Comprehensive Analysis of Rodent-Specific Probasin Gene Reveals Its Evolutionary Origin in Pseudoautosomal Region and Provides Novel Insights into Rodent Phylogeny
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
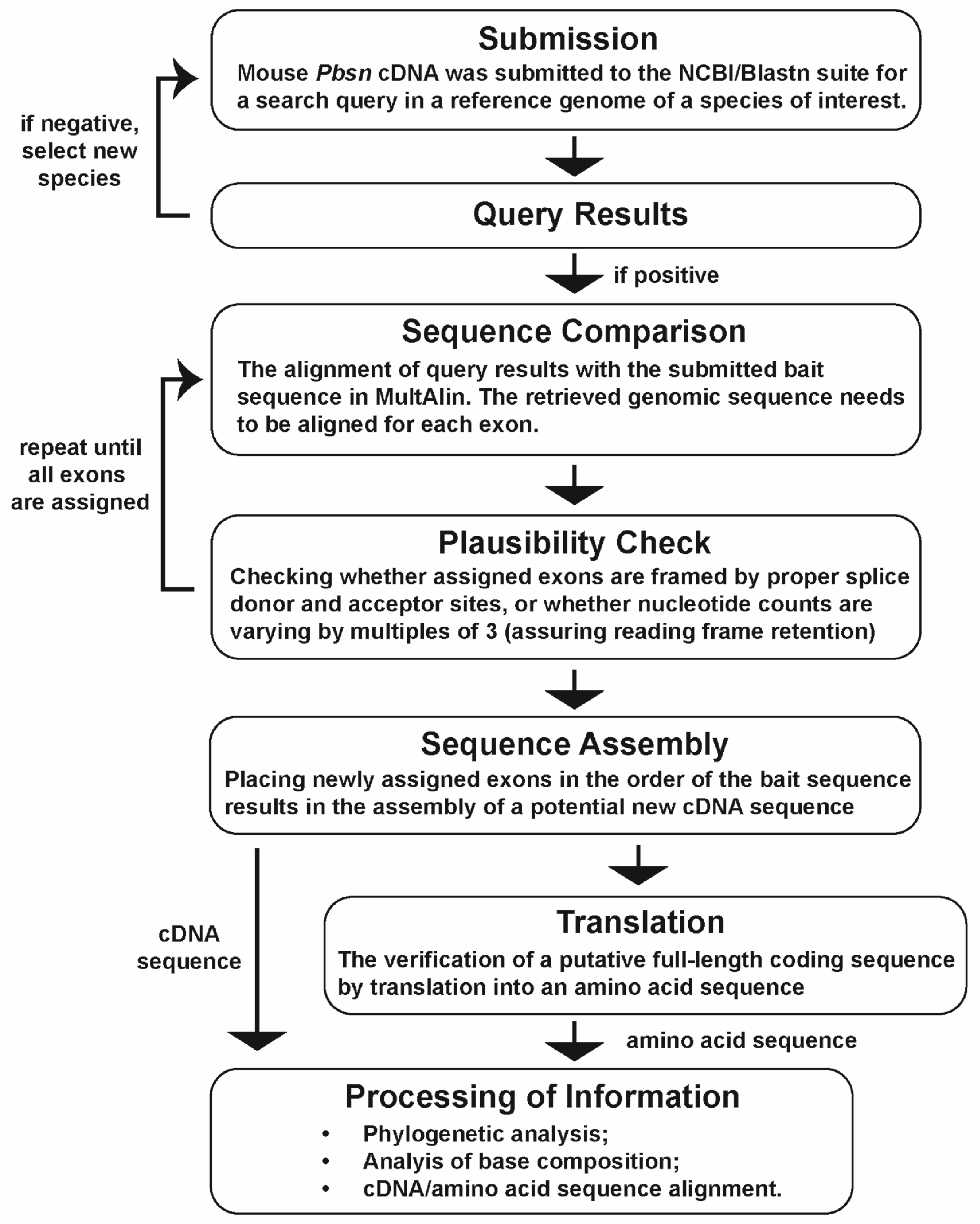
2.1. Bioinformatical Analyses
2.2. RT-PCR Analysis
2.3. Statistical Analyses
3. Results
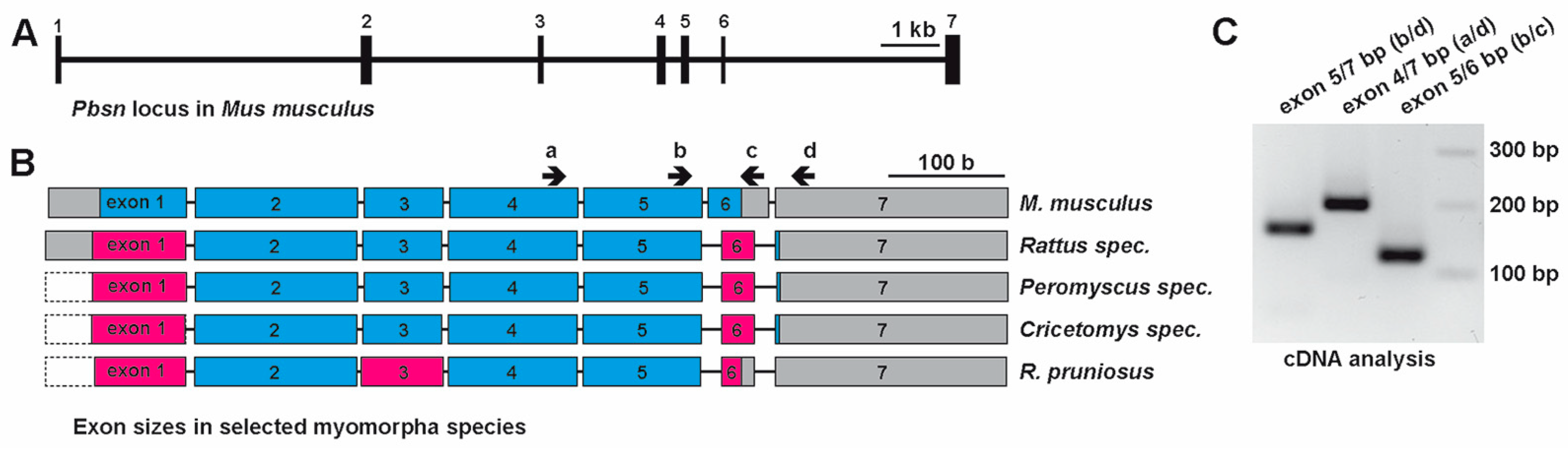
3.1. PBSN Coding Sequences Vary Among Muroidea Families
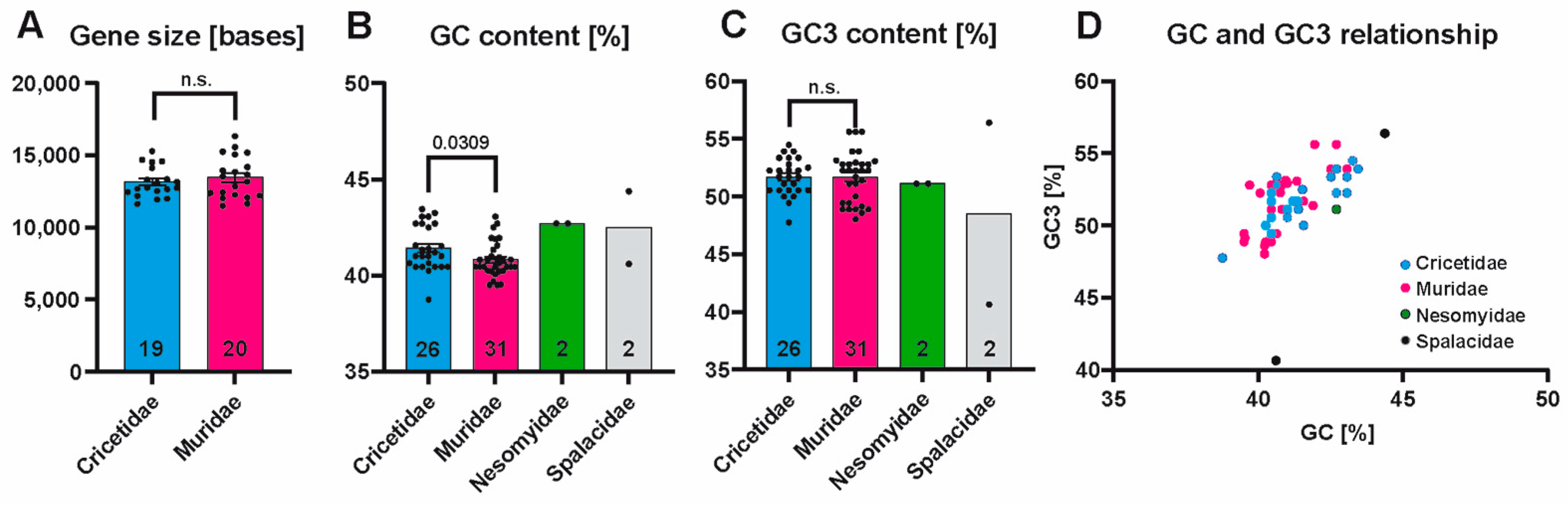
3.2. PBSN Gene Sizes and Nucleotide Compositions Are Comparable Among Muroidea Species
3.3. Phylogenetic Analysis of Probasin-Coding Sequences Outgroups the Spalacida Family
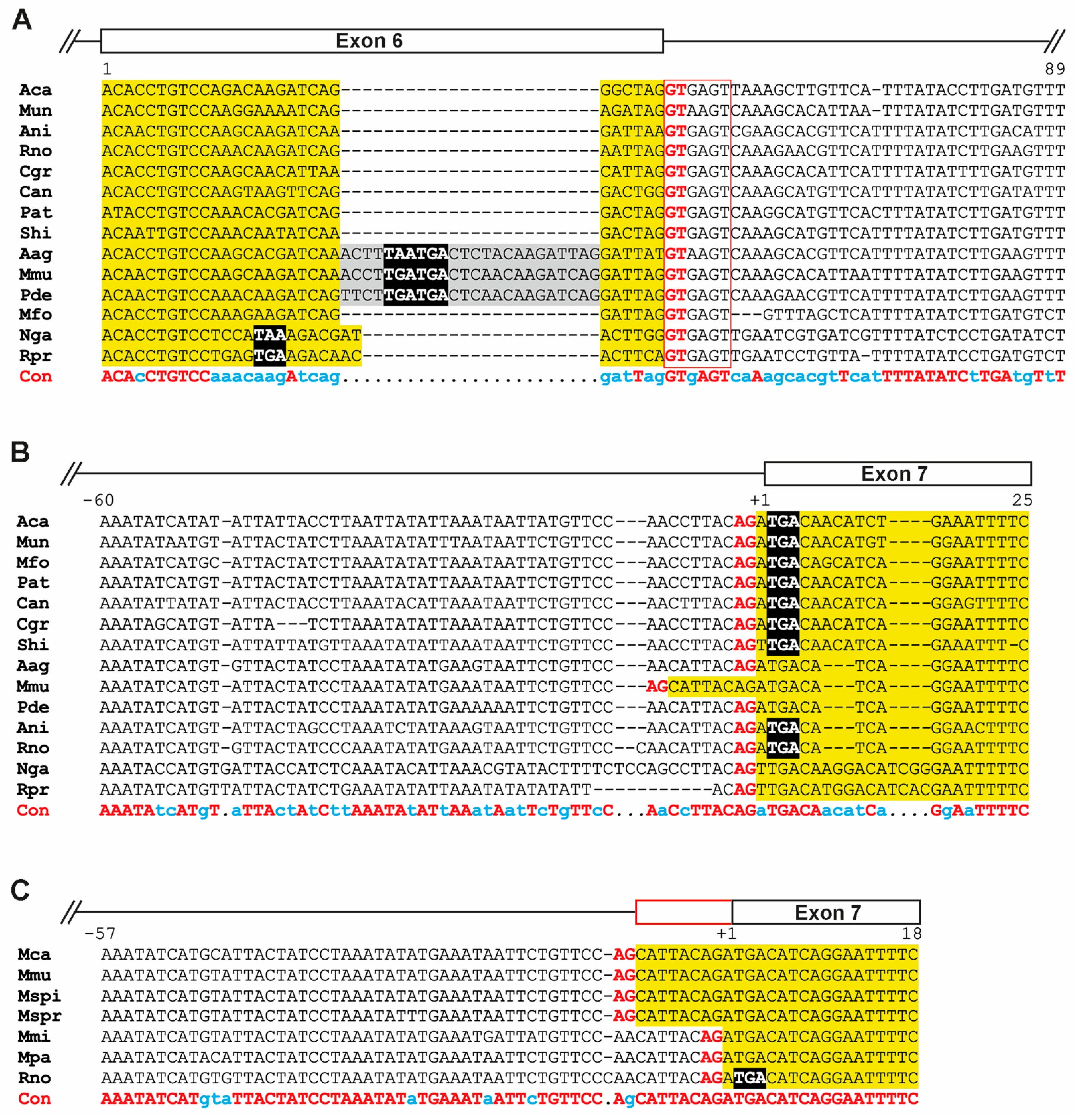
3.4. Genomic Recombination Events Affect PBSN Gene Localization on the X-Chromosome
3.5. Probasin Proteins Retain Structural Features of Lipocalin Protein Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Raudsepp, T.; Chowdhary, B.P. The Eutherian Pseudoautosomal Region. Cytogenet. Genome Res. 2015, 147, 81–94. [Google Scholar] [CrossRef]
- Maxeiner, S.; Gebhardt, S.; Schweizer, F.; Venghaus, A.E.; Krasteva-Christ, G. Of mice and men-and guinea pigs? Ann. Anat. 2021, 238, 151765. [Google Scholar] [CrossRef]
