The Effect of Probiotics on Gut Microbiota Modulation and Its Role in Mitigating Diabetes-Induced Hepatic Damage in Wistar Rats
Simple Summary
Abstract
1. Introduction
Study Objective
2. Methodology
2.1. Experimental Design and Animal Model
2.2. Diabetes Induction and Probiotic Administration
2.3. Blood Sample Collection
2.4. Biochemical and Metabolic Assessments
2.4.1. Body Weight and Fasting Blood Glucose
2.4.2. Glucose Tolerance and Insulin Sensitivity
2.4.3. Antioxidant Enzyme Levels
2.4.4. Serum Lipid Profiles
2.5. Histopathological Analysis of Liver Tissue
2.6. Microbiome Analysis—DNA Extraction and Processing
2.7. Statistical Analysis
3. Results
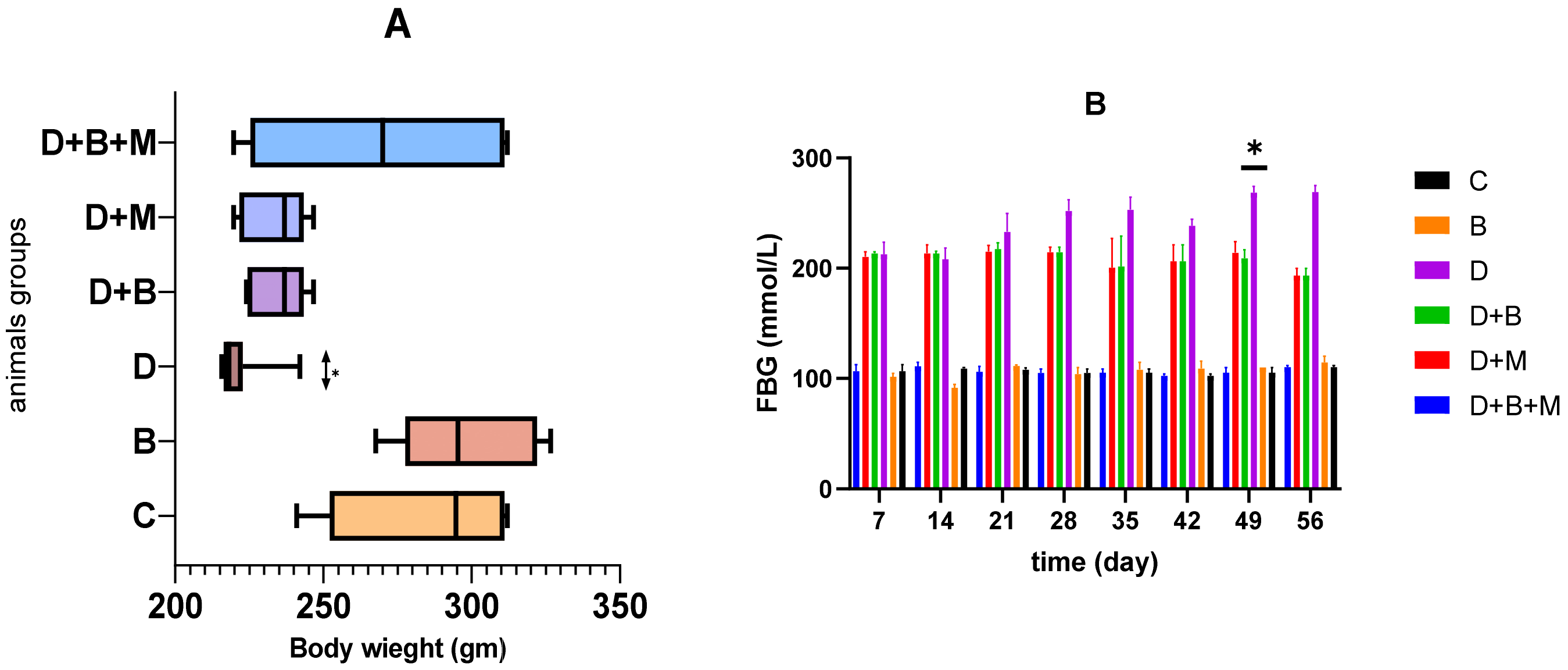
3.1. Effect of Bifidobacterium bifidum on Body Weight and Fasting Blood Glucose Levels in Diabetic Rats Body Weight
3.2. Fasting Blood Glucose (FBG)
