Simple Summary
Ciliates, particularly the genus Frontonia, have been studied to understand their evolutionary history, but challenges remain. This study investigated the evolutionary patterns of Frontonia using genetic and morphological data. Molecular analysis of the SSU rRNA gene revealed four major phylogenetic groups within Frontonia, suggesting its paraphyly. The common ancestor existed approximately 420 million years ago, with distinct groups emerging during the Mesozoic era. Diversification analysis showed higher extinction rates than speciation rates within the genus. Morphological traits, including habitat adaptations, were examined through ancestral state reconstructions, revealing a complex evolutionary history. Habitat transitions were not directly linked to morphological traits such as contractile vacuoles, emphasizing the role of genetic diversity and environmental adaptation. These findings provide valuable insights into the interplay between evolution, extinction, and morphology in ciliates, advancing our understanding of biodiversity and evolutionary biology.
Abstract
Ciliates of the genus Frontonia have been extensively studied to resolve their phylogenetic and evolutionary history, but challenges remain. This study used molecular analyses of SSU rRNA genes, phylogenetic tree reconstruction, molecular dating, and diversification analysis, together with ancestral state reconstruction of morphological traits and habitat preferences. Data included newly sequenced Korean species, GenBank records and published morphological information. Phylogenetic trees revealed paraphyly within Frontonia, identifying four groups that emerged in the Mesozoic era: Group I (~172 mya), Group II (~83 mya), Group III (~115 mya), and Group IV (~190 mya), with a common ancestor dating to ~420 mya in the Palaeozoic era. Diversification analysis revealed higher extinction rates (0.826 and 0.613 species/year) than speciation rates (0.011 and 0.016 species/year). Morphological evolution showed habitat adaptation and plasticity, with habitat transitions unrelated to contractile vacuolar traits. The SSU rRNA gene polymorphism likely contributed to the paraphyletic state of Frontonia. These results highlight the complex evolutionary patterns of the genus, shaped by genetic diversity, morphology, and environmental constraints.
1. Introduction
The research on ciliates, with a particular focus on the genus Frontonia, has indeed advanced significantly due to the integration of molecular and morphological studies. The genus Frontonia established nearly two centuries ago, represents a diverse group of ciliate species. The genus Frontonia comprises free-living ciliates found in freshwater, marine, and brackish environments, characterized by an elongated body, dense ciliation, and a prominent oral apparatus. As heterotrophs, they play a crucial role in microbial food webs and have a close phylogenetic relationship with Paramecium and Apofrontonia, making them valuable for studies on ciliate evolution, physiology, and ecological dynamics [1,2,3,4,5,6,7].
While various research efforts have contributed to the understanding of their morphological traits, the phylogenetic relationships and evolutionary history of many ciliate lineages, including Frontonia, remain unresolved [8]. Phylogenetic analyses indicate that Frontonia is paraphyletic, forming distinct clades with some species grouping with genera like Apofrontonia, Paramecium, Stokesia, and Marituja. This challenges its traditional classification and suggests a need for taxonomic revision. However, the possibility of Frontonia being monophyletic cannot be entirely dismissed [9].
Molecular analyses, particularly of the SSU rRNA gene, have provided crucial insights into the phylogenetic relationships within Frontonia. These analyses have revealed the paraphyletic nature of the genus, with several distinct clades identified. Furthermore, certain species consistently cluster with different genera in the SSU rRNA gene phylogenetic trees, suggesting a complex evolutionary history [8,9,10,11,12].
However, the molecular analysis of the SSU rRNA gene has been the primary tool used to construct phylogenetic trees within Frontonia, with limited integration of morphological data beyond species descriptions. To address this gap, a multifaceted approach is proposed in the current study. This approach involves the integration of molecular data, in particular the SSU rRNA gene, for molecular clock analysis. At the same time, morphological data will be used to reconstruct ancestral states, allowing for a more comprehensive understanding of the phylogenetic relationships and evolutionary history of Frontonia. By combining molecular and morphological data, the study aims to elucidate the evolutionary patterns, phylogenetic relationships, and divergence time of the genus Frontonia. These integrated approaches will provide deeper insights into the diversification patterns and evolutionary processes of the lineages in Frontonia.
2. Materials and Methods
2.1. Sample Collection and Morphological Study
Newly sequenced species of Frontonia were collected from different locations in the Republic of Korea. Detailed information on the collection sites can be found in Table 1. The Frontonia species were cultivated in petri dishes using water collected from their respective habitats. Initial in vivo cell observations were made using both bright field and differential interference contrast microscopy techniques. Subsequent detailed examinations were performed using a compound microscope, with magnifications ranging from 100× to 1000× for both live cells and stained samples. The Protargol-impregnated method was employed to visualize the structure of the buccal area, the oral apparatus, and the ciliary pattern [13].

Table 1.
Frontonia species from the Republic of Korea and their accession number for SSU rRNA gene sequence.
2.2. DNA Extraction, Amplification, and Sequencing
The genomic DNA extraction was carried out using the RED Extract-N-Amp Tissue PCR Kit from Sigma (St. Louis, MO, USA) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) was performed using the forward primer EUKA (5′-GAC CGT CCT AGT TGG TC-3′) [14] or 82F (5′-CTC GGT AAG CGT CAA AG-3′) [9] and the reverse primers D1, D2 rev2 (5′-GAC TGC ACG TTT AGC TAG CA-3′) or D1, D2 rev4 (5′-GTG CCT GGT TCY TCA GAT TG-3′). PCR amplifications were performed using the TaKaRa ExTaq DNA polymerase kit from TaKaRa Bio-medicals (Otsu, Japan) following a specific protocol: an initial denaturation cycle at 94 °C for 2 min, followed by 37 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 40 s, and extension at 72 °C for 4 min. A final extension step was then carried out at 72 °C for 10 min [15].
2.3. Alignment and Phylogenetic Analysis
In this study, 71 sequences from the species of the genus Frontonia obtained from NCBI and newly sequenced in Republic of Korea (Table A1), 20 sequences from related genera (Paranassula, Paramecium, Apofrontonia, Stokesia, Disematosoma, Marituja, Lembadion, Urocentrum, and Tetrahymena) were selected as reference sequences and two species from the genus Tetrahymena were selected as outgroup (T. pyriformis & T. rostrata) (Figure 1). The alignment for this data set was performed by using MAFFT version 7.0 [16]. Subsequent refinement and masking of the alignments were carried out using G-blocks version 0.91b [17]. To determine the best evolutionary models, jModeltest version 2.1.7 [18,19] was employed. Further analysis included maximum likelihood (ML) using PhyML 3.0 software with 1000 non-parametric bootstrap replicates [20]. In addition, Bayesian Inference (BI) analysis was performed using MrBayes version 3.1 [21] with the best parameter from Jmodeltest result; best model GTR + I + G, p-inv 0.3980, and gamma shape 0.4040.