- Maxeiner, S.; Benseler, F.; Krasteva-Christ, G.; Brose, N.; Südhof, T.C. Evolution of the Autism-Associated Neuroligin-4 Gene Reveals Broad Erosion of Pseudoautosomal Regions in Rodents. Mol. Biol. Evol. 2020, 37, 1243–1258. [Google Scholar] [CrossRef] [PubMed]
- Maxeiner, S.; Benseler, F.; Brose, N.; Krasteva-Christ, G. Of Humans and Gerbils- Independent Diversification of Neuroligin-4 Into X- and Y-Specific Genes in Primates and Rodents. Front. Mol. Neurosci. 2022, 15, 838262. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef]
- Burgin, C.J.; Colella, J.P.; Kahn, P.L.; Upham, N.S. How many species of mammals are there? J. Mammal. 2018, 99, 1–14. [Google Scholar] [CrossRef]
- Matuo, Y.; Nishi, N.; Muguruma, Y.; Yoshitake, Y.; Kurata, N.; Wada, F. Localization of prostatic basic protein (“probasin”) in the rat prostates by use of monoclonal antibody. Biochem. Biophys. Res. Commun. 1985, 130, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Kasper, S.; Matusik, R.J. Rat probasin: Structure and function of an outlier lipocalin. Biochim. Biophys. Acta 2000, 