3.3. Effect of Bifidobacterium bifidum and Metformin, Alone and in Combination, on Glucose Tolerance and Insulin Sensitivity in Diabetic Rats
3.4. Effect of Bifidobacterium bifidum and Metformin on Antioxidant Enzyme Levels in Diabetic Rats
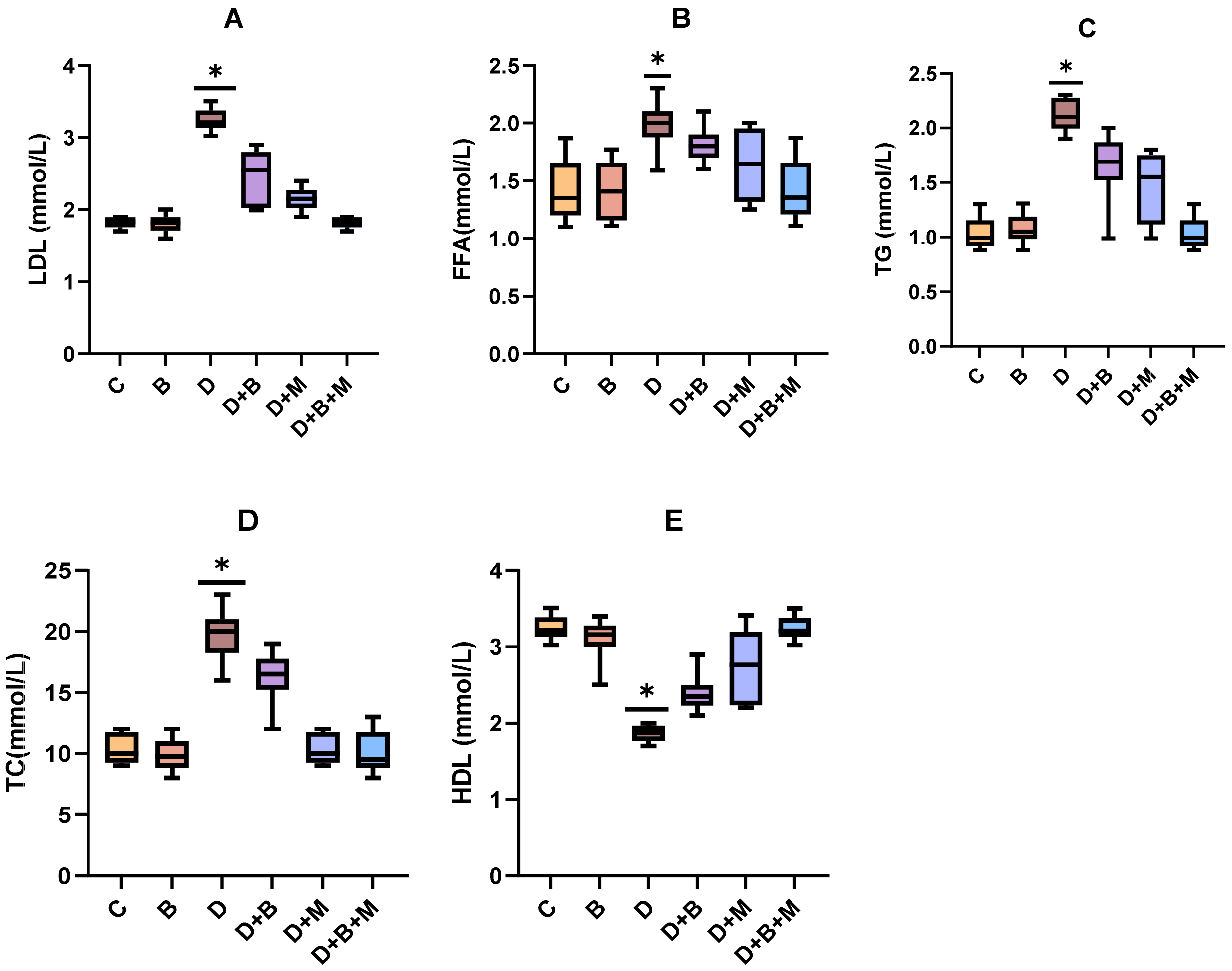