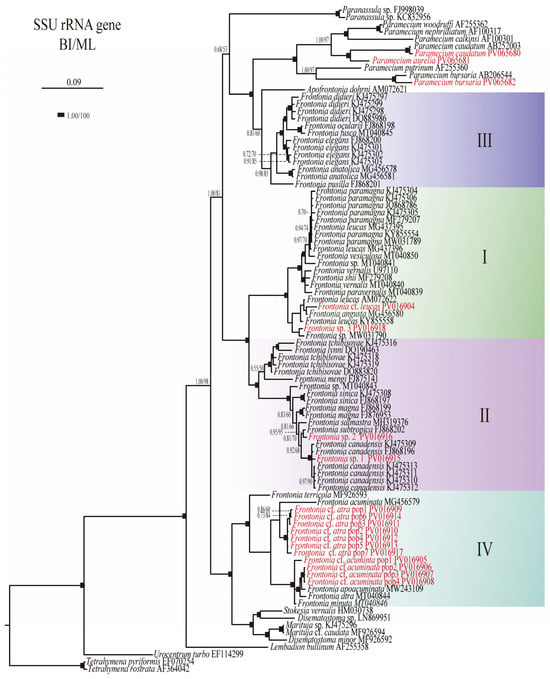
Figure 1.
Phylogenetic tree generated from Bayesian Inference (BI) using Mr Bayes ver. 3.1 software and Maximum Likelihood (ML) from PHYML ver. 3.0 based on the SSU rRNA (top) and SSU-ITS-LSU rRNA (bottom) gene sequences of the genus Frontonia and related genera. The scale bar indicates the number of base changes per 1000 nucleotide positions in BI analysis. Node support is represented as follows: BI posterior probability/ML bootstrap. The newly sequenced species of the genus Frontonia are shown in red. Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV.
2.4. Molecular Dating and Diversification Analysis
The analysis used 33 sequences from representative species of the genus Frontonia, selected on the basis of the availability of both molecular and morphological information (Table A1). In addition, 18 sequences from species of related genera were included in the analysis. Divergence times were estimated using a Bayesian framework that was implemented in BEAST ver. 2.4.5 [22,23]. The software BEAUti ver. 2.4.5 [23] was used to generate the BEAST software input XML file with the following settings; (i) calibrated Yule model, (ii) GTR + I (=0.389) + Γ (=0.412), (iii) four gamma categories for substitution rate heterogeneity, (iv) strict molecular clock, (v) clock rate prior assuming a normal distribution with a mean of 3.88 × 10−4 nucleotide substitutions per site per one million years and a 95% credibility interval ranging from 1.24 × 10−4 to 9.14 × 10−4 [24] and (vi) Yule birth rate set with shape parameter set to 0.001 and scale parameter set to 1000 (gamma shape parameter).
For the estimation of the divergence time, calibration nodes from the genera Paramecium and Tetrahymena were used. These nodes were based on microfossil evidence from Paramecium triassicum and Tetrahymena rostrata-like species found in an amber Triassic slab during the Upper Triassic period, approximately 220–230 million years ago (Mya) [25]. The mean divergence time for these calibration nodes was estimated to be 225 Mya. Markov Chain Monte Carlo (MCMC) analyses started with a random seed and ran for 6,000,000 generations, with trees and all other parameters saved every 10,000th iteration. The quality of the MCMC analysis was assessed using Tracer version 1.6 [26] to ensure convergence and adequate burn-in. The final maximum credibility tree was generated using TreeAnnotator ver. 1.8.1 [22] after discarding the first 10% of sampled trees.
To analyze the diversification dynamics of Frontonia, we used DivBayes and SubT [27], two Bayesian-based approaches for estimating speciation and extinction rates. DivBayes infers diversification rates by integrating ancestral divergence times with the observed and predicted species numbers, assuming a birth-death process. The method utilizes Bayesian posterior distributions to estimate net diversification, accounting for rate variation over time. SubT estimates speciation rates using 95% height data from chronograms generated by BEAST2, incorporating ultrametric tree node depths. To correct for taxon sampling biases, we included predicted species numbers where actual data were unavailable (Table A2), improving accuracy in rate estimation. These methods were chosen due to their robustness in handling incomplete taxon sampling and their ability to infer diversification trends over evolutionary timescales.
2.5. Reconstruction of Ancestral Morphologies
For the prediction of ancestral states, 33 species of the genus Frontonia and Apofrontonia dohrni were selected as representative taxa. The selection was based on the availability and credibility of their molecular and morphological data, as shown in Table A2. To perform the ancestral state reconstruction, 12 important and significant characters were selected within the genus Frontonia. These characters included the number of ciliary rows in peroral membranes (PM), the number of vestibular kineties (VK), the number of postoral kineties (PK), the number of ciliary rows in peniculi 1–3, the structure of P3 (shortened and linearity with P1 and P2), the habitat, the number of contractile vacuoles (CV), the number of contractile vacuolar pores (CVP) and the presence of contractile canals (CC). The character matrix and ancestral state reconstruction were performed using the parsimony model in Mesquite software ver. 3.70 [28]. The reconstruction of ancestral states was based on the topology of the best likelihood tree obtained from the RAxML analysis performed on CIPRES [29].
3. Results
3.1. Phylogenetic Analyses
The phylogenetic analyses based on SSU rRNA gene sequences have provided insights into the evolutionary relationships within the genus Frontonia, revealing the existence of four main groups: Group I: This group includes species such as F. canadensis, F. subtropica, F. salmastra, F. sinica, F. magna, F. mengi, F. tchibisovae, and F. lynni. Both Bayesian Inference (BI) and Maximum Likelihood (ML) analyses strongly support this group, with node support values of 1.00/100%. Group II: Comprising species such as F. paramagna, F. leucas, F. vesiculosa, F. vernalis, F. shii, F. paravernalis, F. leucas, F. angusta, and F. cf. leucas, Group II also shows robust node support values of 1.00/100% for both BI and ML. Group III: This group includes species such as F. didieri, F. ocularis, F. elegans, F. pusilla, and F. anatolica, with slightly lower node support values of 0.98/85% for BI and ML compared to Groups I and II. Group IV: Including species such as F. terricola, F. cf. acuminata, F. acuminata, F. atra, F. apoacuminata, F. minuta, and F. cf. atra, Group IV shows moderate node support (BI/ML, 1.00/88%) (Figure 1).
