Leontodon albanicus subsp. acroceraunicus (Asteraceae, Cichorieae): A New Subspecies from Southern Albania
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphometric Analyses
2.2. AFLPseq Fingerprinting
3. Results
3.1. Morphometric Analyses
3.2. AFLPseq Fingerprinting
4. Discussion
5. Conclusions
Taxonomic Treatment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Samples | Taxon | Locality | Collectors | Voucher Specimens |
|---|---|---|---|---|
| A1297 | albanicus | ALB, Nemërçkë, Maja e Papingut, 2180 m | Conti & Manilla s.n. | APP 48886 |
| A1298 | albanicus | ALB, Nemërçkë, 500–120 m | Conti et al. s.n. | APP 57114 |
| A1299 | albanicus | ALB, Nemërçkë, 500–120 m | Conti et al. s.n. | APP 57115 |
| A1300 | albanicus | ALB, Nemercka—Diopala. | Lakušić et al. s.n. | APP 56301 |
| A1301 | albanicus | ALB, Poliçan—Nemërçkë, 1200–2480 m | Conti et al. s.n. | APP 56996 |
| A1306 | acroceraunicus | ALB, Llogarasa pass, 1700–2000 m | Conti & Manilla s.n. | APP 49368 |
| A1307 | acroceraunicus | ALB, Llogarasa pass, 1700–2000 m | Conti & Manilla s.n. | APP 49368 |
| A1308 | acroceraunicus | ALB, Llogarasa pass, 1700–2000 m | Conti & Manilla s.n. | APP 49368 |
| A1309 | acroceraunicus | ALB, Llogarasa pass, 1700–2000 m | Conti & Manilla s.n. | APP 49385 |
References
- Lack, H.W.; Kilian, N. Leontodon. In Mountain Flora of Greece; Strid, A., Tan, K., Eds.; Edinburgh University Press: Edinburgh, UK, 1991; pp. 526–531. [Google Scholar]
- Meusel, H.; Jäger, E. Vergleichende Chorologie der Zentraleuropeischen Flora, III; Gustav Fischer Verlag: Jena, Germany; Stuttgart, Germany; New York, NY, USA, 1992; pp. 1: ix + 333, 2: ix + 266. [Google Scholar]
- Zidorn, C. Leontodon and Scorzoneroides (Asteraceae, Cichorieae) in Italy. Plant Biosyst. 2012, 146 (Suppl. S1), 41–51. [Google Scholar] [CrossRef]
- Samuel, R.; Gutermann, W.; Stuessy, T.F.; Ruas, C.F.; Lack, H.-W.; Tremetsberger, K.; Talavera, S.; Hermanowski, B.; Ehrendorfer, F. Molecular phylogenetics reveals Leontodon (Asteraceae, Lactuceae) to be diphyletic. Amer. J. Bot. 2006, 93, 1193–1205. [Google Scholar] [CrossRef]
- Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. Available online: https://powo.science.kew.org/ (accessed on 20 January 2025).
- Conti, F. A new combination in Leontodon (Asteraceae, Cichorieae). Phytotaxa 2018, 360, 287–291. [Google Scholar] [CrossRef]
- New York Botanical Garden’s Virtual Herbarium. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. Available online: http://sweetgum.nybg.org/science/ih (accessed on 20 January 2025).