1482, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Hernandez, I.; Wie, Y.; Greenberg, N. Isolation and characterization of mouse probasin: An androgen-regulated protein specifically expressed in the differentiated prostate. Prostate 2000, 43, 255–262. [Google Scholar] [CrossRef]
- Charkoftaki, G.; Wang, Y.; McAndrews, M.; Bruford, E.A.; Thompson, D.C.; Vasiliou, V.; Nebert, D.W. Update on the human and mouse lipocalin (LCN) gene family; including evidence the mouse Mup cluster is result of an “evolutionary bloom”. Hum. Genom. 2019, 13, 11. [Google Scholar] [CrossRef]
- Pelosi, P.; Knoll, W. Odorant-binding proteins of mammals. Biol. Rev. Camb. Philos. Soc. 2022, 97, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, N.M.; DeMayo, F.; Finegold, M.J.; Medina, D.; Tilley, W.D.; Aspinall, J.O.; Cunha, G.R.; Donjacour, A.A.; Matusik, R.J.; Rosen, J.M. Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. USA 1995, 92, 3439–3443. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Szabolcs, M.; Terwilliger, J.D.; Efstratiadis, A. Prostatic intraepithelial neoplasia and adenocarcinoma in mice expressing a probasin-Neu oncogenic transgene. Carcinogenesis 2006, 27, 1054–1067. [Google Scholar] [CrossRef]