Furthermore, Group III shows a close relationship with the genus Apofrontonia, albeit with lower node support (BI/ML, 0.81/60%), and together this sister clade formed a clade with the genus Paramecium with full support. Group IV forms a cluster with the genera Stokesia, Marituja, and Disematostoma, with strong node support (BI/ML, 1.00/100%). These phylogenetic relationships shed light on the evolutionary dynamics and diversification patterns within the genus Frontonia highlighting both strong and moderate support for the identified groups and their relationships with related taxa (Figure 1).
3.2. Estimation of Divergence Times and Diversification Using the SSU rRNA Gene
The divergence time within the genus Frontonia can be estimated using two methods: relaxed clock and strict clock. A relaxed clock is commonly used at higher taxonomic levels such as family or order, while a strict clock is favored for intraspecific level analyses where low rates of variation between branches are expected [30,31,32,33]. In the case of the genus Frontonia, the strict clock approach is more suitable for estimating divergence time due to the limited data set within the genus level, and the minimal variation between branches.
The analyses of divergence time in the genus Frontonia performed in this study show that Peniculia is estimated to have originated approximately 750 million years ago (Mya), confirming the previous findings of Rataj and Vďačný (2018) [24] (Figure 2). Using the strict clock approach, the SSU rRNA gene showed a mean clock rate of 2.36 × 10−4 per year. Node calibrations for the Tetrahymena clade and Paramecium clades indicate their emergence at 223 Mya and 226 Mya, respectively. The common ancestor of the genus Frontonia appeared around 420 Mya and serves as an ancestral point for all members of the Penicullida. Notably, the emergence of three clades within the genus Frontonia occurred relatively recently: Group I around 172 Mya, Group II around 83 Mya, Group III around 115 Mya, and Group IV around 190 Mya (Figure 2).
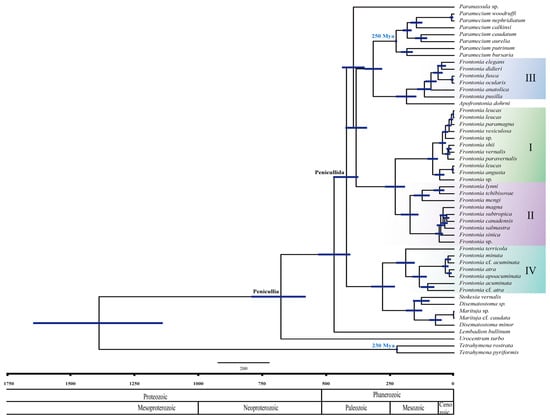
Figure 2.
Maximum credibility tree showing posterior means of divergence times of the genus Frontonia and related genera obtained using Bayesian relaxed molecular dating in BEAST. The 95% credibility intervals are shown as bars for all nodes. The horizontal axis represents the time scale in millions of years. Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV.
The results obtained from Divbayes and SubT for speciation and extinction indicate a higher extinction rate compared to the speciation rate with genus Frontonia: 0.826 species/year over speciation rate: 0.011 species/year (DivBayes 1.1) and extinction rate: 0.613 species/year over speciation rate: 0.016 species/year (SubT1.1).
3.3. Reconstruction of Ancestral Character State
The analysis of ancestral states in the genus Frontonia reveals intriguing evolutionary patterns across several morphological characters;
- Vestibular kineties (VK): Group III retains the ancestral state of three rows of VK, whereas Group I and II show increases to four and five rows, respectively. Notably, F. canadensis shows transitions from three to five and back to three rows, consistent with the ancestral state of the genus Frontonia. Group IV undergoes significant changes, expanding to four, five, and even six rows (Figure 3A).
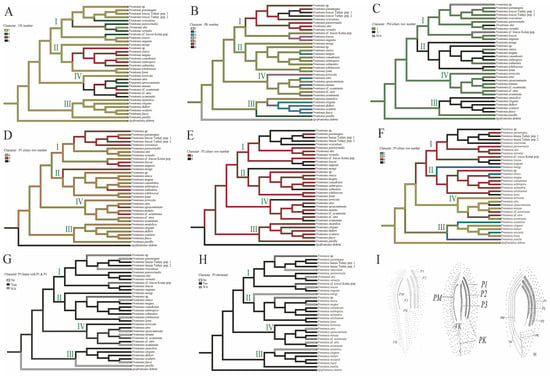 Figure 3. Ancestral state reconstruction of morphological characteristics; (A) vestibular kinety (VK) number, (B) postoral kinety (PK) number, (C) peroral membrane (PM) row number, (D) peniculi 1 (P1) row number, (E) peniculi 2 (P2) row number, (F) peniculi 3 (P3) row number, (G) linearity of peniculi 3 (P3) in relation to peniculi 1 (P1) and peniculi 2 (P2), (H) length of peniculi 3 (P3) in relation to peniculi 1 (P1) and peniculi 2 (P2) (shortened), (I) F. shii [7] (right) as an example of species in which P3 has the same structure as P1 and P2, F. paramagna [7] (middle) as an example of species in which P3 does not have the same structure as P1 and P2, and F. canadensis [5] (left) as an example of species in which P3 is shortened compared to P1 and P2. 0: no, 1: yes. Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV.
Figure 3. Ancestral state reconstruction of morphological characteristics; (A) vestibular kinety (VK) number, (B) postoral kinety (PK) number, (C) peroral membrane (PM) row number, (D) peniculi 1 (P1) row number, (E) peniculi 2 (P2) row number, (F) peniculi 3 (P3) row number, (G) linearity of peniculi 3 (P3) in relation to peniculi 1 (P1) and peniculi 2 (P2), (H) length of peniculi 3 (P3) in relation to peniculi 1 (P1) and peniculi 2 (P2) (shortened), (I) F. shii [7] (right) as an example of species in which P3 has the same structure as P1 and P2, F. paramagna [7] (middle) as an example of species in which P3 does not have the same structure as P1 and P2, and F. canadensis [5] (left) as an example of species in which P3 is shortened compared to P1 and P2. 0: no, 1: yes. Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV. - Postoral kineties (PK): Group I shows various increases in PK number (six to eight rows), Group II retains the ancestral state PK number except for F. magna, Group III shows a decrease in PK number except for F, anatolica, whereas Group IV shows both decreasing and increasing tendencies, with some species reaching up to six rows (Figure 3B).
