The Main Factors Affecting the Minimum Sampling Area Determination Method: Based on Research of the Shrub Layer in Island Pinus massoniana Forests
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Area Overview
2.2. Community Survey
2.3. Species–Area Relationship Models and Minimum Sampling Area Equations
2.4. Evaluation of Fit for Species–Area Relationship Models and Determination of Minimum Sampling Area
3. Results
3.1. Species–Area Relationship in the Shrub Layer
3.2. Minimum Sampling Area in the Shrub Layer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Sample Site No. | Longitude | Latitude | Elevation/(m) | Slope Aspect | Slope/(°) | Slope Position | Crown Density |
|---|---|---|---|---|---|---|---|
| S1 | 119°40′47.66″ | 26°39′07.25″ | 42.4 | SE160 | 30 | Middle | 0.75 |
| S2 | 119°40′47.12″ | 26°39′09.55″ | 81.1 | SE160 | 30 | Up | 0.75 |
| S3 | 119°39′33.26″ | 26°39′53.18″ | 178.4 | N353 | 30 | Up | 0.80 |
| S4 | 119°39′20.11″ | 26°39′51.20″ | 71.9 | NW312 | 30 | Middle | 0.75 |
| S5 | 119°39′32.59″ | 26°39′57.99″ | 123.2 | W261 | 37 | Up | 0.68 |
| S6 | 119°40′22.00″ | 26°39′43.00″ | 175.5 | SE155 | 32 | Middle | 0.80 |
| S7 | 119°40′16.47″ | 26°40′10.93″ | 40.7 | NW289 | 24 | Low | 0.75 |
| S8 | 119°40′10.60″ | 26°40′07.14″ | 96.0 | NE23 | 23 | Low | 0.80 |
Appendix B
| Sample Site No. | Quadrat No. | Number of Species in Different Sampled Areas | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.25 m2 | 1 m2 | 4 m2 | 9 m2 | 16 m2 | 25 m2 | 50 m2 | 100 m2 | 200 m2 | 400 m2 | ||
| S1 | 1 | 4 | 8 | 15 | 18 | 19 | 20 | 21 | 23 | 31 | 38 |
| 2 | 4 | 8 | 15 | 18 | 19 | 20 | 20 | 23 | 29 | 38 | |
| 3 | 3 | 7 | 9 | 16 | 18 | 19 | 20 | 25 | 31 | 38 | |
| 4 | 3 | 7 | 9 | 16 | 18 | 19 | 21 | 25 | 30 | 38 | |
| 5 | 1 | 2 | 5 | 9 | 10 | 11 | 14 | 18 | 30 | 38 | |
| 6 | 1 | 2 | 5 | 9 | 10 | 11 | 13 | 18 | 30 | 38 | |
| 7 | 2 | 4 | 6 | 8 | 11 | 13 | 18 | 25 | 29 | 38 | |
| 8 | 2 | 4 | 6 | 8 | 11 | 13 | 19 | 25 | 30 | 38 | |
| Mean | 2.50 | 5.25 | 8.75 | 12.75 | 14.50 | 15.75 | 18.25 | 22.75 | 30.00 | 38.00 | |
| S2 | 1 | 1 | 3 | 5 | 7 | 9 | 9 | 11 | 13 | 24 | 27 |
| 2 | 1 | 3 | 5 | 7 | 9 | 9 | 13 | 13 | 18 | 27 | |
| 3 | 2 | 5 | 7 | 10 | 13 | 13 | 16 | 22 | 24 | 27 | |
| 4 | 2 | 5 | 7 | 10 | 13 | 13 | 17 | 22 | 23 | 27 | |
| 5 | 0 | 2 | 3 | 4 | 6 | 8 | 10 | 11 | 23 | 27 | |
