.; Wang, G.; Ai, L. Research progress on detection of foodborne pathogens: The more rapid and accurate answer to food safety. Food Res. Int. 2024, 193, 114767. [Google Scholar] [CrossRef]
- Yu, M.; Jiang, C.; Meng, Y.; Wang, F.; Qian, J.; Fei, F.; Yin, Z.; Zhao, W.; Zhao, Y.; Liu, H. Effect of low temperature on the resistance of Listeria monocytogenes and Escherichia coli O157:H7 to acid electrolyzed water. Food Res. Int. 2023, 168, 112776. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Chen, S.; Wang, H.; Zhang, J.; Xu, X.; Wang, H. Advances in understanding the predominance, phenotypes, and mechanisms of bacteria related to meat spoilage. Trends Food Sci. Technol. 2021, 118, 822–832. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, C.; Pu, Y.; Chen, S.; Liu, L.; Cui, Z.; Zhong, Y. Recent Advances in pH-Responsive Freshness Indicators Using Natural Food Colorants to Monitor Food Freshness. Foods 2022, 11, 1884. [Google Scholar] [CrossRef]
- Yang, H.; Meng, Y.; Han, X.; Meng, X.; Yang, B.; Zhang, C.; Wang, X.; Yu, J.; Al-Asmari, F.; Dablool, A.S.; et al. Changes in the ability of Listeria monocytogenes to resist thermal treatment and simulated gastric condition after exposure to sequential stresses in minced meat. Food Res. Int. 2024, 192, 114765. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Russell, T.L.; Berardi, R.R.; Barnett, J.L.; Dermentzoglou, L.C.; Jarvenpaa, K.M.; Schmaltz, S.P.; Dressman, J.B. Upper Gastrointestinal pH in Seventy-Nine Healthy, Elderly, North American Men and Women. Pharm. Res. 1993, 10, 187–196. [Google Scholar] [CrossRef]
- Hershey, D.M. Integrated control of surface adaptation by the bacterial flagellum. Curr. Opin. Microbiol. 2021, 61, 1–7. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, X.; Ji, L.; Yan, W.; Chen, L.; Lu, Z.; Xu, D.; Zha, Y.; Xu, D.; Dong, F. Prevalence and virulence of Vibrio parahaemolyticus isolated from clinical and environmental samples in Huzhou, China. BMC Genomics 2024, 25, 1187. [Google Scholar] [CrossRef]
- Chen, D.; Wang, Z.; Li, X.; Du, H.; Zhang, K.; Cao, S.; Lu, J.; Zhao, S.; Wang, H.; Li, Y. Biological properties of Vibrio parahaemolyticus lytic phages and transcriptome analysis of their interactions with the host. Aquac. Rep. 2024, 39, 102450. [Google Scholar] [CrossRef]
- Zheng, J.; Shi, B.; Sun, J.; Pan, Y.; Ding, Y.; Shi, X.; Zhang, J.; Zhang, H.; He, J.; Zhang, K.; et al. Global phylogeography and genomic characterization of Vibrio parahaemolyticus infections in Jilin province, China (2016–2022). Int. J. Food Microbiol. 2025, 428, 110993. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Tabassum, N.; Anand, R.; Kim, Y.-M. Motility of Vibrio spp.: Regulation and controlling strategies. Appl. Microbiol. Biotechnol. 2020, 104, 8187–8208. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Gao, M.; Li, X.; Zhong, X.; Zhang, X.; Zhou, X.; Yang, M. Vibrio parahaemolyticus Down-Regulates the Intracellular c-di-GMP Level to Promote Swarming Motility by Sensing Surface. Curr. Microbiol. 2024, 82, 32. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F.; Boziaris, I.S. Fish, Antibiotic Resistance, and Food Safety at the Global Level. In Handbook of Public Health Nutrition: International, National, and Regional Perspectives; Preedy, V.R., Patel, V.B., Eds.; Springer: Cham, Switzerland, 2025; pp. 1–21. [Google Scholar]