- Peroral membrane (PM): Groups II and III show an increase in PM ciliary rows, although species such as F. sinica, F. salmastra, and F. lynni revert from two rows to one (Figure 3C).
- Peniculi 1 and 2 (P1 and P2): Group I shows a marked increase from four to five rows, while other groups, with a few exceptions, retain the ancestral state (Figure 3D,E).
- Peniculi 3 evolution: Groups I and II show an increase in ciliary rows from three to four or five, whereas F. mengi and F. sinica uniquely decrease to two rows. Group IV members retain the ancestral state with several species increasing to four or five rows, while Group III decreases from the three ancestral ciliary rows to two. Most Frontonia species retain the ancestral linear structure of peniculi 3, with a shortening of peniculi 3 considered an ancestral character (Figure 3F–H).
- Habitat adaptation: The genus Frontonia originates from brackish habitats before adapting to freshwater environments. Group III remains in the brackish environment and adapts to marine habitats for several species, while Group I and IV retain a freshwater adaptation, except for F. acuminata, which adapts to a marine environment. Group II evolves from freshwater to marine and brackish habitats (Figure 4).
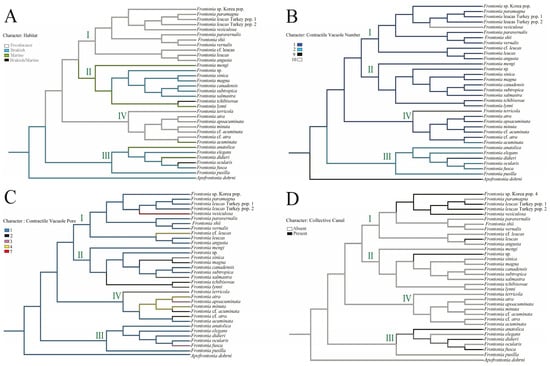 Figure 4. Reconstruction of ancestral state of Frontonia habitat (A) and morphological characteristics; (B) number of contractile vacuole (CV), (C) number of contractile vacuolar pore (CVP) and (D) presence of contractile canal (CC). Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV.
Figure 4. Reconstruction of ancestral state of Frontonia habitat (A) and morphological characteristics; (B) number of contractile vacuole (CV), (C) number of contractile vacuolar pore (CVP) and (D) presence of contractile canal (CC). Each clade or group of genus Frontonia in phylogenetic tree represent by I–IV. - Contractile vacuole (CV): The ancestral analysis suggests that the genus originally possessed a single CV, a character often retained. Group III, excluding some species, increases from one to two CVs, while unique species in Group IV show up to 10 CVs per individual (Figure 4).
- Contractile vacuolar pores (CVP) and collecting canals (CC): The evolution of CVPs and CCs is not closely correlated with the evolution of CVs (Figure 4).
These results shed light on the evolutionary history and adaptive strategies of Frontonia species, providing valuable insights into morphological evolution and habitat preferences.
4. Discussion
4.1. Phylogenetic Relationships and Morphological Distinctions in Frontonia
In this study, a paraphyletic phylogenetic tree was obtained, aligning with previous findings. While Groups III and IV are consistently separated from Groups I and II, it remains premature to conclude that they do not belong to the genus Frontonia. Morphological analysis reveals that both groups share multiple synapomorphic traits with Groups I and II, suggesting that redefining them as distinct genera would require a comprehensive taxonomic revision of Frontonia.
Group III and the genus Apofrontonia form a sister clade closely related to the genus Paramecium clade. However, their phylogenetic proximity is not mirrored in their morphological characteristics. For instance, Frontonia Group III possesses a peniculi 3 structure similar to peniculi 1 and 2, whereas genus Apofrontonia exhibits a distinct structure, and the genus Paramecium features quadrulus instead of peniculi 3. Likewise, Frontonia Group IV and its sister clade, which includes the genera Marituja and Stokesia, exhibit notable differences in peniculi 3 morphology, further distinguishing them from peniculi I and II. These findings underscore the complex interplay between phylogenetic relationships and morphological evolution within Frontonia [34].
4.2. Evolutionary Patterns of Morphological Characters in the Genus Frontonia
The evolution of morphological traits in Frontonia reflects both stable characteristics and dynamic modifications, influenced by selective pressures and developmental constraints. Key traits of the oral apparatus—including the number of vestibular kineties (VK), postoral kineties (PK), peroral membrane (PM) ciliary rows, and peniculi structures—exhibit distinct evolutionary trajectories [8]
4.2.1. Oral Apparatus Evolution: Patterns and Constraints
Vestibular kineties show a general increasing trend in Frontonia, suggesting a functional or adaptive advantage. However, occasional reversals, such as in F. canadensis, imply a potential evolutionary limit, possibly due to structural constraints or trade-offs in ciliary coordination. Meanwhile, postoral kineties demonstrate a more variable pattern, reflecting relaxed selection or species-specific functional adaptations.
The peroral membrane exhibits both doubling trends and reversals within Group II and III members, highlighting its plasticity. The reversion to a single-rowed PM in F. sinica and related species suggests that the ancestral state remains functionally viable, possibly due to energetic efficiency or redundancy in feeding structures. This aligns with the concept that trait reversibility may be constrained by ecological pressures and developmental flexibility.
Peniculi structures further emphasize differential evolutionary stability. Peniculi 1 and 2 remain conserved across species, whereas peniculi 3 undergoes frequent modifications in number, length, and structure. This suggests that while peniculi 1 and 2 perform essential functions limiting their variability, peniculi 3 may provide additional structural flexibility, allowing species-specific adaptations.
4.2.2. Contractile Vacuole Evolution: Stability vs. Plasticity
The contractile vacuole (CV) system, responsible for osmoregulation, is often shaped by habitat conditions in cilates such as Frontonia [35,36,37,38]. However, ancestral state reconstruction indicates no strict correlation between habitat and CV number, nor its associated traits (contractile vacuolar pores and contraction canals). This suggests that osmotic regulation in Frontonia is governed by factors beyond direct environmental influence, such as cellular physiology, metabolic constraints, or phylogenetic inheritance.