| 6 | 0 | 2 | 3 | 4 | 6 | 8 | 9 | 10 | 16 | 27 | |
| 7 | 1 | 2 | 3 | 6 | 9 | 9 | 10 | 11 | 18 | 27 | |
| 8 | 1 | 2 | 3 | 6 | 9 | 9 | 10 | 11 | 16 | 27 | |
| Mean | 1.00 | 3.00 | 4.50 | 6.75 | 9.25 | 9.75 | 12.00 | 14.13 | 20.25 | 27.00 | |
| S3 | 1 | 2 | 4 | 6 | 10 | 13 | 17 | 23 | 35 | 39 | 52 |
| 2 | 2 | 4 | 6 | 10 | 13 | 17 | 14 | 35 | 44 | 52 | |
| 3 | 2 | 6 | 14 | 19 | 20 | 22 | 25 | 28 | 39 | 52 | |
| 4 | 2 | 6 | 14 | 19 | 20 | 22 | 26 | 28 | 42 | 52 | |
| 5 | 1 | 3 | 5 | 8 | 13 | 19 | 28 | 36 | 42 | 52 | |
| 6 | 1 | 3 | 5 | 8 | 13 | 19 | 27 | 36 | 49 | 52 | |
| 7 | 2 | 6 | 11 | 17 | 20 | 23 | 31 | 40 | 44 | 52 | |
| 8 | 2 | 6 | 11 | 17 | 20 | 23 | 31 | 40 | 49 | 52 | |
| Mean | 1.75 | 4.75 | 9.00 | 13.50 | 16.50 | 20.25 | 25.63 | 34.75 | 43.50 | 52.00 | |
| S4 | 1 | 1 | 7 | 9 | 14 | 15 | 17 | 21 | 29 | 37 | 52 |
| 2 | 1 | 7 | 9 | 14 | 15 | 17 | 23 | 29 | 45 | 52 | |
| 3 | 2 | 4 | 7 | 10 | 13 | 17 | 20 | 24 | 37 | 52 | |
| 4 | 2 | 4 | 7 | 10 | 13 | 17 | 20 | 24 | 34 | 52 | |
| 5 | 4 | 5 | 7 | 12 | 13 | 13 | 17 | 22 | 34 | 52 | |
| 6 | 4 | 5 | 7 | 12 | 13 | 13 | 17 | 22 | 37 | 52 | |
| 7 | 2 | 3 | 7 | 11 | 15 | 18 | 23 | 32 | 45 | 52 | |
| 8 | 2 | 3 | 7 | 11 | 15 | 18 | 27 | 32 | 37 | 52 | |
| Mean | 2.25 | 4.75 | 7.50 | 11.75 | 14.00 | 16.25 | 21.00 | 26.75 | 38.25 | 52.00 | |
| S5 | 1 | 1 | 2 | 4 | 7 | 8 | 9 | 15 | 17 | 37 | 41 |
| 2 | 1 | 2 | 4 | 7 | 8 | 9 | 11 | 17 | 24 | 41 | |
| 3 | 2 | 4 | 8 | 10 | 15 | 22 | 23 | 34 | 37 | 41 | |
| 4 | 2 | 4 | 8 | 10 | 15 | 22 | 31 | 34 | 38 | 41 | |
| 5 | 4 | 7 | 14 | 15 | 17 | 19 | 21 | 23 | 38 | 41 | |
| 6 | 4 | 7 | 14 | 15 | 17 | 19 | 19 | 23 | 37 | 41 | |
| 7 | 1 | 3 | 6 | 6 | 10 | 14 | 16 | 17 | 24 | 41 | |
| 8 | 1 | 3 | 6 | 6 | 10 | 14 | 14 | 17 | 37 | 41 | |
| Mean | 2.00 | 4.00 | 8.00 | 9.50 | 12.50 | 16.00 | 18.75 | 22.75 | 34.00 | 41.00 | |
| S6 | 1 | 1 | 2 | 8 | 13 | 16 | 22 | 25 | 29 | 41 | 50 |
| 2 | 1 | 2 | 8 | 13 | 16 | 22 | 25 | 29 | 40 | 50 | |
| 3 | 3 | 6 | 8 | 11 | 12 | 14 | 20 | 25 | 36 | 50 | |
| 4 | 3 | 6 | 8 | 11 | 12 | 14 | 17 | 25 | 40 | 50 | |
| 5 | 3 | 6 | 8 | 11 | 12 | 14 | 19 | 24 | 40 | 50 | |
| 6 | 3 | 6 | 8 | 11 | 12 | 14 | 18 | 24 | 44 | 50 | |
| 7 | 1 | 3 | 6 | 8 | 9 | 14 | 20 | 29 | 40 | 50 | |
| 8 | 1 | 3 | 6 | 8 | 9 | 14 | 21 | 29 | 41 | 50 | |
| Mean | 2.00 | 4.25 | 7.50 | 10.75 | 12.25 | 16.00 | 20.63 | 26.75 | 40.25 | 50.00 | |