- Kolmogorov, A. Sulla determinazione empirica di una legge di distribuzione. Ist. Ital. Attuari G. 1933, 4, 89–91. [Google Scholar]
- Student. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: Principles and Practice of Numerical Classification; W.H. Freeman and Company: San Francisco, CA, USA, 1973; pp. 381–409. [Google Scholar]
- Krzanowski, W.J. Principles of Multivariate Analysis; Clarendon Press: Oxford, UK, 1990; pp. 33–85. [Google Scholar]
- Addinsoft. XLSTAT: Data Analysis and Statistical Solution for Microsoft Excel; Addinsoft: Paris, France, 2024. [Google Scholar]
- Dorfner, M.; Ott, T.; Ott, P.; Oberprieler, C. Long-read genotyping with SLANG (Simple Long-read loci Assembly of Nanopore data for Genotyping). Appl. Plant Sci. 2022, 10, e11484. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Dickson, E. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 1987, 36, 715–722. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc. Amer. 1987, 19, 11–15. [Google Scholar] [CrossRef]
- Conti, F.; Oberprieler, C.; Dorfner, M.; Schabel, E.; Nicoara, R.; Bartolucci, F. Adonis fucensis (A. sect. Adonanthe, Ranunculaceae), a new species from the central Apennines (Italy). Biology 2023, 12, 118. [Google Scholar] [CrossRef]
- Conti, F.; Oberprieler, C.; Dorfner, M.; Schabel, E.; Bartolucci, F. Pedicularis rostratospicata subsp. marsica (P. sect. Rostratae, Orobanchaceae), a new subspecies from the central Apennines (Italy). Biology 2023, 12, 2614. [Google Scholar] [CrossRef]