Interestingly, while Group II members maintain a stable CV number despite habitat shifts, Group III exhibits an evolutionary transition from a single CV to two CVs. This divergence suggests that CV evolution may be lineage-specific rather than driven by direct ecological pressures, supporting the idea that some morphological features persist due to historical contingency rather than continuous selective pressure.
4.2.3. Reversals and the Neutral Morphological Theory
The presence of reversals in both peroral membrane structure and vestibular kineties aligns with the neutral morphological theory, which proposes that morphological adaptations evolve within a limited range dictated by functional efficiency rather than continuous directional selection. Similar reversions in ciliary structures across different Frontonia lineages reinforce the idea that some morphologies represent optimal configurations, leading to recurrent evolutionary patterns.
While some lineages exhibit long-term stability in traits, others undergo modifications, suggesting a balance between conserved developmental pathways and selective pressures favoring structural plasticity. Additional studies on genetic regulation and functional morphology could provide further insights into the mechanisms driving these evolutionary patterns [39].
4.3. Evolutionary History of Genus Frontonia
The evolutionary history of the genus Frontonia provides insights into its emergence and divergence over millions of years and sheds light on the environmental factors and evolutionary processes that have shaped its diversity. The common ancestor of the four groups within the genus Frontonia and members of Penicullida emerged and diverged approximately 420 million years ago during the Palaeozoic era (Figure 2) [40]. Each group or clade within Frontonia originated at different geological times. Groups I and II shared a common ancestor at the beginning of the Mesozoic era about 230 million years ago, with Group I diverging about 172 million years ago and Group II diverging about 83 million years ago. Group III, morphologically close to the genus Apofrontonia, shared a common ancestor at the beginning of the Mesozoic era, about 185 million years ago, while Group III itself diverged about 115 million years ago. Group IV appeared about 190 million years ago, making it one of the oldest groups within Frontonia. Group I is the youngest, emerging at the end of the Mesozoic era.
4.3.1. Paleozoic Environmental Influences on Frontonia Evolution
The Paleozoic era, encompassing the Cambrian, Ordovician, Silurian, Devonian, and Carboniferous periods, was marked by significant evolutionary events, including the Cambrian explosion—a period of rapid speciation [40]. Chronogram analysis suggests that the ancestor of Frontonia likely diverged during this era. The warm climatic conditions of the Cambrian facilitated species diversification; however, fossil records indicate significant environmental fluctuations, driven by glacial and deglacial cycles, which influenced sea levels and temperatures.
Two major mass extinctions in the Paleozoic era, occurring at the end of the Devonian and Permian (late Carboniferous period), were linked to abrupt environmental shifts [41,42]. Although direct fossil evidence for ciliates is lacking, these extreme conditions may have constrained Frontonia’s ancestral diversification, as reflected in the chronogram, which shows long branches with minimal speciation events.
Diversification tests suggest a higher extinction rate than the speciation rate in Frontonia, indicating the challenges faced by ancestral populations in maintaining their existence over geological time scales. The surviving Frontonia lineages persisted into the early Mesozoic, where they underwent diversification alongside other microbial taxa, such as dinoflagellates and algae, as global temperatures stabilized [43,44].
4.3.2. Paraphyly and SSU rRNA Gene Polymorphism in Frontonia
The current paraphyly within the genus Frontonia may be due to SSU rRNA gene polymorphism in its ancestor, contributing to the high genetic diversity observed in the past. Harsh environmental conditions during the Palaeozoic era probably subjected ancestral Frontonia populations to natural selection, resulting in the decline of many populations. However, surviving populations retained variations in their SSU rRNA gene, potentially leading to divergent evolutionary trajectories. Over time, during the Mesozoic era, these divergent populations underwent speciation, possibly in response to changing environmental conditions, as supported by recent studies [8,45].
5. Conclusions
Our study provides a comprehensive analysis of the morphological evolution in Frontonia, with a focus on the oral apparatus and contractile vacuole characteristics. Ancestral state reconstruction reveals distinct evolutionary patterns, where the number of vestibular kineties tends to increase, while postoral kineties exhibit greater variability. The peroral membrane demonstrates both expansion and reversal, indicating morphological plasticity within certain lineages. Similarly, peniculi 1 and 2 remain evolutionarily stable, whereas peniculi 3 show greater structural modifications.
Although habitat transitions have been considered a key factor influencing ciliate morphology, our findings suggest that the number of contractile vacuoles and related structures remains largely stable despite environmental shifts. This indicates that habitat alone does not drive their evolution, and other selective pressures or genetic constraints may play a more significant role. The presence of both stable and reversible traits within Frontonia supports the idea of morphological evolution occurring within a constrained framework, aligning with the neutral morphological theory.
The historical perspective of the genus Frontonia traces its origins back to the Cambrian explosion, revealing survival challenges during the Paleozoic era and subsequent diversification throughout the Mesozoic era. The observed paraphyly within Frontonia is attributed to SSU rRNA gene polymorphism in its ancestor, reflecting high genetic diversity and adaptation to changing environmental conditions over time.
Author Contributions
Conceptualization, R.K.W. and M.K.S.; methodology, R.K.W. and R.A.; software, R.K.W.; validation, R.K.W. and M.K.S.; formal analysis, R.K.W.; investigation, R.K.W. and R.A.; resources, M.K.S.; data curation, R.K.W.; writing—original draft preparation, R.K.W.; writing—review and editing, R.K.W., M.K.S. and R.A.; visualization, R.K.W.; supervision, M.K.S.; project administration, M.K.S.; funding acquisition, M.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the NRF Korea funded by the Ministry of Education of the Republic of Korea (RS-2021-NR065806).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Abu Taher (University of Ulsan, University of British Columbia) for providing samples (Frontonia cf. atra) for analysis in this paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Datasheet Genus Frontonia members in this Study.
Table A1.