3.5. Effects of Bifidobacterium bifidum on Serum Lipid Profiles in Diabetic Rats
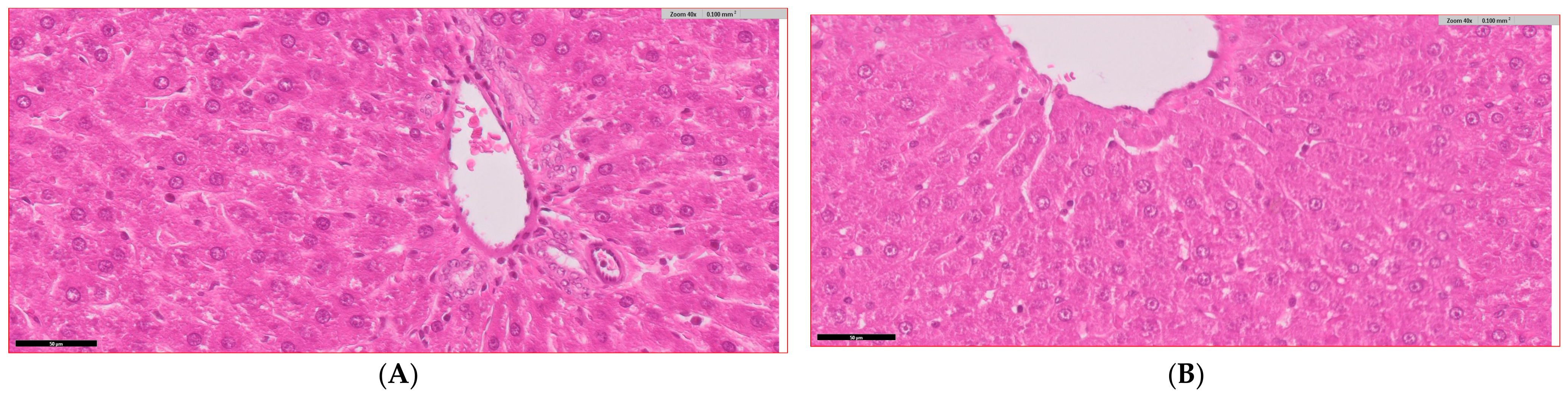
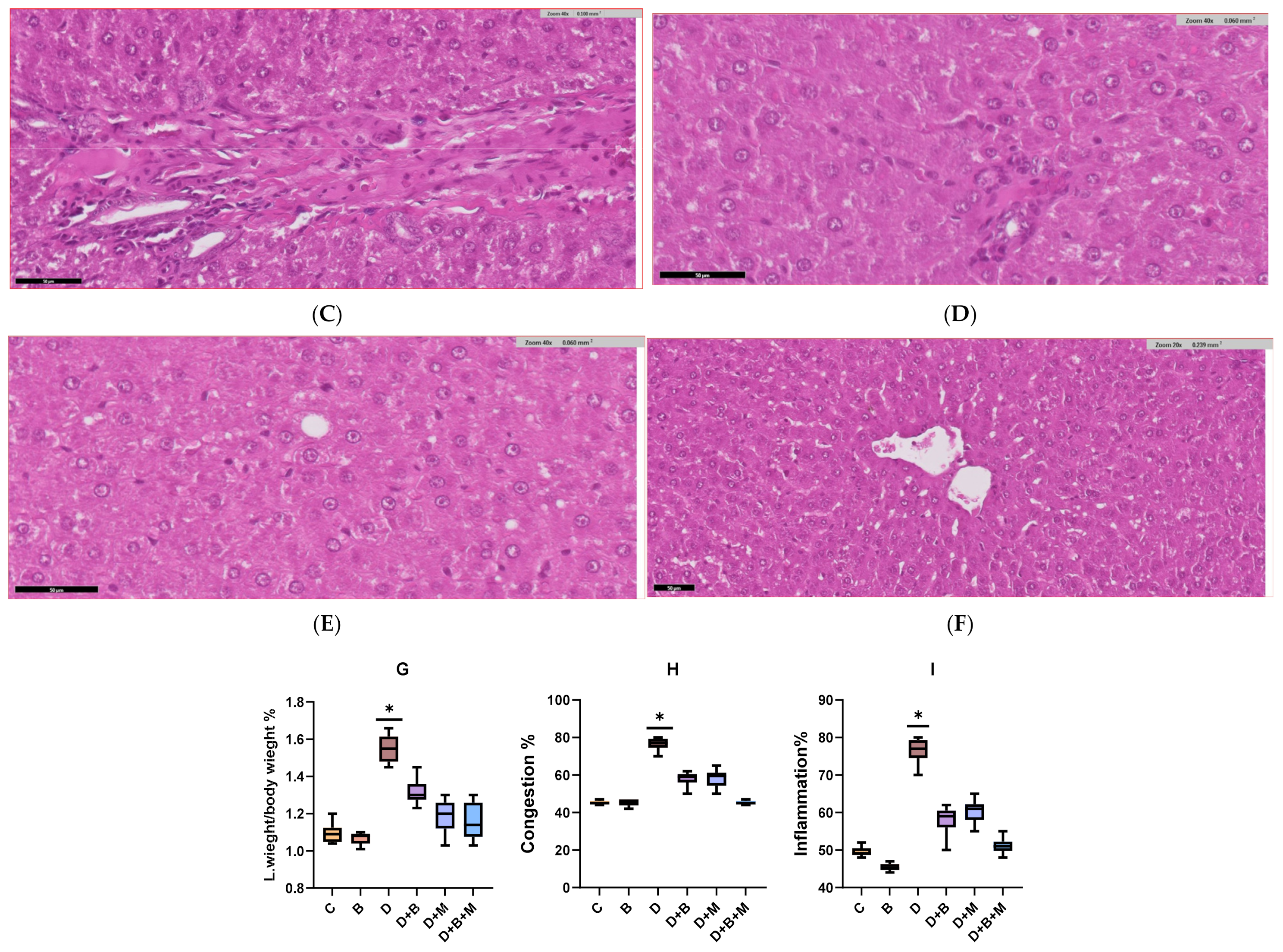
3.6. Histopathological Analysis of Liver Tissue Across Experimental Groups
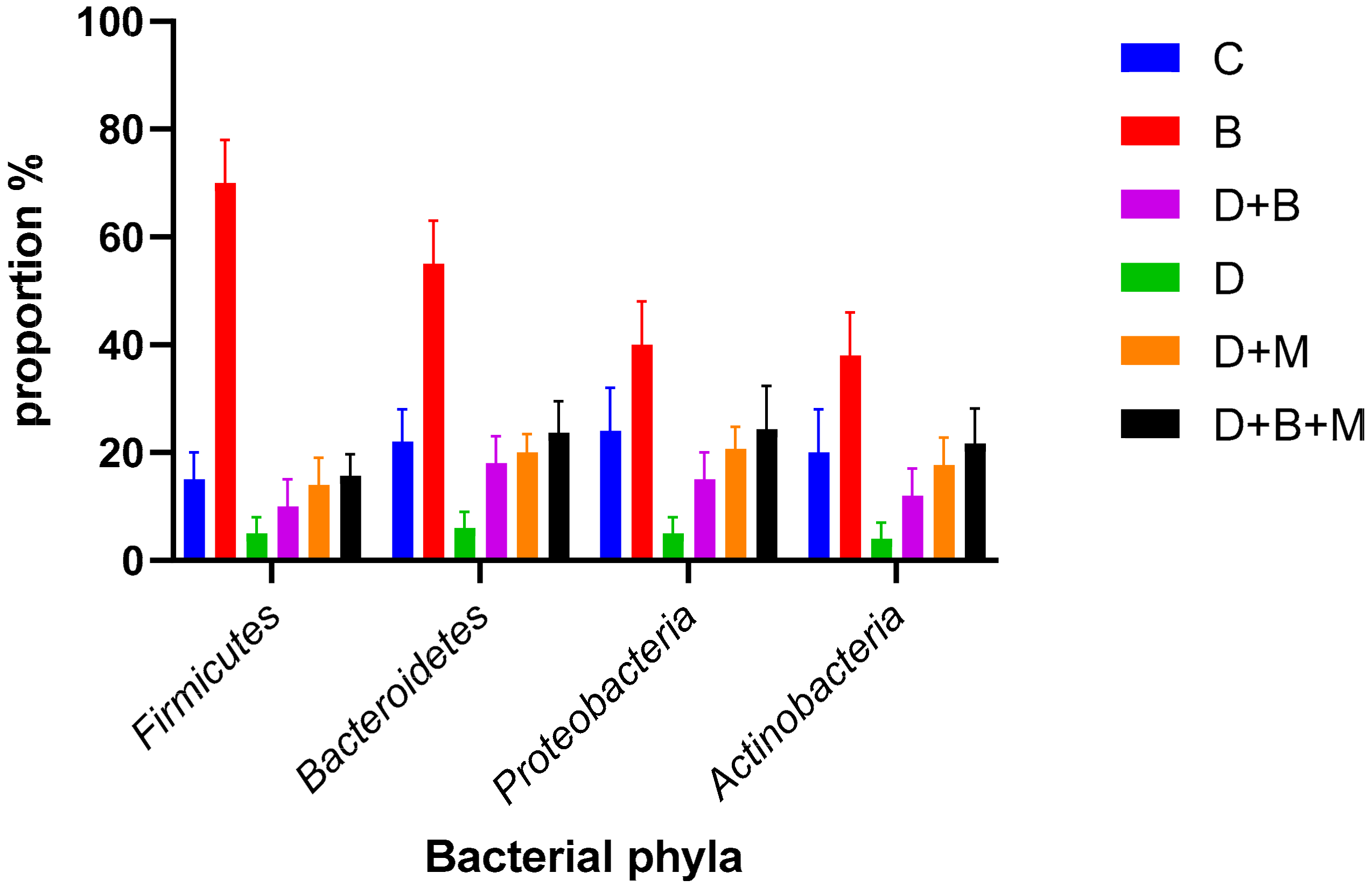
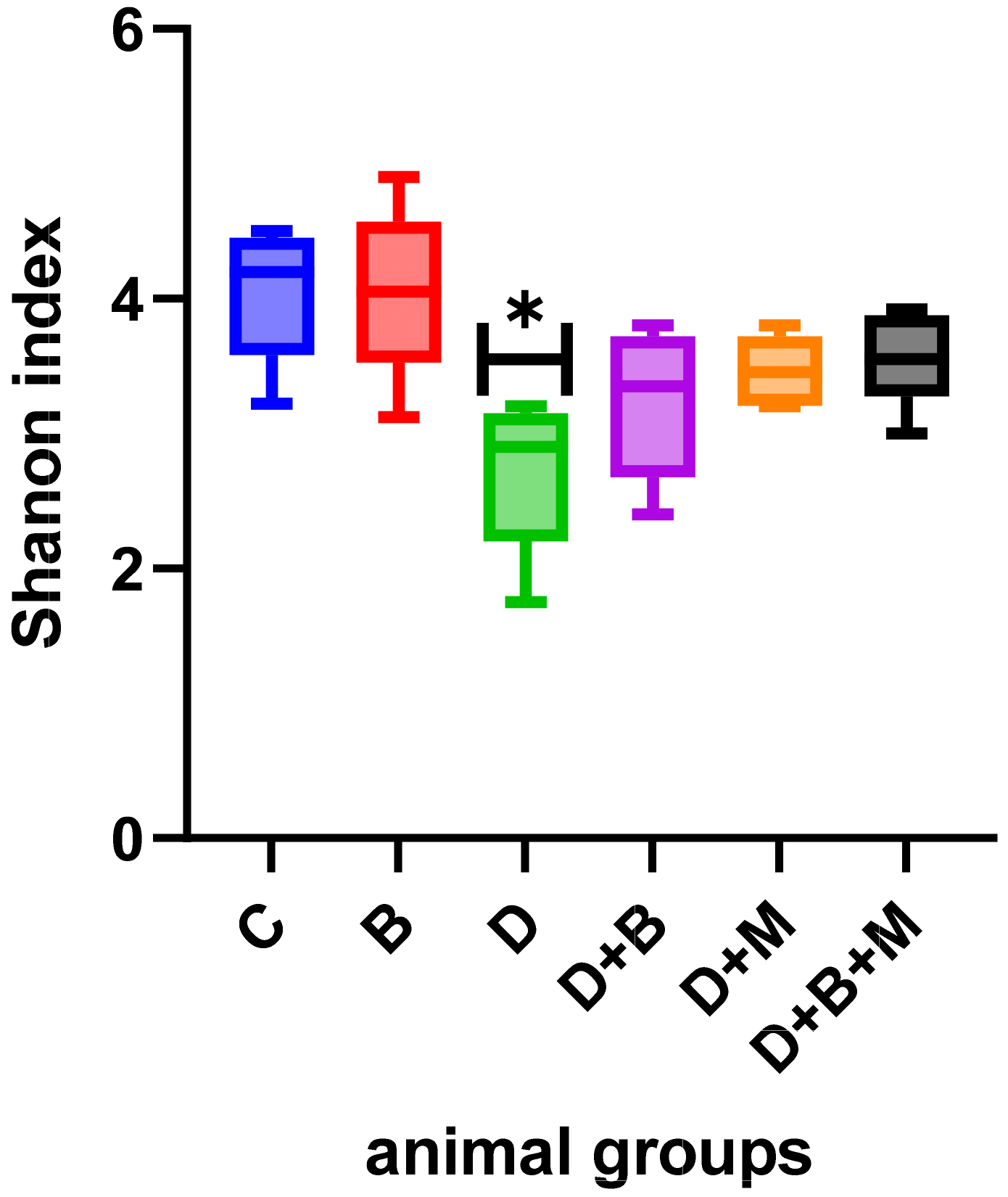