| S7 | 1 | 2 | 3 | 8 | 9 | 14 | 20 | 24 | 26 | 29 | 42 |
| 2 | 2 | 3 | 8 | 9 | 14 | 20 | 23 | 26 | 35 | 42 | |
| 3 | 3 | 5 | 8 | 10 | 12 | 18 | 23 | 25 | 29 | 42 | |
| 4 | 3 | 5 | 8 | 10 | 12 | 18 | 19 | 25 | 36 | 42 | |
| 5 | 2 | 6 | 10 | 14 | 19 | 24 | 27 | 33 | 36 | 42 | |
| 6 | 2 | 6 | 10 | 14 | 19 | 24 | 27 | 33 | 41 | 42 | |
| 7 | 1 | 3 | 6 | 8 | 11 | 16 | 23 | 30 | 35 | 42 | |
| 8 | 1 | 3 | 6 | 8 | 11 | 16 | 22 | 30 | 41 | 42 | |
| Mean | 2.00 | 4.25 | 8.00 | 10.25 | 14.00 | 19.50 | 23.50 | 28.50 | 35.25 | 42.00 | |
| S8 | 1 | 1 | 2 | 4 | 6 | 8 | 10 | 12 | 20 | 28 | 41 |
| 2 | 1 | 2 | 4 | 6 | 8 | 10 | 16 | 20 | 33 | 41 | |
| 3 | 3 | 4 | 5 | 8 | 10 | 11 | 15 | 17 | 29 | 41 | |
| 4 | 3 | 4 | 5 | 8 | 10 | 11 | 15 | 17 | 28 | 41 | |
| 5 | 2 | 6 | 7 | 9 | 12 | 15 | 18 | 23 | 29 | 41 | |
| 6 | 2 | 6 | 7 | 9 | 12 | 15 | 19 | 23 | 35 | 41 | |
| 7 | 4 | 6 | 10 | 12 | 12 | 13 | 18 | 25 | 33 | 41 | |
| 8 | 4 | 6 | 10 | 12 | 12 | 13 | 20 | 25 | 35 | 41 | |
| Mean | 2.50 | 4.50 | 6.50 | 8.75 | 10.50 | 12.25 | 16.63 | 21.25 | 31.25 | 41.00 | |
| Grand mean | 2.00 | 4.34 | 7.47 | 10.50 | 12.94 | 15.72 | 19.55 | 24.70 | 34.09 | 42.88 | |
References
- Zhang, J.T. Quantitative Ecology, 2nd ed.; Science Press: Beijing, China, 2011; pp. 51–57. [Google Scholar]
- Song, Y.C. Vegetation Ecology, 2nd ed.; Higher Education Press: Beijing, China, 2017; pp. 44–45, 73–84. [Google Scholar]
- Keeley, J.E. Relating species abundance distributions to species area curves in two Mediterranean-type shrublands. Divers. Distrib. 2003, 9, 253–259. [Google Scholar] [CrossRef]
- Yang, Z.S.; Li, Y.X.; Liu, W.; Wu, T.D.; Wang, Z.G. Fitting of species-area relationship and determination of minimum sampling area for plant communities on barren hillsides of arid valleys in the upper reaches of the Minjiang River. J. Ecol. Rural Environ. 2010, 26, 227–230. [Google Scholar] [CrossRef]
- Liu, Z.L.; Sun, X.; Gao, J.L.; Ye, X.H. Determination of minimum sampling area for Artemisia ordosica community in Ordos Plateau, Inner Mongolia. Res. Soil Water Conserv. 2013, 20, 185–189+195. [Google Scholar]
- Jiang, J.; Zhang, C.Y.; Zhao, X.H. Plant species-area relationship in a 42 hm2 research plot of coniferous and board-leaved mixed forest in Jiaohe, Jilin Province, China. Chin. J. Plant Ecol. 2012, 36, 30–38. [Google Scholar] [CrossRef]