Datasheet Genus Frontonia members in this Study.
| Species | Accession No. | Origin | Molecular Information (Source by Publication Based on SSU rRNA Gene) | Morphological Information |
|---|---|---|---|---|
| SSU | ||||
| Frontonia anatolica | MG456578 | Turkey | Kizildang & Yildiz, 2019 [10] | Kizildang & Yildiz, 2019 [10], Yildiz & Senler, 2013 [46] |
| Frontonia anatolica | MG456581 | Turkey | ||
| Frontonia acuminata | MG456579 | Turkey | Kizildang & Yildiz, 2019 [10] | Kizildang & Yildiz, 2019 [10], Foissner, et al., 1994 [47] |
| Frontonia angusta | MG456580 | Turkey | Kizildang & Yildiz, 2019 [10] | Kizildang & Yildiz, 2019 [10], Foissner, et al., 2002 [48], Foissner, et al., 1994 [47] |
| Frontonia canadensis | KJ475313 | Shenzen, China | Zhao et al., 2016 [8] | Pan, X., et al., 2013 [5] |
| Frontonia canadensis | KJ475311 | Huizhou, China | ||
| Frontonia canadensis | KJ475309 | Huizhou, China | ||
| Frontonia canadensis | KJ475310 | Nansha, China | ||
| Frontonia canadensis | KJ475312 | Hongkang, China | ||
| Frontonia canadensis | FJ868196 | Nansha, China | Fan et al., 2013 [4] | |
| Frontonia cf. acuminata | PV016905 | Gwangju, Republic of Korea | present study | present study |
| Frontonia cf. acuminata | PV016906 | Ulsan, Republic of Korea | ||
| Frontonia cf. acuminata | PV016907 | Gunsan, Republic of Korea | ||
| Frontonia cf. acuminata | PV016908 | Ulsan, Republic of Korea | ||
| Frontonia cf. atra | PV016909 | Daejon, Republic of Korea | ||
| Frontonia cf. atra | PV016910 | Gunsan, Republic of Korea | ||
| Frontonia cf. atra | PV016911 | Masan, Republic of Korea | ||
| Frontonia cf. atra | PV016912 | Chungcheongbukdo, Republic of Korea | ||
| Frontonia cf. atra | PV016913 | Gyeongsangnamdo, Republic of Korea | ||
| Frontonia cf. atra | PV016914 | Ulsan, Republic of Korea | ||
| Frontonia cf. atra | PV016917 | Gyeongju, Republic of Korea | ||
| Frontonia atra | MT040844 | Serchio river, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12], Foissner, et al., 1994 [47] |
| Gangneung-si | Omar and Jung, 2021 [49] | |||
| Frontonia didieri | KJ475297 | Qingdao, China | Zhao et al., 2016 [8] | Long et al., 2008 [2] |
| Frontonia didieri | KJ475299 | Qingdao, China | ||
| Frontonia didieri | KJ475298 | Shenzen, China | ||
| Frontonia didieri | DQ885986 | Qingdao, China | Long et al., 2008 [2] | |
| Frontonia ocularis | FJ868198 | Guangzhou, China | Pan et al., 2013 [50] | Pan et al., 2013 [50] |
| Gangneung-si | Omar and Jung, 2021 [49] | |||
| Frontonia elegans | FJ868200 | Guangzhou, China | Fan et al., 2013 [4] | Fan et al., 2013 [4] |
| Frontonia elegans | KJ475301 | Huizhou, China | Zhao et al., 2016 [8] | |
| Frontonia elegans | KJ475302 | Huizhou, China | ||
| Frontonia elegans | KJ475303 | Huizhou, China | ||
| Frontonia cf. leucas | PV016904 | Gwangju, Republic of Korea | present study | present study |
| Frontonia leucas | MG437395 | Turkey | Yildiz & Kizildang, 2017 [10] | Yildiz & Kizildang, 2019 [10] |
| Frontonia leucas | MG437396 | |||
| Frontonia leucas | AM072622 | Italy | Fokin et al., 2006 [34] | Serra et al., 2021 [12], Foissner, et al., 1994 [47] |
| Frontonia leucas | KY855558 | Andhra Pradesh, India | Serra et al., 2021 [21] | |
| Frontonia lynni | DQ190463 | Qingdao, China | Long et al., 2008 [2] | Long et al., 2005 [1] |
| Frontonia magna | FJ868199 | Zhuhai, China | Zhao et al., 2016 [8] | Fan et al., 2011 [3] |
| Frontonia magna | FJ876953 | Shenzen, China | Fan et al., 2011 [3] | |
| Frontonia mengi | FJ875141 | Qingdao, China | Fan et al., 2011 [3] | Fan et al., 2011 [3] |
| Frontonia paramagna | KJ475304 | Guangzhou, China | Zhao et al., 2016 [8] | |
| Frontonia paramagna | KJ475306 | Sichuan, China | Zhao et al., 2016 [8] | |
| Frontonia paramagna | JQ868786 | Harbin, China | Chen et al., 2014 [6] | Chen et al., 2014 [6] |
| Frontonia paramagna | KJ475305 | Qingdao, China | Zhao et al., 2016 [8] | |
| Frontonia paramagna | MF279207 | Harbin, China | Cai et al., 2018 [7] | Cai et al., 2018 [7] |
| Frontonia paramagna | KY855554 | Andhra Pradesh, India | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia paramagna | MW031789 | Heilojiang, China | Sun et al.,2021 [9] | Sun et al., 2021 [9] |
| Frontonia shii | MF279208 | Harbin, China | Cai et al., 2018 [7] | Cai et al., 2018 [7] |
| Frontonia sinica | KJ475308 | Qingdao, China | Zhao et al., 2016 [8] | Fan et al., 2013 [4] |
| Frontonia sinica | FJ868197 | Shenzen, China | Fan et al., 2013 [4] | |
| Frontonia subtropica | FJ868202 | Shenzen, China | Fan et al., 2013 [4] | Pan et al., 2013 [4] |
| Frontonia tchibisovae | KJ475318 | Qingdao, China | Zhao et al., 2016 [8] | Long et al., 2008 [2] |
| Frontonia tchibisovae | KJ475316 | Qingdao, China | ||
| Frontonia tchibisovae | KJ475319 | Qingdao, China | ||
| Frontonia tchibisovae | DQ883820 | Yantai, China | Long et al., 2008 [2] | |
| Frontonia terricola | MF926593 | Guangzhou, China | Xu et al., 2018 [51] | Xu et al., 2018 [51], Foissner, et al., 2002 [50] |
| Frontonia pusilla | FJ868201 | Guangzhou, China | Fan et al., 2013 [4] | Fan et al., 2013 [4] |
| Frontonia vernalis | U97110 | UK | Hirt et al., 1997 [12] | |
| Frontonia vernalis | MT040840 | Tuscany, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia paravernalis | MT040839 | Tuscany, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia salmastra | MH319376 | Tuscany, Italy | Fokin et al., 2019 [52] | Fokin et al., 2019 [52] |
| Frontonia vesiculosa | MT040850 | Andhra Pradesh, India | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia minuta | MT040846 | Serchio river, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia fusca | MT040845 | Serchio river, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12], Fokin, 2008 [53] |
| Frontonia sp. | MT040843 | India | Serra et al., 2021 [12] | Serra et al., 2021 [12] |
| Frontonia sp. | MT040841 | Serchio river, Italy | Serra et al., 2021 [12] | Serra et al., 2021 [12] |

Table A2.