- Wu, X.; Wu, J.; Huang, J.; Powell, W.C.; Zhang, J.; Matusik, R.J.; Sangiorgi, F.O.; Maxson, R.E.; Sucov, H.M.; Roy-Burman, P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech. Dev. 2001, 101, 61–69. [Google Scholar] [CrossRef]
- Maxeiner, S.; Krasteva-Christ, G.; Althaus, M. Pitfalls of using sequence databases for heterologous expression studies-a technical review. J. Physiol. 2023, 601, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Hollenhorst, M.I.; Husnik, T.; Zylka, M.; Duda, N.; Flockerzi, V.; Tschernig, T.; Maxeiner, S.; Krasteva-Christ, G. Human airway tuft cells influence the mucociliary clearance through cholinergic signalling. Respir. Res. 2023, 24, 267. [Google Scholar] [CrossRef] [PubMed]
- D’Elía, G.; Fabre, P.H.; Lessa, E.P. Rodent systematics in an age of discovery: Recent advances and prospects. J. Mammal. 2019, 100, 852–871. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Spence, A.M.; Sheppard, P.C.; Davie, J.R.; Matuo, Y.; Nishi, N.; McKeehan, W.L.; Dodd, J.G.; Matusik, R.J. Regulation of a bifunctional mRNA results in synthesis of secreted and nuclear probasin. Proc. Natl. Acad. Sci. USA 1989, 86, 7843–7847. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Kumar, S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 2002, 19, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Steppan, S.; Adkins, R.; Anderson, J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 2004, 53, 533–553. [Google Scholar] [CrossRef]
- Fabre, P.H.; Herrel, A.; Fitriana, Y.; Meslin, L.; Hautier, L. Masticatory muscle architecture in a water-rat from Australasia (Murinae; Hydromys) and ist implication for the evolution of carnivory in rodents. J. Anat. 2017, 231, 380–397. [Google Scholar] [CrossRef]
- Aghová, T.; Kimura, Y.; Bryja, J.; Dobigny, G.; Granjon, L.; Kergoat, G.J. Fossils know it best: Using a new set of fossil calibrations to improve the temporal phylogenetic framework of murid rodents (Rodentia: Muridae). Mol. Phylogenet. Evol. 2018, 128, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Steppan, S.J.; Schenk, J.J. Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS ONE 2017, 12, e0183070. [Google Scholar] [CrossRef]
- Hobolth, A.; Christensen, O.F.; Mailund, T.; Schierup, M.H. Genomic Relationships and Speciation Times of Human, Chimpanzee, and Gorilla Inferred from a Coalescent Hidden Markov Model. PLoS Genet. 2007, 3, e7. [Google Scholar] [CrossRef] [PubMed]
- Matuo, Y.; Adams, P.S.; Nishi, N.; Yasumitsu, H.; Crabb, J.W.; Matusik, R.J.; McKeehan, W.L. The androgen-dependent rat prostate protein, probasin, is a heparin-binding protein that co-purifies with heparin-binding growth factor-1. In Vitro Cell. Dev. Biol. 1989, 25, 581–584. [Google Scholar]
- Roberts, S.A.; Prescott, M.C.; Davidson, A.J.; McLean, L.; Beynon, R.J.; Hurst, J.L. Individual odour signatures that mice learn are shaped by involatile major urinary proteins (MUPs). BMC Biol. 2018, 16, 48. [Google Scholar] [CrossRef]

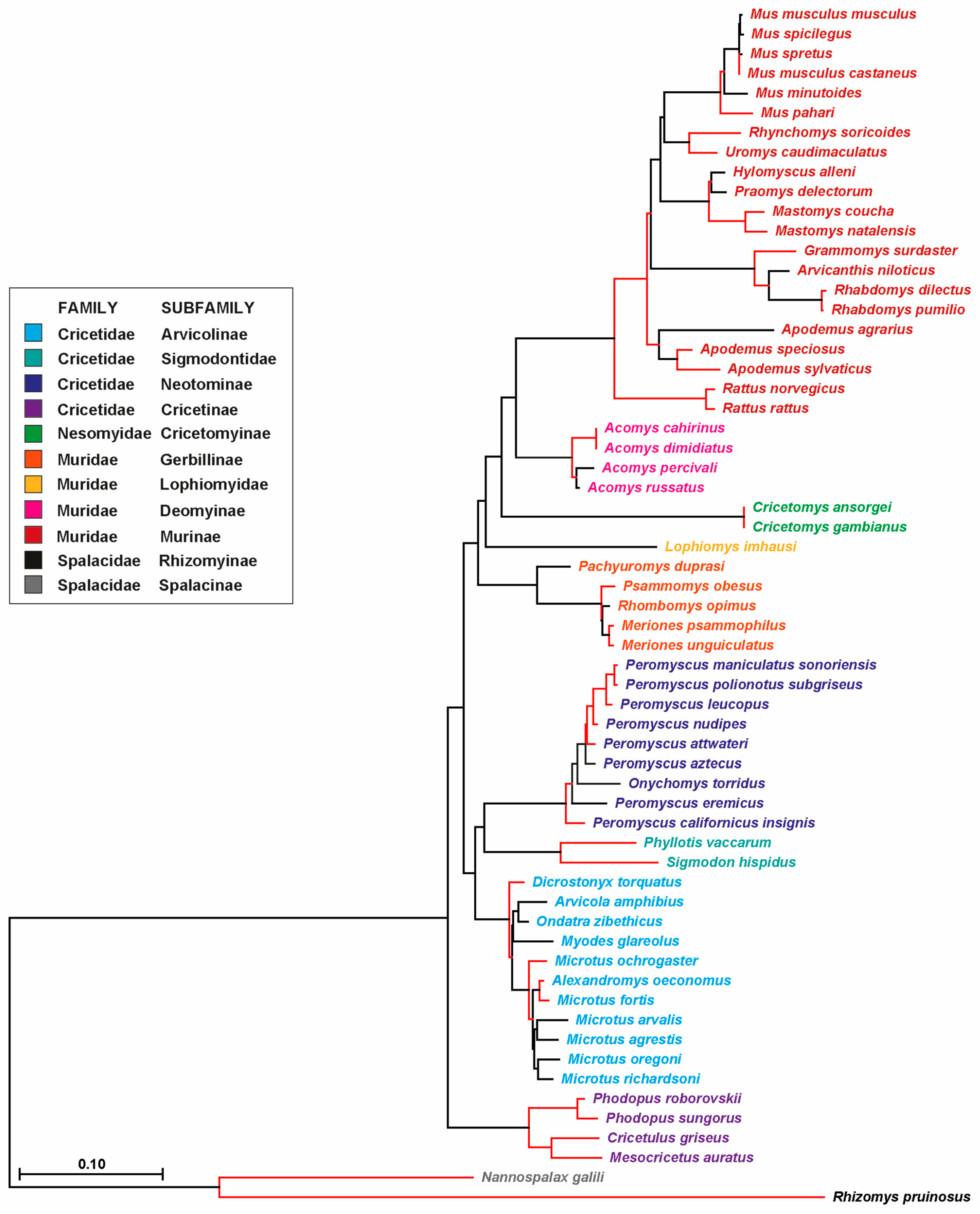
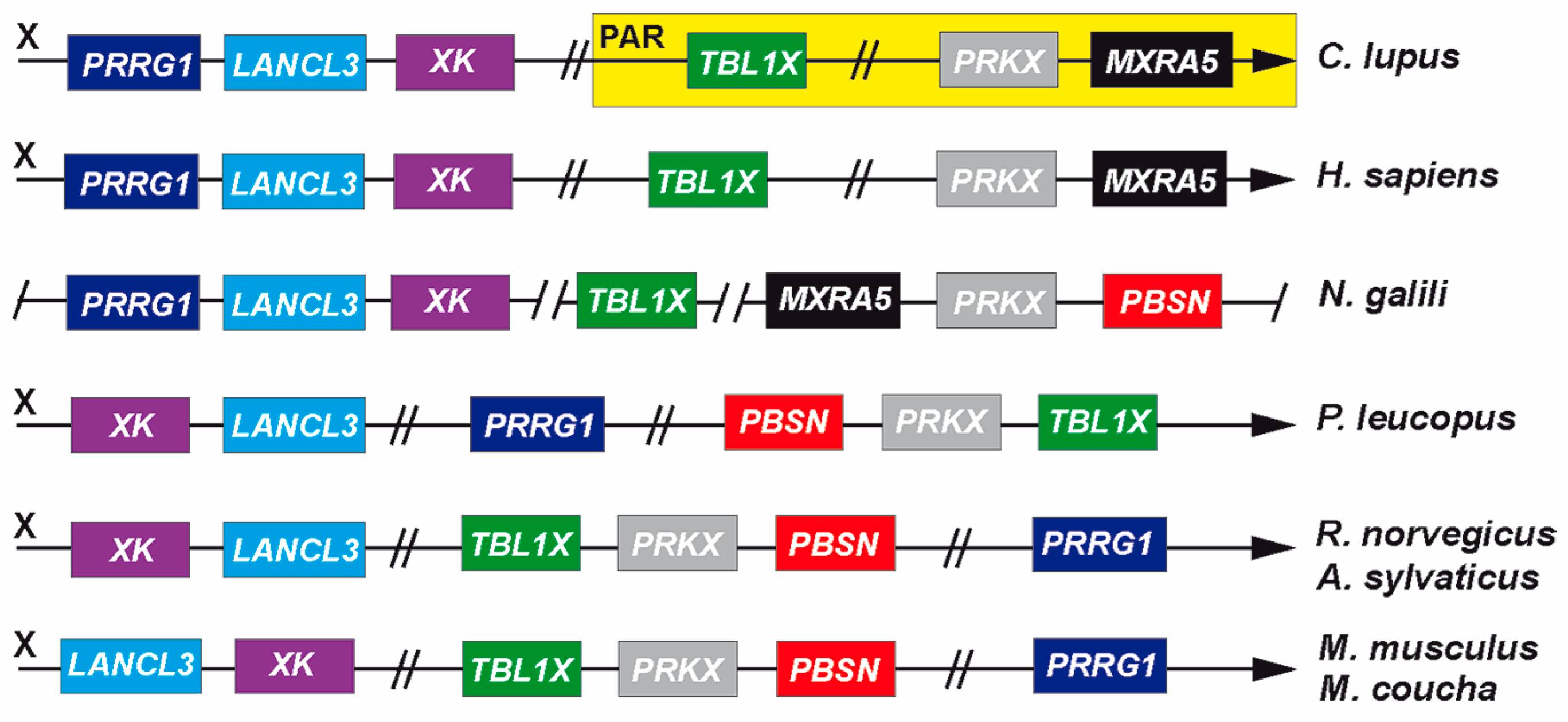
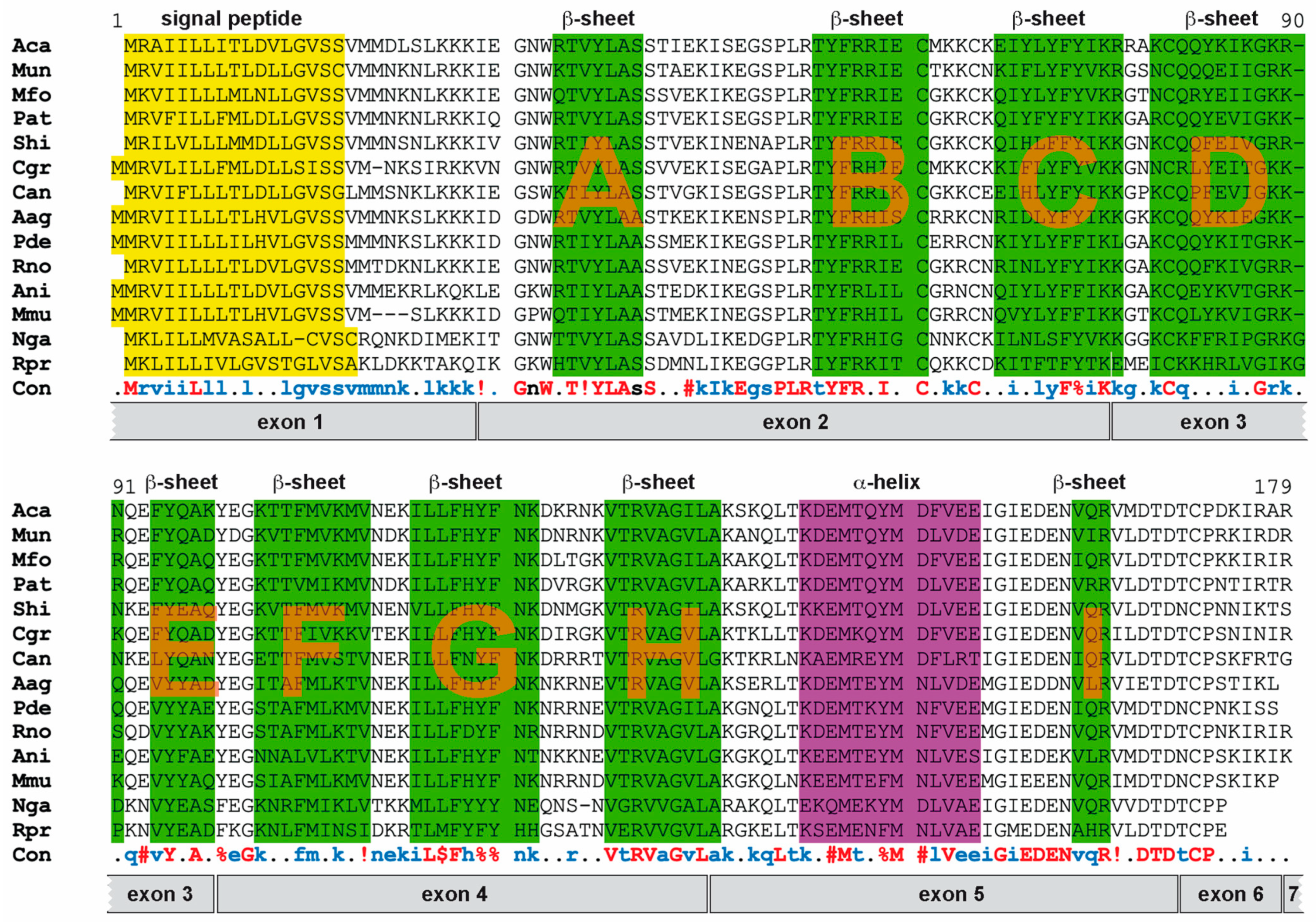
| Family | Species |
|---|---|
| Cricetidae | Alexandromys oeconomus 1, Arvicola amphibius 2, Cricetulus griseus 2, Dicrostonyx torquatus 1, Mesocricetus auratus 1, Microtus agrestis 1, M. arvalis 1, M. fortis 2, M. ochrogaster 1, M. oregoni 2, M. richardsoni 1, Myodes