- Yang, L.M.; Han, M.; Zhao, S.C. Studies on sampling intensity of plant diversity of grassland communities in the Songnen Plain of China. J. Jilin Agric. Univ. 1996, 18, 33–36. [Google Scholar] [CrossRef]
- Wang, X.L.; Wang, A.; Shi, H.H.; Peng, S.T.; Gong, L.P.; Qin, X.B. Carbon storage of Pinus thunbergii and Robinia pseudoacacia plantations on Nanchangshan Island, Changdao County of Shandong Province, China. Chin. J. Appl. Ecol. 2013, 24, 1263–1268. [Google Scholar] [CrossRef]
- Cui, N.J.; Chen, X.H.; Liu, Y.; Zhang, J.; Yang, W.Q. Shrub and herb diversity at different ages of Pinus massoniana plantation. Acta Ecol. Sin. 2014, 34, 4313–4323. [Google Scholar] [CrossRef][Green Version]
- Xiao, J.H.; Lai, M.Y.; Li, M.; Wang, C.X.; Zhong, Z.F.; Wen, Q.Y.; Qian, Z.J.; Luo, Z.; Wang, Y.X.; Deng, Q.Y.; et al. Characteristics of Pinus massoniana forest community and species diversity of island in Sandu Gulf, Ningde, Fujian Province. Acta Ecol. Sin. 2023, 43, 9827–9843. [Google Scholar] [CrossRef]
- Xiao, J.; Zhong, Z.; Wang, C.; Li, M.; Wen, Q.; Lin, X.; Luo, Z.; Qian, Z.; Li, M.; Deng, C. Rare species are significant in harsh environments and unstable communities: Based on the changes of species richness and community stability in different sub-assemblages. Sustainability 2023, 15, 13994. [Google Scholar] [CrossRef]
- Xiao, J.H.; Wen, Q.Y.; Zhong, Z.F.; Lin, X.T.; Wang, Y.X.; Xie, Y.Q.; Weng, F.F.; Deng, Q.Y.; Ding, G.C.; Deng, C.Y. Interspecific association and environmental interpretation of dominant species in shrub layer of Pinus massoniana community on Chinese islands. Ecol. Evol. 2024, 14, e70647. [Google Scholar] [CrossRef]
- Yao, Y.Q.; Zhang, Z.H.; Liang, S.C.; Bi, X.L.; Li, G.R.; Hu, G. Structure of Cyclobalanopsis glauca population on karst hills of Guilin. J. Zhejiang For. Sci. Technol. 2008, 28, 8–11. [Google Scholar] [CrossRef]
- Yu, F.H.; Krüsi, B.; Schütz, M.; Schneller, J.J.; Wildi, O. Plant communities affect the species-area relationship on Carex sempervirens tussocks. Flora 2008, 203, 197–203. [Google Scholar] [CrossRef]
- Bonet, A. Secondary succession of semi-arid Mediterranean old-fields in south-eastern Spain: Insights for conservation and restoration of degraded lands. J. Arid Environ. 2004, 56, 213–233. [Google Scholar] [CrossRef]