Datasheet that was input into speciation rate analysis software.
Table A2.
Datasheet that was input into speciation rate analysis software.
| DivBAyes 1.1 | SubT 1.1 | ||
|---|---|---|---|
| Estimated Taxa a | Diversification Date | Data of Taxa b | Substitution Taxa c |
| 69 species | 450 Mya | 33 species | 36 species |
a Estimated taxa based on Genus Frontonia species on database and published paper. b Taxa that currently SSU rRNA gene available and used in the divergent time analysis. c Taxa that SSU rRNA gene not available and not used in the divergent time analysis.
References
- Long, H.; Song, W.; Gong, J.; Hu, X.; Ma, H.; Zhu, M.; Wang, M. Frontonia lynni n. sp., a new marine ciliate (Protozoa, Ciliophora, Hymenostomatida) from Qingdao, China. Zootaxa 2005, 1003, 57–64. [Google Scholar] [CrossRef]
- Long, H.; Song, W.; Al-Rasheid, K.A.S.; Wang, Y.; Yi, Z.; Al-Quraishy, S.A.; Lin, X.; Al-Farraj, S.A. Taxonomic studies on three marine species of Frontonia from Northern China: F. didieri n. sp., F. multinucleata n. sp. and F. tchibisovae Burkovsky, 1970 (Ciliophora: Peniculida). Zootaxa 2008, 50, 35–50. [Google Scholar] [CrossRef]
- Fan, X.; Chen, X.; Song, W.; Al-Rasheid, K.A.S.; Warren, A. Two novel marine Frontonia species, Frontonia mengi spec. nov. and Frontonia magna spec. nov. (Protozoa; Ciliophora), with Notes on Their Phylogeny Based on Small-Subunit RRNA Gene Sequence Data. Int. J. Syst. Evol. Microbiol. 2011, 61, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Lin, X.; Liu, W.; Xu, Y.; Al-Farraj, S.A.; Al-Rasheid, K.A.S.; Warren, A. Morphology of three new marine Frontonia species (Ciliophora; Peniculida) with note on the phylogeny of this genus. Eur. J. Protistol. 2013, 49, 312–323. [Google Scholar] [CrossRef]
- Pan, X.; Gao, F.; Liu, W.; Fan, X.; Warren, A.; Song, W. Morphology and SSU RRNA gene sequences of three Frontonia species, including a description of F. subtropica spec. nov. (Ciliophora, Peniculida). Eur. J. Protistol. 2013, 49, 67–77. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Pan, X.; Ding, W.; Al-Rasheid, K.A.S.; Qiu, Z. Morphology and phylogeny of a new Frontonia ciliate, F. paramagna spec. nov. (Ciliophora, Peniculida) from Harbin, Ortheast China. Zootaxa 2014, 3827, 375–386. [Google Scholar] [CrossRef]
- Cai, X.; Wang, C.; Pan, X.; El-Serehy, H.A.; Mu, W.; Gao, F.; Qiu, Z. Morphology and systematics of two freshwater Frontonia species (Ciliophora, Peniculida) from Northeastern China, with comparisons among the Ffeshwater Frontonia spp. Eur. J. Protistol. 2018, 63, 105–116. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Gentekaki, E.; Zhan, A.; Al-Farraj, S.A.; Song, W. Utility of combining morphological characters, nuclear and mitochondrial genes: An attempt to resolve the conflicts of species identification for ciliated protists. Mol. Phylogenet. Evol. 2016, 94, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Li, Y.; Cai, X.; Liu, Y.; Chen, Y.; Pan, X. Further insights into the phylogeny of peniculid ciliates (Ciliophora, Oligohymenophorea) based on multigene data. Mol. Phylogenet. Evol. 2021, 154, 107003. [Google Scholar] [CrossRef] [PubMed]
- Kizildag, S.; Yildiz, I. Morphology and molecular phylogeny of four Frontonia species from Turkey (Protista, Ciliophora). Zootaxa 2019, 4609, 548–564. [Google Scholar] [CrossRef]
- Li, T.; Pan, X.; Lu, B.; Miao, M.; Liu, M. Taxonomy and molecular phylogeny of a new freshwater ciliate Frontonia apoacuminata sp. nov. (Protista, Ciliophora, Oligohymenophorea) from Qingdao, PR China. Int. J. Syst. Evol. Microbiol. 2021, 71, 005071. [Google Scholar] [CrossRef]
- Serra, V.; D’Alessandro, A.; Nitla, V.; Gammuto, L.; Modeo, L.; Petroni, G.; Fokin, S.I. The neotypification of Frontonia vernalis (Ehrenberg, 1833) Ehrenberg, 1838 and the description of Frontonia paravernalis sp. nov. trigger a critical revision of frontoniid systematics. BMC Zool. 2021, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Bourland, W.A.; Song, W. Protargol synthesis: An in-house protocol. J. Eukaryot. Microbiol. 2013, 60, 609–614. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Choi, J.K.; Dolan, J.R.; Shin, H.C.; Lee, S.; Yang, E.J. Morphological and ribosomal DNA-based characterization of six Antarctic ciliate morphospecies from the Amundsen Sea with phylogenetic analyses. J. Eukaryot. Microbiol. 2013, 60, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and high-performance computing Europe PMC Funders Group. Nat. Methods 2015, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Rataj, M.; Vďačný, P. dawn of astome ciliates in light of morphology and time-calibrated phylogeny of Haptophrya Planariarum, an obligate endosymbiont of freshwater turbellarians. Eur. J. Protistol. 2018, 64, 54–71. [Google Scholar] [CrossRef]
- Schönborn, W.; Dörfelt, H.; Foissner, W.; Krienitz, L.; Schäfer, U. A fossilized microcenosis in triassic amber. J. Eukaryot. Microbiol. 1999, 46, 571–584. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Ryberg, M.; Nilsson, R.H.; Brandon Matheny, P. Divbayes and SubT: Exploring species diversification using bayesian statistics. Bioinformatics 2011, 27, 2439–2440. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. 2019. Available online: http://www.mesquiteproject.org (accessed on 1 June 2022).