- Oberprieler, C. The Wettstein tesseract: A tool for conceptualising species-rank decisions and illustrating speciation trajectories. Taxon 2023, 72, 1–7. [Google Scholar] [CrossRef]
- Baldacci, A. Rivista critica della collezione botanica fatta nel 1892 in Albania. Malpighia 1894, 8, 159–192. [Google Scholar]
- Barina, Z.; Somogyi, G.; Pifkó, D.; Rakaj, M. Checklist of vascular plants of Albania. Phytotaxa 2018, 378, 1–339. [Google Scholar] [CrossRef]
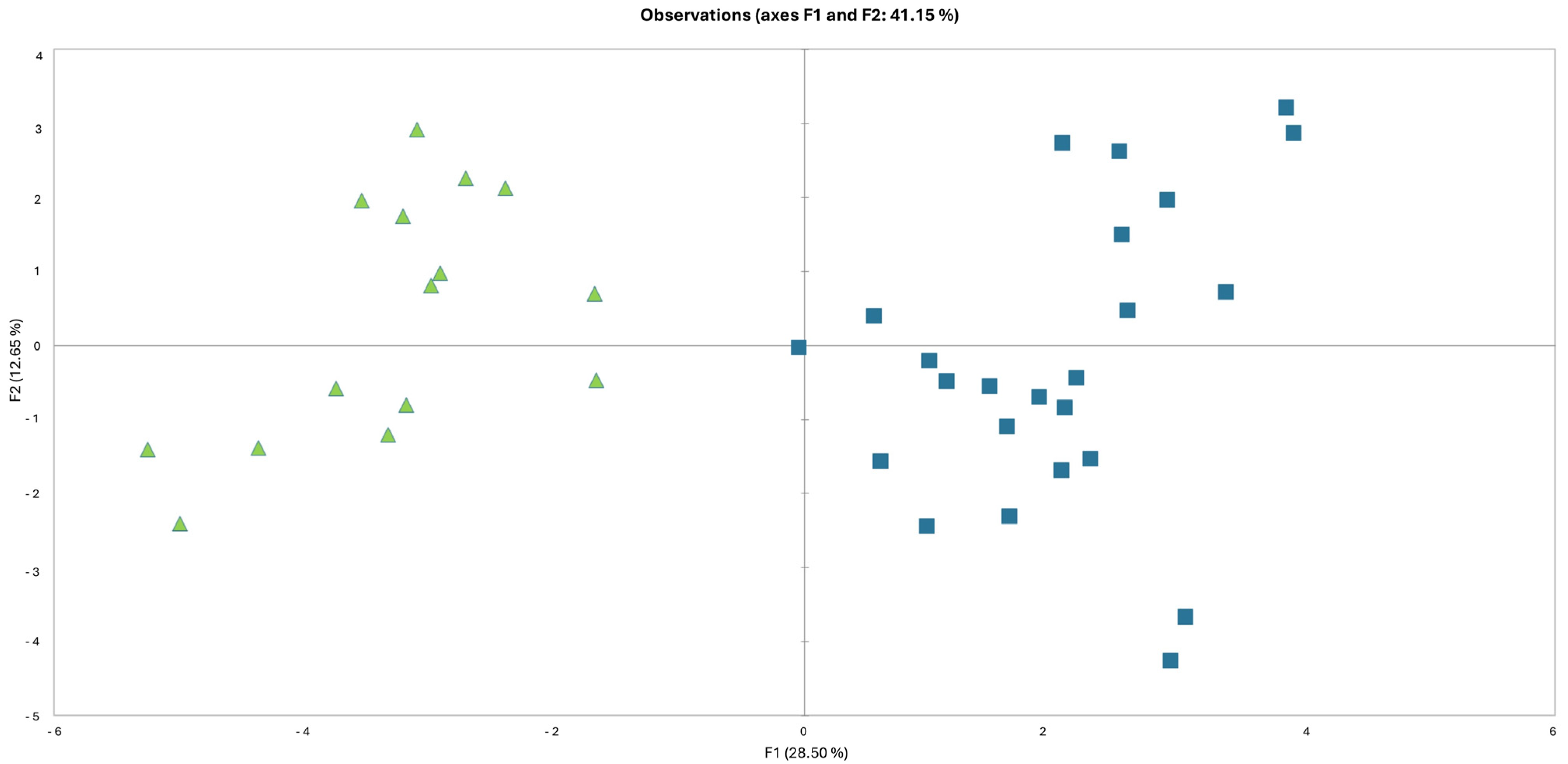
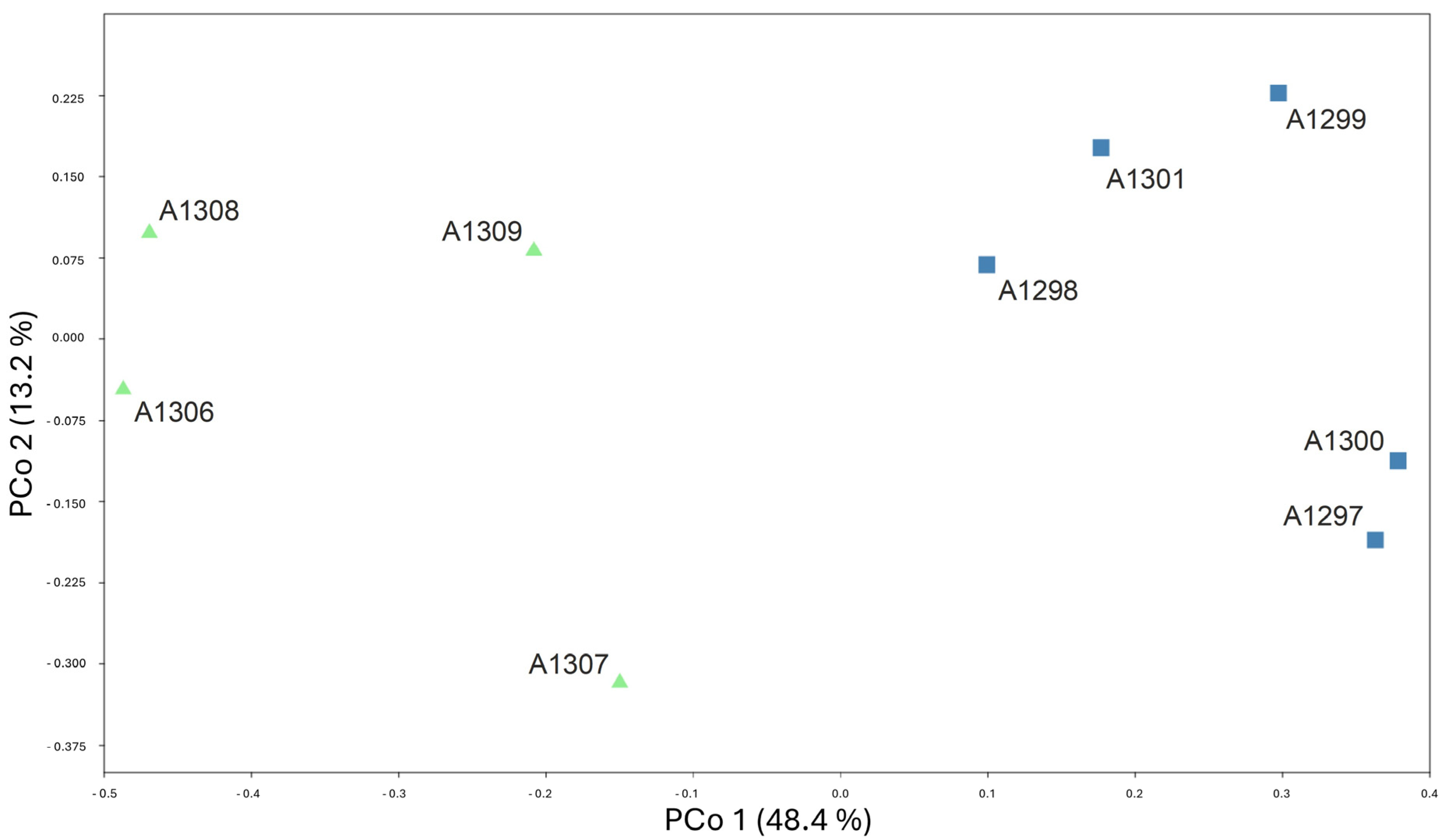



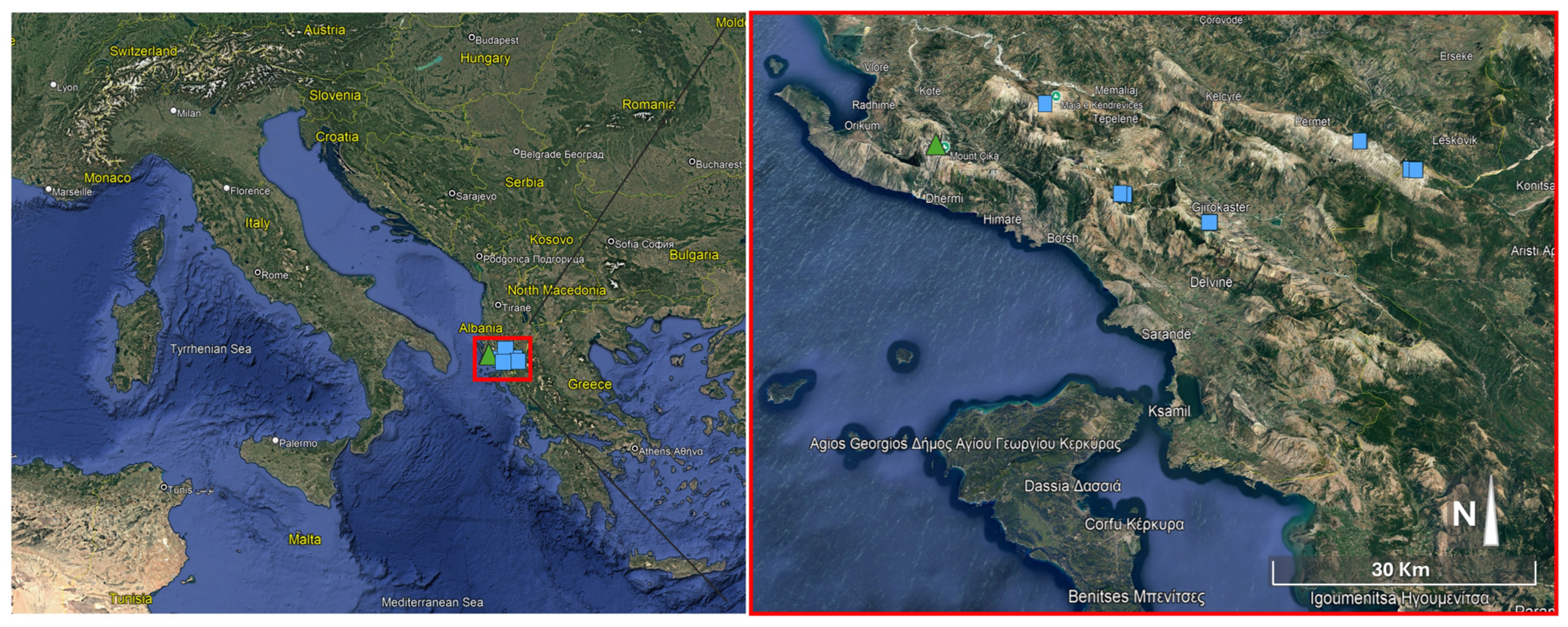
| Group 1 (L. albanicus subsp. albanicus) | Group 2 (L. albanicus subsp. acroceraunicus) | ||||||
|---|---|---|---|---|---|---|---|
| Character | Mean | Min | Max | Mean | Min | Max | p-Value |
| Plant height (cm) * | 132.17 | 76 | 280 | 110.28 | 65 | 170 | 0.162 |
| Leaf length (mm) * | 64.62 | 31 | 125 | 53.47 | 22 | 95 | 0.121 |
| Leaf width (mm) * | 11.67 | 4 | 20 | 8.67 | 5 | 13 | 0.011 |
| Number of teeth along each leaf side ** | 3.21 | 1 | 7 | 3.87 | 2 | 7 | 0.144 |
| Width of leaf teeth (mm) ** | 1.77 | 0.3 | 4.2 | 2.41 | 0.8 | 7 | 0.265 |
| Number of hairs per 1 mm2 on leaf surface * | 31.33 | 15 | 47 | 47.38 | 31 | 95 | 0.001 |
| Leaf hairs: most common number of distal rays ** | 7.71 | 6 | 8 | 4.13 | 4 | 5 | <0.001 |
| Leaf hairs: minimum number of distal rays ** | 5.29 | 4 | 6 | 3.33 | 3 | 4 | <0.001 |