- Miao, L.Y.; Wang, X.A.; Wang, Z.G. Relationships between species diversity indices of Larix chinensis communities and environmental factors. Acta Bot. Boreal.-Occid. Sin. 2004, 24, 1888–1894. [Google Scholar]
- Wang, Z.G.; Wu, G.L.; Duan, R.Y.; Zhang, Z.X. Relationships betwen different life from species and its possessive area in Duozhijian, Dabieshan mountain, west Anhui pronvince. J. Shaanxi Norm. Univ. (Nat. Sci. Ed.) 2016, 44, 4. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.X.; Ren, K.X.; Arzigul; Wei, X.C.; Cheng, S.; Wang, L. Species-area curves of the three methods to expand comparative plot area. J. Capital Norm. Univ. (Nat. Sci. Ed.) 2014, 35, 60–63. [Google Scholar] [CrossRef]
- He, F.L.; Legendre, P. On species-area relations. Am. Nat. 1996, 148, 719. [Google Scholar] [CrossRef]
- He, F.L.; Legendre, P. Species diversity patterns derived from species-area models. Ecology 2002, 83, 1185–1198. [Google Scholar] [CrossRef]
- Tjørve, E. Shapes and functions of species-area curves(II): A review of new models and parameterizations. J. Biogeogr. 2009, 36, 1435–1445. [Google Scholar] [CrossRef]
- Schoereder, J.H.; Galbiati, C.; Ribas, C.R.; Sobrinho, T.G.; Sperber, C.F.; DeSouza, O.; Lopes-Andrade, C. Should we use proportional sampling for species-area studies? J. Biogeogr. 2004, 31, 1219–1226. [Google Scholar] [CrossRef]
- Williams, M.R.; Lamont, B.B.; Henstridge, J.D. Species-area functions revisited. J. Biogeogr. 2009, 36, 1994–2004. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer: New York, NY, USA, 2002; pp. 927–936. [Google Scholar]
- Ren, X.M.; Yang, G.H.; Wang, D.X.; Qin, X.W.; Liu, Z.X.; Zhao, S.X.; Bai, Y. Minimum sampling areas for shrub and herb layers of several types of plant communities in Taibai Mountain. Acta Bot. Boreal.-Occid. Sin. 2011, 31, 1024–1031. [Google Scholar]
- Xiao, J.H.; Zhang, L.T.; Wang, C.X.; Lai, M.Y.; Xiong, K.H.; Wen, Q.Y.; Deng, C.Y. Minimum sample area of three forest communities in Langqi Island. Plant Sci. J. 2023, 41, 279–291. [Google Scholar] [CrossRef]
- Archibald, E.E.A. The specific character of plant communities: II. A quantitative approach. J. Ecol. 1949, 37, 274–288. [Google Scholar] [CrossRef]