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. No TitleThe CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, UT, USA, 18–21 July 2011; pp. 1–8. [Google Scholar]
- Ho, S.Y.W.; Duchêne, S. Molecular-clock methods for estimating evolutionary rates and timescales. Mol. Ecol. 2014, 23, 5947–5965. [Google Scholar] [CrossRef]
- Rajter, Ľ.; Vďačný, P. Rapid radiation, gradual extinction and parallel evolution challenge generic classification of spathidiid ciliates (Protista, Ciliophora). Zool. Scr. 2016, 45, 200–223. [Google Scholar] [CrossRef]
- Vďačný, P. Estimation of divergence times in litostomatean ciliates (Ciliophora: Intramacronucleata), using Bayesian relaxed clock and 18S rRNA gene. Eur. J. Protistol. 2015, 51, 321–334. [Google Scholar] [CrossRef]
- Vďačný, P.; Breiner, H.-W.; Yashchenko, V.; Dunthorn, M.; Stoeck, T.; Foissner, W. The Chaos Prevails: Molecular Phylogeny of the Haptoria (Ciliophora, Litostomatea). Protist 2014, 165, 93–111. [Google Scholar] [CrossRef] [PubMed]
- Fokin, S.; Andreoli, I.; Verni, F.; Petroni, G. Apofrontonia dohrni sp. n. and the phylogenetic relationships within Peniculia (Protista, Ciliophora, Oligohymenophorea). Zool. Scr. 2006, 35, 289–300. [Google Scholar] [CrossRef]
- Patterson, D.J. Contractile Vacuoles and Associated Structures: Their Organization and Function. Biol. Rev. 1980, 55, 1–46. [Google Scholar] [CrossRef]
- Sun, P.; Clamp, J.; Xu, D.; Huang, B.; Shin, M.K. An integrative approach to phylogeny reveals patterns of environmental distribution and novel evolutionary relationships in a major group of ciliates. Sci. Rep. 2016, 6, 21695. [Google Scholar] [CrossRef]
- Syberg-Olsen, M.J.; Irwin, N.A.T.; Vannini, C.; Erra, F.; Di Giuseppe, G.; Boscaro, V.; Keeling, P.J. Biogeography and character evolution of the ciliate genus Euplotes (Spirotrichea, Euplotia), with description of Euplotes curdsi sp. nov. PLoS ONE 2016, 11, e0165442. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Warren, A.; Song, W.B. Species delimitation for the molecular taxonomy and ecology of the widely distributed microbial eukaryote genus euplotes (Alveolata, Ciliophora). Proc. R. Soc. B Biol. Sci. 2018, 285, 20172159. [Google Scholar] [CrossRef] [PubMed]
- Lahr, D.J.G.; Laughinghouse, H.D.; Oliverio, A.M.; Gao, F.; Katz, L.A. How discordant morphological and molecular evolution among microorganisms can revise our notions of biodiversity on Earth. BioEssays 2014, 36, 950–959. [Google Scholar] [CrossRef]
- Babcock, L.E.; Peng, S.C.; Brett, C.E.; Zhu, M.Y.; Ahlberg, P.; Bevis, M.; Robison, R.A. Global climate, sea level cycles, and biotic events in the Cambrian Period. Palaeoworld 2015, 24, 5–15. [Google Scholar] [CrossRef]
- Sepkoski, J.J. Environmental Trends in Extinction During the Paleozoic; American Association for the Advancement of Science: Washington, DC, USA, 2010; Volume 235, pp. 64–66. Available online: http://www.jstor.org/stable/1698912 (accessed on 1 June 2024).
- Stanley, S.M.; Yang, X. A double mass extinction at the end of the paleozoic era. Science 1994, 266, 1340–1344. [Google Scholar] [CrossRef]
- Fensome, R.A.; MacRae, R.A.; Moldowan, J.M.; Taylor, F.J.R.; Williams, G.L. The early Mesozoic radiation of dinoflagellates. Paleobiology 1996, 22, 329–338. [Google Scholar] [CrossRef]
- Grzebyk, D.; Schofield, O.; Vetriani, C.; Falkowski, P.G. The Mesozoic Radiation of Eukaryotic Algae: The Portable Plastid Hypothesis. J. Phycol. 2003, 39, 259–267. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, T.; Wang, Y.; Katz, L.A.; Gao, F.; Song, W. Disentangling sources of variation in SSU RDNA sequences from single cell analyses of ciliates: Impact of copy number variation and experimental error. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170425. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, I.; Şenler, N.G. Frontonia anatolica n. sp., a new peniculid ciliate (Protista, Ciliophora) from Lake Van, Turkey. Turkish J. Zool. 2013, 37, 24–30. [Google Scholar] [CrossRef]
- Foissner, W. Progress in taxonomy of planktonic freshwater ciliates. Mar. Microb. Food Webs 1994, 8, 9–35. [Google Scholar]
- Foissner, W.; Agatha, S.; Berger, H. Soil Ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with Emphasis on Two Contrasting Environments, the Etosha Region and the Namib Desert. Part I: Text and Line Drawings; Biologiezentrum der Oberösterreichischen Landesmuseums: Linz, Austria, 2002; Volume 5, pp. 1–1063. [Google Scholar]
- Omar, A.; Jung, J. New record of 21 ciliate species (Protozoa, Ciliophora) from South Korea. J. Species Res. 2021, 10, 301–320. [Google Scholar] [CrossRef]
- Pan, X.; Liu, W.; Yi, Z.; Fan, X.; Al-Rasheid, K.A.S.; Lin, X. Studies on three diverse Frontonia species (Ciliophora, Peniculida), with brief notes on 14 marine or brackish congeners. Acta Protozool. 2013, 52, 35–49. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, F.; Fan, X. Reconsideration of the systematics of Peniculida (Protista, Ciliophora) based on SSU rRNA gene sequences and new morphological features of Marituja and Disematostoma. Hydrobiologia 2018, 806, 313–331. [Google Scholar] [CrossRef]
- Fokin, S.I.; Serra, V.; Ferrantini, F.; Modeo, L.; Petroni, G. “Candidatus Hafkinia Simulans” gen. nov., sp. nov., a novel Holospora-like Bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora): Multidisciplinary characterization of the new endosymbiont and Its Host. Microb. Ecol. 2019, 77, 1092–1106. [Google Scholar] [CrossRef]
- Fokin, S.I. Rediscovery and characterisation of Frontonia fusca (QUENNERSTEDT, 1869) KAHL, 1931 (Ciliophora, Peniculia). Denisia 2008, 23, 251–259. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).