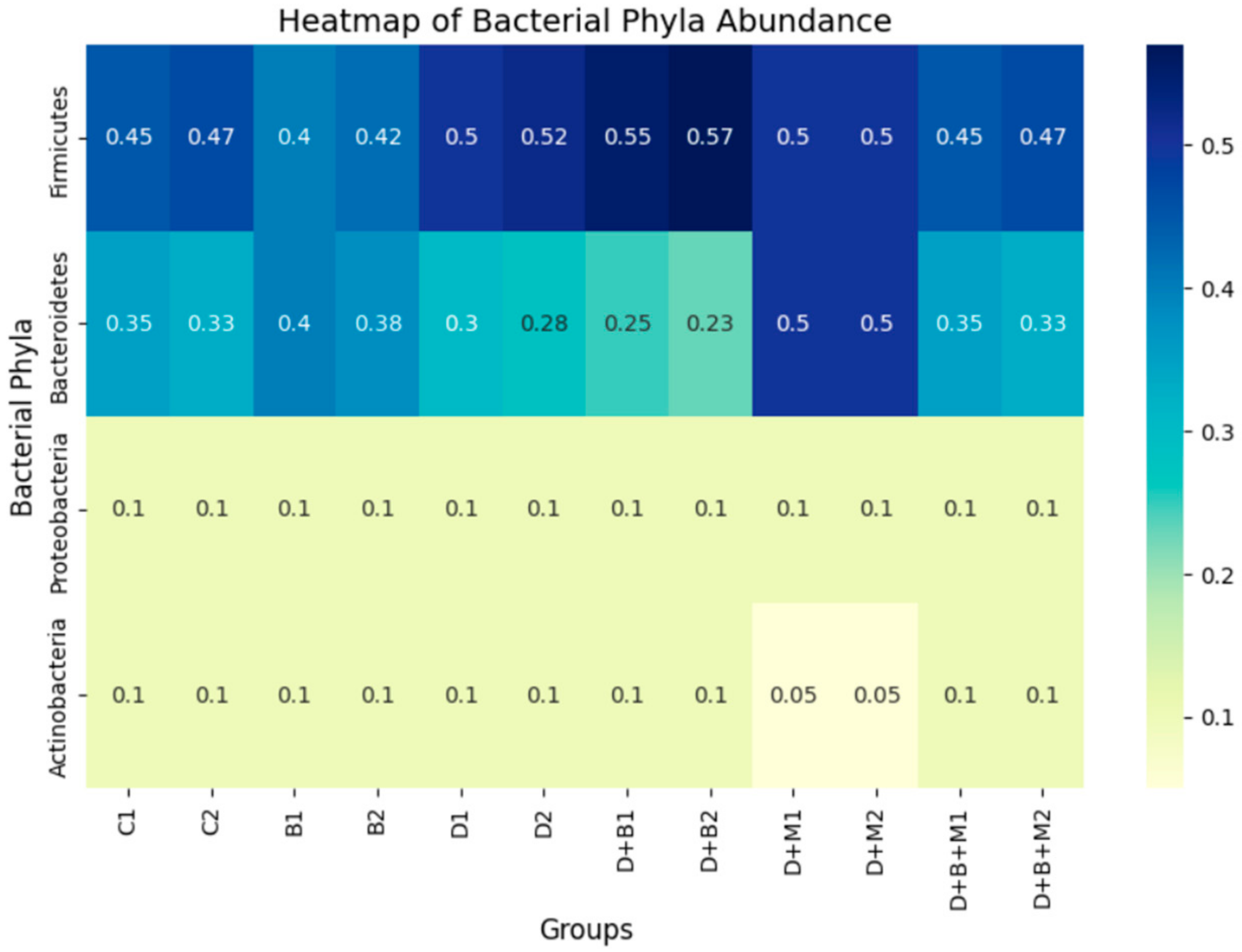
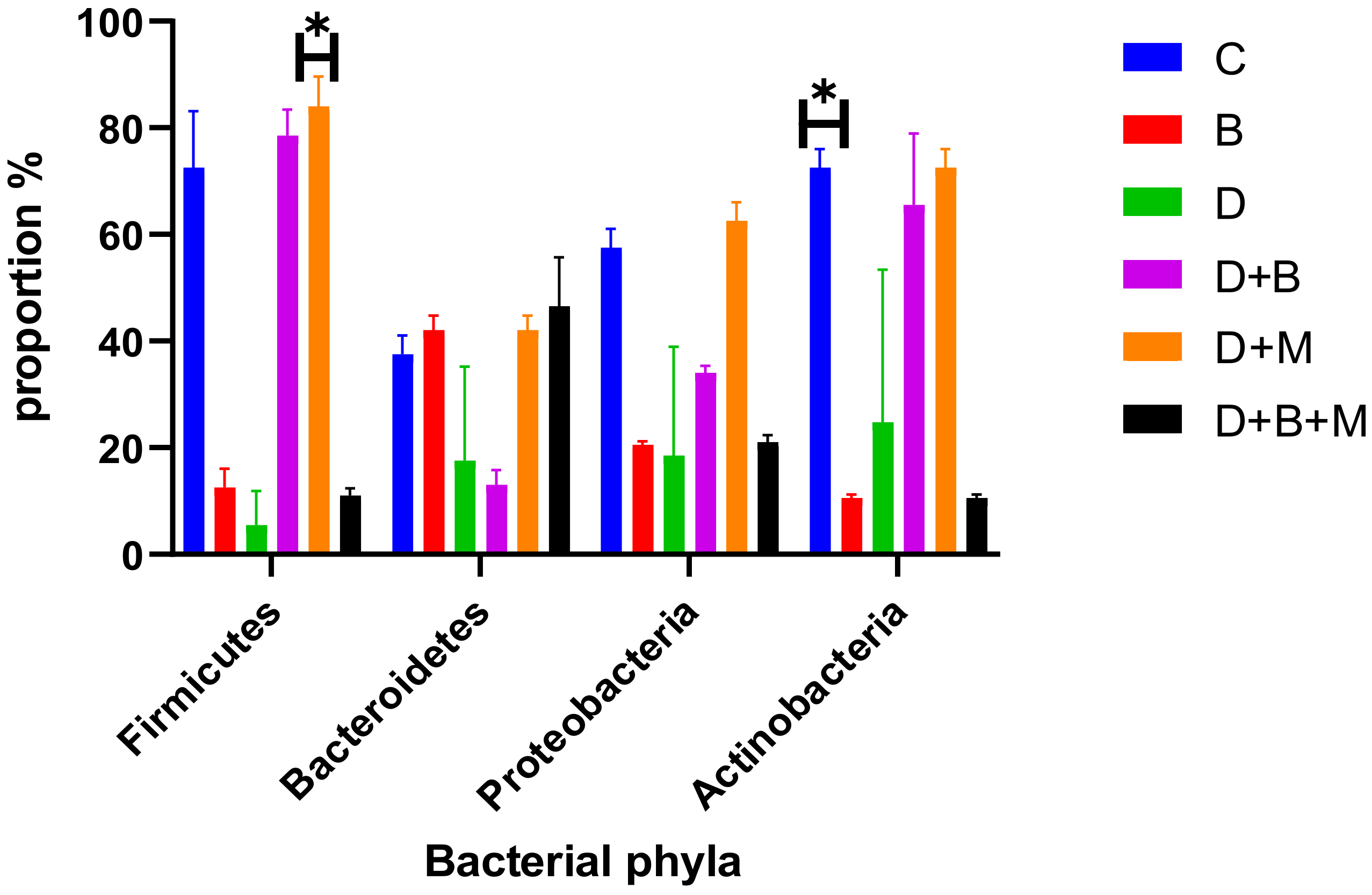
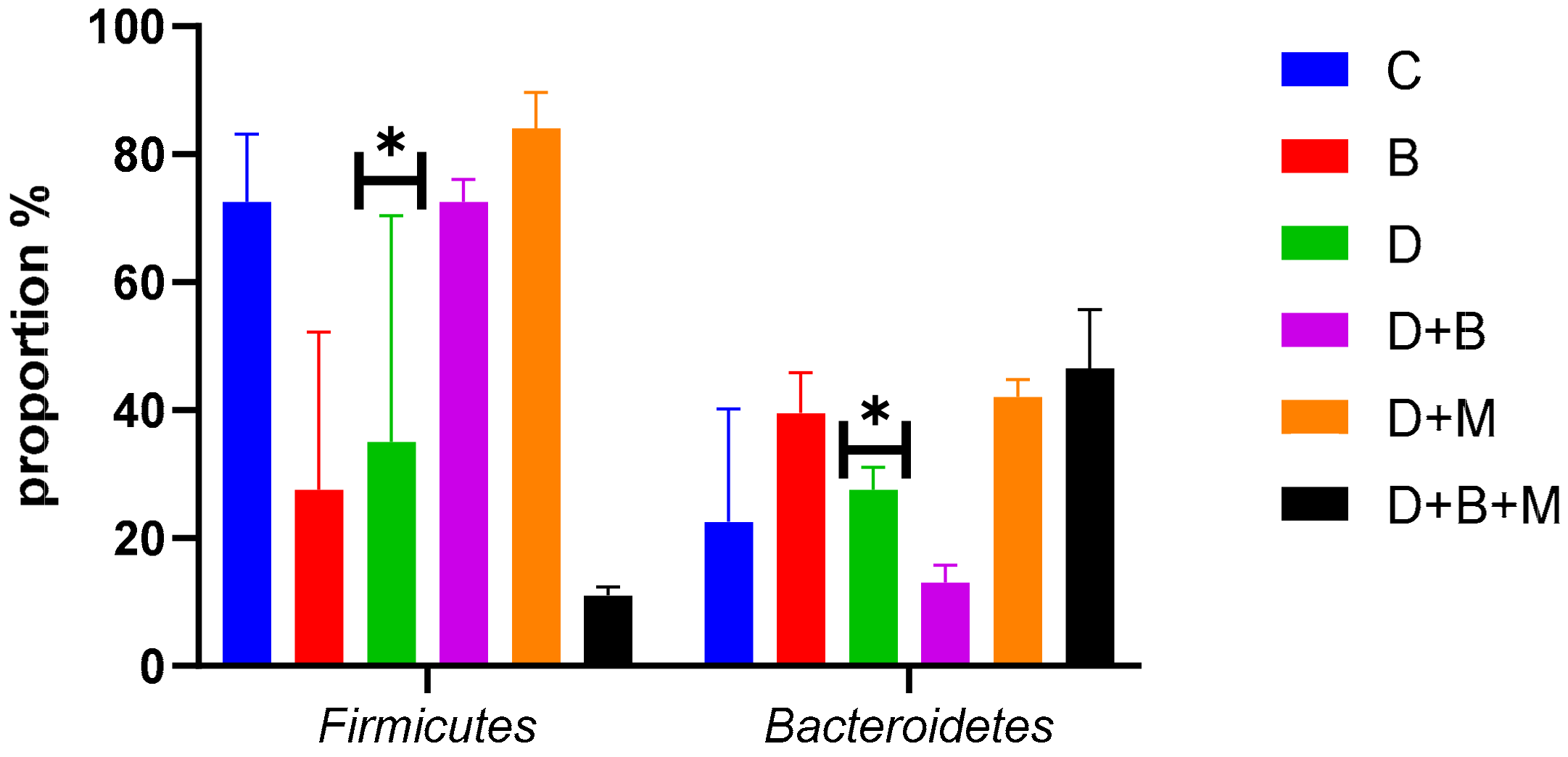
3.7. Microbiome Composition and Modulation by Bifidobacterium bifidum in Diabetic Rats
4. Discussion
5. Significance and Future Research Perspectives
- 1.