- Dong, S.K.; Tang, L.; Wang, X.X.; Liu, Y.H.; Liu, S.L.; Liu, Q.R.; Wu, Y.; Li, Y.Y.; Su, X.K.; Zhao, C. Minimum plot size for estimating plant biodiversity of the alpine grasslands on the Qinghai-Tibetan Plateau. Biodivers. Sci. 2013, 21, 651–657. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Cui, Y.; Zheng, Q.Z.; Yang, N.; Zhang, Y.; Yue, H.T.; Wang, X.X. The minimum sampling size for estimating species richness of 2 types of typical alpine meadows in Shangri-La. J. Southwest For. Univ. 2021, 41, 125–132. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, D.X. The method of grey modeling for the equation of Logistic. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2004, 33, 535–537. [Google Scholar] [CrossRef]
- Williams, C.B. Area and number of species. Nature 1943, 152, 264–267. [Google Scholar] [CrossRef]
- Worthen, W.B. Community composition and nested-subset analyses: Basic descriptors for community ecology. Oikos 1996, 76, 417–426. [Google Scholar] [CrossRef]
- Zhang, W.H.; Chen, C.; Huang, D.; Arshad, M.N.; Wang, L.X. Relationship of plant species diversity and sampling area under different grazing intensities in the steppe of northern China. Rangel. Ecol. Manag. 2023, 87, 150–157. [Google Scholar] [CrossRef]
- DeMalach, N.; Saiz, H.; Zaady, E.; Maestre, F.T. Plant species-area relationships are determined by evenness, cover and aggregation in drylands worldwide. Glob. Ecol. Biogeogr. 2018, 28, 290–299. [Google Scholar] [CrossRef]
- He, C.Q.; Fan, F.; Qiao, X.J.; Zhou, Z.; Xu, H.; Li, S.; Zhu, J.L.; Wang, S.P.; Tang, Z.Y.; Fang, J.Y. Sampling origins and directions affect the minimum sampling area in forest plots. J. Veg. Sci. 2024, 35, e13232. [Google Scholar] [CrossRef]


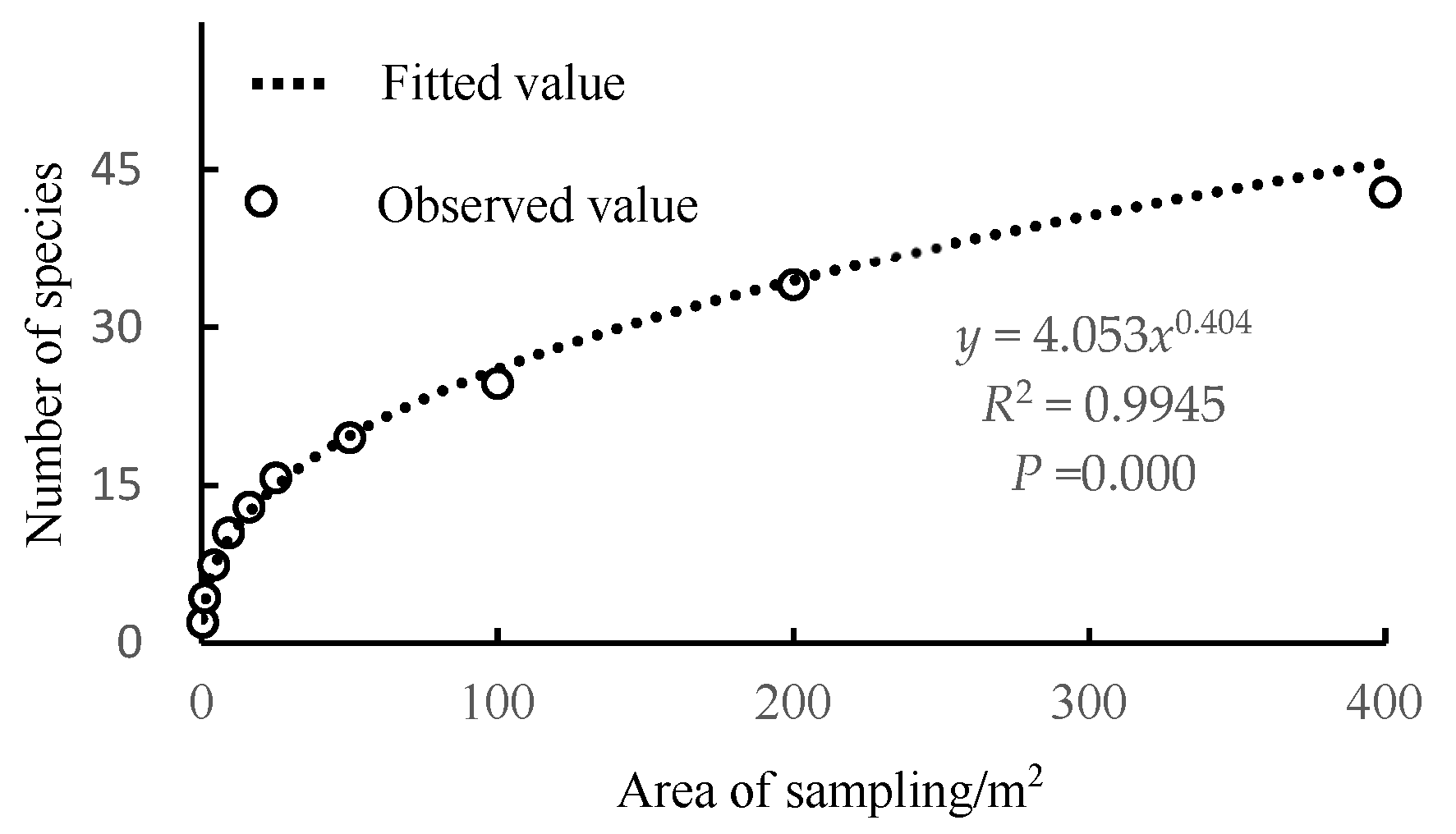

| Function | Species–Area Fitting Function Model | Minimum Sampling Area Equation |
|---|---|---|
| Logarithm | (1) S = a + blnA | (4) A = exp[(ρSt − a)/b] |
| Power | (2) S = aAb | (5) A = (ρSt/a)1/b |
| Logistic | (3) S = c/(1 + ae−bA) | (6) A = −{ln[(c/ρSt − 1)/a]}/b |
| Plot No. | Function | Parameter | R2 | p | AIC | Plot No. | Function | Parameter | R2 | p | AIC | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | a | b | c | ||||||||||
| S1 | (1) | 4.314 | 4.478 | - | 0.901 | 0.000 | 58.14 | S6 | (1) | 1.810 | 6.154 | - | 0.827 | 0.000 | 70.96 |
| (2) | 4.976 | 0.348 | - | 0.982 | 0.000 | −10.66 | (2) | 3.956 | 0.428 | - | 0.997 | 0.000 | −23.78 | ||