- Okaiyeto, S.A.; Sutar, P.P.; Chen, C.; Ni, J.-B.; Wang, J.; Mujumdar, A.S.; Zhang, J.-S.; Xu, M.-Q.; Fang, X.-M.; Zhang, C.; et al. Antibiotic resistant bacteria in food systems: Current status, resistance mechanisms, and mitigation strategies. Agr. Commun. 2024, 2, 100027. [Google Scholar] [CrossRef]
- Anupama, K.P.; Deeksha, K.; Deeksha, A.; Karunasagar, I.; Karunasagar, I.; Maiti, B. Comparative performance of TCBS and TSA for the enumeration of trh+ Vibrio parahaemolyticus by direct colony hybridization. J. Microbiol. Methods 2019, 157, 37–42. [Google Scholar] [CrossRef]
- Han, S.-J.; Kim, D.-K. Synergistic effect of naringenin and mild heat for inactivation of E. coli O157:H7, S. Typhimurium, L. monocytogenes, and S. aureus in peptone water and cold brew coffee. Int. J. Food Microbiol. 2025, 430, 111051. [Google Scholar] [CrossRef]
- Han, A.; Paek, J.; Lee, S.-Y. Thermal resistance of Escherichia coli O157:H7 in laboratory media, milk, and beef extracts during non-isothermal processing at various heating rates. Food Microbiol. 2023, 110, 104187. [Google Scholar] [CrossRef]
- Zhang, K.; Sze, C.W.; Zhao, H.; Liu, J.; Li, C. Borrelia burgdorferi serine protease HtrA is a pleiotropic regulator of stress response, motility, flagellar hemostasis, and infectivity. Commun. Biol. 2025, 8, 341. [Google Scholar] [CrossRef]
- Flores-Villaseñor, H.; Velázquez-Román, J.; León-Sicairos, N.; Angulo-Zamudio, U.A.; Lira-Morales, C.; Martínez-García, J.J.; Acosta-Smith, E.; Valdés-Flores, J.; Tapia-Pastrana, G.; Canizalez-Román, A. Serodiversity, antibiotic resistance, and virulence genes of Vibrio parahaemolyticus in oysters collected in coastal areas of northwestern Mexico between 2012 and 2020. Food Microbiol. 2024, 123, 104567. [Google Scholar] [CrossRef]
- Giannakara, M.; Koumandou, V.L. Evolution of two-component quorum sensing systems. Access Microbiol 2022, 4, 000303. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.F.; Georgellis, D. Environmental adaptation and diversification of bacterial two-component systems. Curr. Opin. Microbiol. 2023, 76, 102399. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, Y.; Suh, D.H.; Lee, C.H.; Yoo, S.M.; Lee, S.Y.; Yoon, S.H. Heat-responsive and time-resolved transcriptome and metabolome analyses of Escherichia coli uncover thermo-tolerant mechanisms. Sci. Rep. 2020, 10, 17715. [Google Scholar] [CrossRef] [PubMed]
- Bickle, T.A.; Howard, G.A.; Traut, R.R. Ribosome Heterogeneity: The Nonuniform Distribution of Specific Ribosomal Proteins Among Different Functional Classes of Ribosomes. J. Biol. Chem. 1973, 248, 4862–4864. [Google Scholar] [CrossRef]