- Clinical Translation and Long-Term Effects
- o
- While preclinical findings are promising, controlled clinical trials in diabetic patients are essential to validate the therapeutic potential of Bifidobacterium bifidum and establish standardized dosing regimens. Future studies should assess its long-term effects on hepatic and metabolic outcomes in diverse patient populations.
- 2.
- Microbiome-Driven Therapeutic Strategies
- o
- Investigating the synergistic effects of Bifidobacterium bifidum with other probiotic strains or dietary interventions could enhance its efficacy. Multi-omics approaches, including metagenomics and metabolomics, should be employed to map host–microbiome interactions at a systems level.
- 3.
- Synbiotic and Postbiotic Applications
- o
- The development of synbiotic formulations combining Bifidobacterium bifidum with prebiotics that selectively promote its growth may optimize its therapeutic benefits. Additionally, exploring the potential of postbiotics—bioactive compounds derived from probiotics—could offer novel treatment strategies.
- 4.
- Comparative Efficacy with Conventional Therapies
- o
- Future studies should compare Bifidobacterium bifidum supplementation with conventional anti-diabetic and hepatoprotective treatments to determine its relative efficacy and potential for integration into current treatment protocols.
- 5.
- Mechanistic Elucidation Using Advanced Models
- o
- Employing humanized gut microbiota models and in vitro hepatic organoid systems may provide deeper mechanistic insights into the host–microbe interactions driving Bifidobacterium bifidum’s protective effects.
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021. [Google Scholar]
- Wu, X.; Chen, Y.; Li, Z.; Cheng, Y.; Zhang, J.; Zhao, Y.; Zhao, X.; Liu, Y.; Wang, C.; Huang, H.; et al. Streptozotocin-induced diabetic models in rodents. J. Endocrinol. 2020, 245, R1–R12. [Google Scholar] [CrossRef]
- Li, Q.; Cao, L.; Tian, Y.; Zhang, Y.; Wang, X.; Wu, H.; Liu, J.; Chen, M.; Wang, Y.; Sun, W.; et al. Gut microbiota alterations in diabetic complications. Trends Endocrinol. Metab. 2021, 32, 125–141. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Gibson, G.R.; et al. Gut microbiota and metabolic disorders: From obesity to NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 150–166. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R.; Roden, M.; Meinitzer, A.; Fauler, G.; Vogel, W.; Stadlbauer, V.; Puspok, A.; Stojakovic, T.; Eder, M.; et al. The role of gut-liver axis in metabolic diseases. Gastroenterology 2022, 162, 558–575. [Google Scholar] [CrossRef]
- Wang, P.; Li, T.; Zhao, Y.; Zhu, Y.; Chen, W.; Li, L.; Gao, M.; Zhang, X.; Wang, X.; Yang, Y.; et al. Probiotics in liver diseases: Mechanistic insights. Hepatology 2023, 78, 344–358. [Google Scholar] [CrossRef]
- Cani, P.D.; Jordan, B.F.; Holst, J.J.; Delzenne, N.M. The gut microbiota as a therapeutic target in metabolic disease: Focus on diabetes. Nat. Rev. Endocrinol. 2019, 15, 615–631. [Google Scholar] [CrossRef]
- Chang, C.J.; Lin, C.S.; Lu, C.C.; Martel, J.; Ko, Y.F.; Ojcius, D.M.; Tseng, S.F.; Chung, Y.C.; Hsu, Y.S.; Lai, H.C.; et al. Bifidobacterium bifidum in NAFLD: A randomized controlled trial. Clin. Nutr. 2023, 42, 345–356. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, J.; Yao, H.; Hu, H.; He, J.; Chen, Y.; Liang, X.; Wang, Q.; Liu, S.; Zhang, H.; et al. Prebiotics and metabolic disorders. Gut Microbes 2023, 15, 1985451. [Google Scholar] [CrossRef]
- Zhou, Z.; Ye, X.; Wang, L.; Jin, L.; Wang, D.; Zhang, X.; Liu, S.; Wang, Q.; Chen, Y.; Liu, Y.; et al. Gut microbiota modulation for metabolic disorders: From functional foods to targeted therapies. Nutrients 2023, 15, 1321. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y.; Jafari, M.; Zarei, M.; et al. Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xu, C.; Zhang, C.; Jin, M.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M.; et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Kobyliak, N.; Conte, C.; Cammarota, G.; Haley, A.P.; Styriak, I.; Gaspar, L.; Fusek, J.; Rodrigo, L.; Kruzliak, P. Probiotics in prevention and treatment of obesity: A critical view. Nutr. Metab. 2016, 13, 14. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Cover, T.M.; Thomas, J.A. Elements of Information Theory, 2nd ed.; Wiley-Interscience: Hoboken, NJ, USA, 2006. [Google Scholar]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Bai, X.; Ni, J.; Beretov, J.; Wasinger, V.C.; Wang, S.; Zhu, Y.; Huang, L.; Li, Y.; Lu, L.; Xu, Z.; et al. Activation of the eIF2α/ATF4 axis drives triple-negative breast cancer radioresistance by promoting glutathione biosynthesis. Redox Biol. 2021, 43, 101993. [Google Scholar] [CrossRef]
- Peng, Y.H.; Sweet, D.H.; Lin, S.P.; Yu, C.P.; Chao, P.D.L.; Hou, Y.C. Green tea inhibited the elimination of nephro-cardiovascular toxins and deteriorated the renal function in rats with renal failure. Sci. Rep. 2015, 5, 16226. [Google Scholar] [CrossRef]
- Wang, P.; Li, D.; Hu, X.; Chen, F. Gut microbiota and metabolic disorders: From obesity to NAFLD. Front. Microbiol. 2021, 12, 710138. [Google Scholar] [CrossRef]
- STZ-100 Streptozotocin; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/s0130 (accessed on 19 March 2025).