| Leaf hairs: maximum number of distal rays ** | 9.33 | 8 | 11 | 5.8 | 5 | 8 | <0.001 |
| Leaf hairs: stalk length (mm) * | 0.17 | 0.1 | 0.27 | 0.19 | 0.06 | 0.32 | 0.269 |
| Leaf hairs: maximum stalk length (mm) * | 0.26 | 0.14 | 0.37 | 0.34 | 0.16 | 0.55 | 0.006 |
| Leaf hairs: ray length (mm) * | 0.19 | 0.13 | 0.18 | 0.21 | 0.12 | 0.27 | 0.158 |
| Involucre: length (mm) * | 14.33 | 12 | 19 | 12.15 | 10 | 16 | 0.002 |
| Involucre: external bract length (mm) * | 5.43 | 3.5 | 7 | 3.71 | 2.5 | 5.5 | <0.001 |
| Stellate hairs on external involucral bracts: number of rays ** | 4.95 | 3 | 8 | 2.78 | 1 | 4 | <0.001 |
| Hairs on external involucral bracts: hair length (mm) * | 1.58 | 0.9 | 3 | 1.35 | 0.83 | 2 | 0.113 |
| Hairs on external and/or middle involucral bracts: length of simple moniliformis hairs (mm) * | 0.2 | 0.1 | 0.38 | 0.18 | 0.12 | 0.31 | 0.251 |
| Stellate hairs on external involucral bract margins: number of rays ** | 5.05 | 3 | 8 | 2.71 | 1 | 5 | <0.001 |
| Stellate hairs on external involucral bract margins: stalk length (mm) ** | 0.46 | 0 | 2 | 0.13 | 0.06 | 0.34 | 0.129 |
| Stellate hairs on external involucral bract margins: ray length (mm) * | 0.21 | 0.1 | 0.5 | 0.1 | 0 | 0.23 | 0.001 |
| Stellate hairs on middle involucral bract: number of rays * | 4.05 | 0 | 5 | 2.21 | 1 | 3 | <0.001 |
| Stellate hairs on middle involucral bract: stalk length (mm) * | 1.05 | 0 | 2.3 | 1.13 | 0.18 | 2.2 | 0.698 |