| (3) | 0.112 | 0.981 | 38.04 | 0.927 | 0.000 | 26.41 | (3) | 0.134 | 0.980 | 50.10 | 0.944 | 0.000 | 23.84 | ||
| S2 | (1) | 1.791 | 3.205 | - | 0.863 | 0.000 | 55.08 | S7 | (1) | 3.256 | 5.526 | - | 0.913 | 0.000 | 60.87 |
| (2) | 2.438 | 0.412 | - | 0.973 | 0.000 | −3.41 | (2) | 4.195 | 0.415 | - | 0.985 | 0.000 | −9.50 | ||
| (3) | 0.239 | 0.978 | 27.01 | 0.917 | 0.000 | 30.70 | (3) | 0.127 | 0.977 | 42.01 | 0.931 | 0.000 | 29.66 | ||
| S3 | (1) | 3.098 | 6.810 | - | 0.899 | 0.000 | 66.79 | S8 | (1) | 2.144 | 4.775 | - | 0.804 | 0.000 | 67.39 |
| (2) | 4.353 | 0.448 | - | 0.981 | 0.000 | −5.30 | (2) | 4.043 | 0.370 | - | 0.993 | 0.000 | −19.07 | ||
| (3) | 0.122 | 0.976 | 52.01 | 0.931 | 0.000 | 30.41 | (3) | 0.148 | 0.981 | 41.10 | 0.955 | 0.000 | 20.37 | ||
| S4 | (1) | 2.291 | 6.129 | - | 0.825 | 0.000 | 70.97 | All | (1) | 2.670 | 5.268 | - | 0.867 | 0.000 | 64.68 |
| (2) | 4.352 | 0.411 | - | 0.996 | 0.000 | −22.83 | (2) | 4.053 | 0.404 | - | 0.994 | 0.000 | −19.81 | ||
| (3) | 0.133 | 0.980 | 52.06 | 0.935 | 0.000 | 26.14 | (3) | 0.141 | 0.978 | 42.90 | 0.937 | 0.000 | 23.12 | ||
| S5 | (1) | 2.657 | 5.071 | - | 0.863 | 0.000 | 64.26 | ||||||||
| (2) | 3.965 | 0.401 | - | 0.992 | 0.000 | −15.93 | |||||||||
| (3) | 0.145 | 0.978 | 41.02 | 0.942 | 0.000 | 26.56 | |||||||||
| Plot No. | Different Estimation Precision | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ = 0.5 | ρ = 0.6 | ρ = 0.7 | ρ = 0.75 | ρ = 0.8 | ρ = 0.85 | ρ = 0.9 | ρ = 0.95 | |||||||||
| Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | Area/ m2 | Slope | |
| S1 | 47.0 | 0.141 | 79.4 | 0.100 | 123.6 | 0.075 | 150.7 | 0.066 | 181.4 | 0.058 | 215.9 | 0.052 | 254.4 | 0.047 | 297.2 | 0.042 |
| S2 | 63.7 | 0.087 | 99.2 | 0.067 | 144.1 | 0.054 | 170.4 | 0.049 | 199.3 | 0.045 | 230.9 | 0.041 | 265.3 | 0.038 | 302.5 | 0.035 |
| S3 | 54.0 | 0.216 | 81.2 | 0.172 | 114.5 | 0.142 | 133.5 | 0.131 | 154.2 | 0.121 | 176.6 | 0.112 | 200.6 | 0.104 | 226.3 | 0.098 |
| S4 | 77.4 | 0.138 | 120.6 | 0.106 | 175.5 | 0.085 | 207.6 | 0.077 | 242.9 | 0.070 | 281.5 | 0.065 | 323.5 | 0.059 | 369.0 | 0.055 |
| S5 | 60.2 | 0.137 | 94.8 | 0.104 | 139.2 | 0.083 | 165.4 | 0.075 | 194.2 | 0.068 | 226.0 | 0.062 | 260.6 | 0.057 | 298.2 | 0.052 |
| S6 | 74.3 | 0.144 | 113.7 | 0.112 | 163.0 | 0.092 | 191.5 | 0.084 | 222.7 | 0.077 | 256.6 | 0.071 | 293.2 | 0.066 | 332.7 | 0.061 |