- Moll, I.; Engelberg-Kulka, H. Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 2012, 37, 493–498. [Google Scholar] [CrossRef]
- Dai, X.; Zhu, M.; Warren, M.; Balakrishnan, R.; Patsalo, V.; Okano, H.; Williamson, J.R.; Fredrick, K.; Wang, Y.-P.; Hwa, T. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016, 2, 16231. [Google Scholar] [CrossRef]
- MacLean, A.; Legendre, F.; Appanna, V.D. The tricarboxylic acid (TCA) cycle: A malleable metabolic network to counter cellular stress. Crit. Rev. Biochem. Mol. Biol. 2023, 58, 81–97. [Google Scholar] [CrossRef]
- Ahmad, M.; Aduru, S.V.; Smith, R.P.; Zhao, Z.; Lopatkin, A.J. The role of bacterial metabolism in antimicrobial resistance. Nat. Rev. Microbiol. 2025, 136, 1–16. [Google Scholar] [CrossRef]
- Clifton, B.E.; Alcolombri, U.; Uechi, G.-I.; Jackson, C.J.; Laurino, P. The ultra-high affinity transport proteins of ubiquitous marine bacteria. Nature 2024, 634, 721–728. [Google Scholar] [CrossRef]
- Plaza, N.; Pérez-Reytor, D.; Corsini, G.; García, K.; Urrutia, Í.M. Contribution of the Type III Secretion System (T3SS2) of Vibrio parahaemolyticus in Mitochondrial Stress in Human Intestinal Cells. Microorganisms 2024, 12, 813. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhang, Y. Structure, function and regulation of the thermostable direct hemolysin (TDH) in pandemic Vibrio parahaemolyticus. Microb. Pathog. 2018, 123, 242–245. [Google Scholar] [CrossRef]
- Habich, A.; Chaves Vargas, V.; Robinson, L.A.; Allsopp, L.P.; Unterweger, D. Distribution of the four type VI secretion systems in Pseudomonas aeruginosa and classification of their core and accessory effectors. Nat. Commun. 2025, 16, 888. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Meng, H.; Gu, D.; Li, Y.; Jia, M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 2019, 222, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Kaval, K.G.; Chimalapati, S.; Siegel, S.D.; Garcia, N.; Jaishankar, J.; Dalia, A.B.; Orth, K. Membrane-localized expression, production and assembly of Vibrio parahaemolyticus T3SS2 provides evidence for transertion. Nat. Commun. 2023, 14, 1178. [Google Scholar] [CrossRef]
- Zou, A.J.; Kinch, L.; Chimalapati, S.; Garcia, N.; Tomchick, D.R.; Orth, K. Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus. J. Biol. Chem. 2023, 299, 104591. [Google Scholar] [CrossRef]
- Pazhani, G.P.; Chowdhury, G.; Ramamurthy, T. Adaptations of Vibrio parahaemolyticus to Stress During Environmental Survival, Host Colonization, and Infection. Front. Microbiol. 2021, 12, 737299. [Google Scholar] [CrossRef]
- Gonçalves, N.S.; Ghaem-Maghami, M.; Monteleone, G.; Frankel, G.; Dougan, G.; Lewis, D.J.; Simmons, C.P.; MacDonald, T.T. Critical role for tumor necrosis factor alpha in controlling the number of lumenal pathogenic bacteria and immunopathology in infectious colitis. Infect. Immun. 2001, 69, 6651–6659. [Google Scholar] [CrossRef]