| Stellate hairs on middle involucral bract: ray length (mm) ** | 0.26 | 0 | 0.8 | 0.3 | 0 | 1.1 | 0.913 |
| Achene: length (mm) * | 5.19 | 4 | 6.3 | 4.9 | 3.5 | 6 | 0.152 |
| Pappus: ray length (mm) * | 9.38 | 7.3 | 11.5 | 8,05 | 5.9 | 11 | 0.001 |
| Pappus: plume length (mm) * | 0.28 | 0 | 0.48 | 0.3 | 0 | 0.55 | 0.52 |
| Pappus: denticule length (mm) * | 0.05 | 0.03 | 0.08 | 0.04 | 0.02 | 0.07 | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conti, F.; Bracchetti, L.; Dorfner, M.; Benda, N.; Oberprieler, C. Leontodon albanicus subsp. acroceraunicus (Asteraceae, Cichorieae): A New Subspecies from Southern Albania. Biology 2025, 14, 259. https://doi.org/10.3390/biology14030259
Conti F, Bracchetti L, Dorfner M, Benda N, Oberprieler C. Leontodon albanicus subsp. acroceraunicus (Asteraceae, Cichorieae): A New Subspecies from Southern Albania. Biology. 2025; 14(3):259. https://doi.org/10.3390/biology14030259
Chicago/Turabian StyleConti, Fabio, Luca Bracchetti, Marco Dorfner, Nadine Benda, and Christoph Oberprieler. 2025. "Leontodon albanicus subsp. acroceraunicus (Asteraceae, Cichorieae): A New Subspecies from Southern Albania" Biology 14, no. 3: 259. https://doi.org/10.3390/biology14030259
APA StyleConti, F., Bracchetti, L., Dorfner, M., Benda, N., & Oberprieler, C. (2025). Leontodon albanicus subsp. acroceraunicus (Asteraceae, Cichorieae): A New Subspecies from Southern Albania. Biology, 14(3), 259. https://doi.org/10.3390/biology14030259