- DS-200 Digital Scale; Ohaus: Parsippany, NJ, USA, 2023; Available online: https://us.ohaus.com/en-US/Products/Balances/Portable-Balances/Scout-STX (accessed on 19 March 2025).
- GLU-001 Enzymatic Assay Kit for Glucose Measurement; Roche Diagnostics: Rotkreuz, Switzerland, 2023; Available online: https://diagnostics.roche.com/global/en/products/params/glucose-assay.html (accessed on 19 March 2025).
- COBAS Integra 400 Automated Biochemical Analyzer; Roche Diagnostics: Rotkreuz, Switzerland, 2023; Available online: https://diagnostics.roche.com/global/en/products/instruments/cobas-integra-400-plus.html (accessed on 19 March 2025).
- INS-101 ELISA Kit for Insulin Quantification; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/rab0328 (accessed on 19 March 2025).
- CAT-001 Catalase Assay Kit; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/cat100 (accessed on 19 March 2025).
- SOD-101 Superoxide Dismutase Assay Kit; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/19160 (accessed on 19 March 2025).
- GPx-202 Glutathione Peroxidase Assay Kit; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/cs0260 (accessed on 19 March 2025).
- GST-303 Glutathione S-Transferase Assay Kit; Roche Diagnostics: Rotkreuz, Switzerland, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/cs0410 (accessed on 19 March 2025).
- Bradford-1000 Protein Assay Instrument; Bio-Rad: Hercules, CA, USA, 2023; Available online: https://www.bio-rad.com/en-us/product/bradford-protein-assay (accessed on 19 March 2025).
- X-123 Enzymatic Assay Kits for LDL, FFA, TG, TC; H-101 HDL Kit; Roche Diagnostics: Rotkreuz, Switzerland, 2023; Available online: https://diagnostics.roche.com/global/en/products/params/ldl-cholesterol-gen3.html (accessed on 19 March 2025).
- F-10 10% Formalin Solution; Sigma-Aldrich: St. Louis, MO, USA, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/sigma/ht501128 (accessed on 19 March 2025).
- P-20 Paraffin for Tissue Embedding; Merck: Darmstadt, Germany, 2023; Available online: https://www.sigmaaldrich.com/US/en/product/mm/104300 (accessed on 19 March 2025).
- Leica RM2255 Microtome; Leica Biosystems: Nußloch, Germany, 2023; Available online: https://www.leicabiosystems.com/us/products/microtomes/rotary-microtomes/details/product/leica-rm2255-fully-automated-rotary-microtome/ (accessed on 19 March 2025).
- Olympus BX51 Light Microscope; Olympus Corporation: Tokyo, Japan, 2023; Available online: https://www.olympus-lifescience.com/en/microscopes/compound/bx51/ (accessed on 19 March 2025).
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.H.; Hsieh, T.C. Proposing a resolution to debates on diversity partitioning. Ecology 2012, 93, 2037–2051. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project: Exploring the microbial part of ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qumsani, A.T. The Effect of Probiotics on Gut Microbiota Modulation and Its Role in Mitigating Diabetes-Induced Hepatic Damage in Wistar Rats. Biology 2025, 14, 323. https://doi.org/10.3390/biology14040323
Qumsani AT. The Effect of Probiotics on Gut Microbiota Modulation and Its Role in Mitigating Diabetes-Induced Hepatic Damage in Wistar Rats. Biology. 2025; 14(4):323. https://doi.org/10.3390/biology14040323
Chicago/Turabian StyleQumsani, Alaa Talal. 2025. "The Effect of Probiotics on Gut Microbiota Modulation and Its Role in Mitigating Diabetes-Induced Hepatic Damage in Wistar Rats" Biology 14, no. 4: 323. https://doi.org/10.3390/biology14040323
APA StyleQumsani, A. T. (2025). The Effect of Probiotics on Gut Microbiota Modulation and Its Role in Mitigating Diabetes-Induced Hepatic Damage in Wistar Rats. Biology, 14(4), 323. https://doi.org/10.3390/biology14040323