- Yu, X.; Liang, Y.; Zhou, Y.; He, L.; Liu, Y.; Fu, L.; Lin, H.; Zhang, Y.; Lu, D. 23S rRNA from Vibrio parahaemolyticus regulates the innate immune response via recognition by TLR13 in orange-spotted grouper (Epinephelus coioides). Dev. Comp. Immunol. 2021, 114, 103837. [Google Scholar] [CrossRef]
| Gene | Sequence (5′ to 3′) |
|---|---|
| 16S rRNA-F | TATCCTTGTTTGCCAGCGAG |
| 16S rRNA-R | CTACGACGCACTTTTTGGGA |
| fdoI-F | GCGTTAGAAGGCATGGTGACTG |
| fdoI-R | TGTTTCGCCCATCGCTTCTTTAG |
| rplC-F | CTGGTACATCTAAGGGTAAAGGTTTCC |
| rplC-R | GAGCACGGTGAGACAATGAGTTAC |
| torA-F | TTGTTCCAATCTAAGCCTGACTTCC |
| torA-R | ATCCACTCCATCTCGCTCATACC |
| hisA -F | TCTTCAGGTGGTATCGGCTCATTAG |
| hisA-R | TGCCTCCTCTGCTGTGAACTTAC |
| cheY-F | CCTGCTTCGTGACTTGGGTTTC |
| cheY-R | TGGCATATTCCAGTCGGTTACAAC |
| gorA-F | CGCAGGCTACATCGCAGTTG |
| gorA-R | GCTACGCAGTGGTGACTCTTTAC |
| trxB-F | TCGGCGGCGGTAACACAG |
| trxB-R | ATCTTCTCAGCACGGAATGAATCAC |
| fliC-F | GATCATTGATGCGGCACTGAAATAC |
| fliC-R | CGTTCTCGTTAATGTTGTCCAAGTTG |
| pfkA-F | CACTGTGGTGACCTAACTCTGATG |
| pfkA-R | GCGATGCCGTCTTGGATGTTG |
| pckA-F | GGATGACGATGGTGTCTTCAACTTC |
| pckA-R | TGTAGATGTCTGGCTCCGCTTC |
| glpA-F | GCAAGCGATCACACGAGACTAC |
| glpA-R | TGCCACCGAATACTGACAACAATG |
| tdh-F | TTGAAGATGTAATGGCTGAACTAGGC |
| tdh-R | CGACCACCATGATTCATTGTTGTTAG |
| vscF-F | AACTTCAGCACGCTATCAACAAATG |
| vscF-R | AGGATCGACTGCATCACATCTTTG |
| ectB-F | CTCTTGCGGACGGTCAGTTG |
| ectB-R | TAAGTGGTGGAAGGAAGCGAATAAC |
| pyrC-F | CCTCGCTCCTATCGTTAATGACTTC |
| pyrC-R | ATGGTAGCGGCAACATTATCAGAC |
| potD-F | CTACCTAACCGCCGTTACAGAATC |
| potD-R | CACCAACCGAGTCTTGCCATTC |
| pH of the Gastric Acid | The Bacterial Count (log10 CFU/mL) | |||
|---|---|---|---|---|
| VPE5 | VPE7 | VPE27 | VPE28 | |
| pH 2.5 | 0 | 0 | 0 | 0 |
| pH 3.5 | 0 | 0 | 6.05 ± 0.08 | 7.71 ± 0.08 |
| pH 4.5 | 7.86 ± 0.02 | 8.22 ± 0.04 | 7.76 ± 0.20 | 8.03 ± 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, S.; Jiang, J.; Song, Z.; Zhou, Y.; Chen, L.; Tang, S.; Pan, Y.; Zhao, Y.; Liu, H. Transcriptome Sequencing Reveals Survival Strategies and Pathogenic Potential of Vibrio parahaemolyticus Under Gastric Acid Stress. Biology 2025, 14, 396. https://doi.org/10.3390/biology14040396
Ji S, Jiang J, Song Z, Zhou Y, Chen L, Tang S, Pan Y, Zhao Y, Liu H. Transcriptome Sequencing Reveals Survival Strategies and Pathogenic Potential of Vibrio parahaemolyticus Under Gastric Acid Stress. Biology. 2025; 14(4):396. https://doi.org/10.3390/biology14040396
Chicago/Turabian StyleJi, Shiying, Jinlin Jiang, Zhiyong Song, Yu Zhou, Lu Chen, Shiying Tang, Yingjie Pan, Yong Zhao, and Haiquan Liu. 2025. "Transcriptome Sequencing Reveals Survival Strategies and Pathogenic Potential of Vibrio parahaemolyticus Under Gastric Acid Stress" Biology 14, no. 4: 396. https://doi.org/10.3390/biology14040396
APA StyleJi, S., Jiang, J., Song, Z., Zhou, Y., Chen, L., Tang, S., Pan, Y., Zhao, Y., & Liu, H. (2025). Transcriptome Sequencing Reveals Survival Strategies and Pathogenic Potential of Vibrio parahaemolyticus Under Gastric Acid Stress. Biology, 14(4), 396. https://doi.org/10.3390/biology14040396