| S7 | 48.5 | 0.180 | 75.2 | 0.139 | 109.1 | 0.112 | 128.8 | 0.102 | 150.4 | 0.093 | 174.1 | 0.085 | 199.8 | 0.079 | 227.6 | 0.073 |
| S8 | 80.5 | 0.094 | 131.7 | 0.069 | 199.7 | 0.053 | 240.7 | 0.047 | 286.6 | 0.042 | 337.6 | 0.038 | 394.0 | 0.035 | 456.0 | 0.032 |
| All | 61.8 | 0.140 | 97.0 | 0.107 | 142.0 | 0.085 | 168.5 | 0.077 | 197.7 | 0.070 | 229.7 | 0.064 | 264.6 | 0.059 | 302.5 | 0.054 |
| Plot No. | Minima Area | Number of Estimates Species | Number of Actual Species | Total Number of Species |
|---|---|---|---|---|
| S1 | 79.4 | 22.8 | 18.25~22.75 | 38.00 |
| S2 | 50.6 | 12.3 | 12.00~14.13 | 27.00 |
| S3 | 217.4 | 48.5 | 43.50~52.00 | 52.00 |
| S4 | 133.9 | 32.6 | 26.75~38.25 | 52.00 |
| S5 | 101.4 | 25.3 | 22.75~34.00 | 41.00 |
| S6 | 140.7 | 32.9 | 26.75~40.25 | 50.00 |
| S7 | 132.2 | 31.8 | 28.50~35.25 | 42.00 |
| S8 | 73.3 | 19.8 | 16.63~21.25 | 41.00 |
| All | 109.0 | 27.0 | 24.70~34.09 | 42.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Wen, Q.; Zhong, Z.; Xie, Y.; Wang, Y.; Cai, X.; Lin, Y.; Weng, F.; Ding, G.; Deng, C. The Main Factors Affecting the Minimum Sampling Area Determination Method: Based on Research of the Shrub Layer in Island Pinus massoniana Forests. Biology 2025, 14, 372. https://doi.org/10.3390/biology14040372
Xiao J, Wen Q, Zhong Z, Xie Y, Wang Y, Cai X, Lin Y, Weng F, Ding G, Deng C. The Main Factors Affecting the Minimum Sampling Area Determination Method: Based on Research of the Shrub Layer in Island Pinus massoniana Forests. Biology. 2025; 14(4):372. https://doi.org/10.3390/biology14040372
Chicago/Turabian StyleXiao, Jihong, Qingyan Wen, Zhifei Zhong, Yanqiu Xie, Yingxue Wang, Xing Cai, Yuchen Lin, Feifan Weng, Guochang Ding, and Chuanyuan Deng. 2025. "The Main Factors Affecting the Minimum Sampling Area Determination Method: Based on Research of the Shrub Layer in Island Pinus massoniana Forests" Biology 14, no. 4: 372. https://doi.org/10.3390/biology14040372
APA StyleXiao, J., Wen, Q., Zhong, Z., Xie, Y., Wang, Y., Cai, X., Lin, Y., Weng, F., Ding, G., & Deng, C. (2025). The Main Factors Affecting the Minimum Sampling Area Determination Method: Based on Research of the Shrub Layer in Island Pinus massoniana Forests. Biology, 14(4), 372. https://doi.org/10.3390/biology14040372