glareolus 2, Ondatra zibethicus 1, Onychomys torridus 2, Peromyscus attwateri 1, P. aztecus 1, P. californicus insignis 2, P. emericus 2, P. leucopus, 2 P. manniculatus sonoriensis 2, P. nudipes 1, P. polionotus subgriseus 1, Phodopus roborovskii 2, P. sungorus 1, Phyllotis vaccarum 1, Sigmodon hispidus 1 |
| Muridae | Acomys cahirinus 1, A. dimidiatus 1, A. percivali 1, A. russatus 2, Apodemus agrarius 1, A. speciosus 1, A. sylvaticus 2, Arvicanthis niloticus 2, Grammomys surdaster 2, Hylomyscus alleni 1, Lophiomys imhaus 1, Mastomys coucha 2, M. natalensis 1, Meriones psammophilus 1, M. unguiculatus 2, Mus minutoides 1, M. musculus 2,3, M. musculus castaneus 1, M. pahari 2, M. spicilegus 1, M. spretus1, Pachyuromys duprasi 1, Praomys delectorum 1, Psammomys obesus 2, Rattus norvegicus 2, R. rattus 2, Rhabdomys dilectus 1, R. pumilio 1, Rhombomys opimus 1, Rhynchomys soricoides 1, Uromys caudimaculus 1 |
| Nesomyidae | Cricetomys ansorgei 1, C. gambianus 1 |
| Spalacidae | Nannospalax galili 2, Rhizomys pruinosus 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maxeiner, S.; Walter, L.; Zeitz, S.L.; Krasteva-Christ, G. Comprehensive Analysis of Rodent-Specific Probasin Gene Reveals Its Evolutionary Origin in Pseudoautosomal Region and Provides Novel Insights into Rodent Phylogeny. Biology 2025, 14, 239. https://doi.org/10.3390/biology14030239
Maxeiner S, Walter L, Zeitz SL, Krasteva-Christ G. Comprehensive Analysis of Rodent-Specific Probasin Gene Reveals Its Evolutionary Origin in Pseudoautosomal Region and Provides Novel Insights into Rodent Phylogeny. Biology. 2025; 14(3):239. https://doi.org/10.3390/biology14030239
Chicago/Turabian StyleMaxeiner, Stephan, Lukas Walter, Samuel Luca Zeitz, and Gabriela Krasteva-Christ. 2025. "Comprehensive Analysis of Rodent-Specific Probasin Gene Reveals Its Evolutionary Origin in Pseudoautosomal Region and Provides Novel Insights into Rodent Phylogeny" Biology 14, no. 3: 239. https://doi.org/10.3390/biology14030239
APA StyleMaxeiner, S., Walter, L., Zeitz, S. L., & Krasteva-Christ, G. (2025). Comprehensive Analysis of Rodent-Specific Probasin Gene Reveals Its Evolutionary Origin in Pseudoautosomal Region and Provides Novel Insights into Rodent Phylogeny. Biology, 14(3), 239. https://doi.org/10.3390/biology14030239






