Repetitive DNA Dynamics, Phylogenetic Relationships and Divergence Times in Andean Ctenomys (Rodentia: Ctenomyidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chromosomal Preparations and Genomic DNA Extraction
2.2. Cytogenetic Hybridization Techniques
2.3. Phylogenetic Analysis and Divergence Time Estimation
3. Results
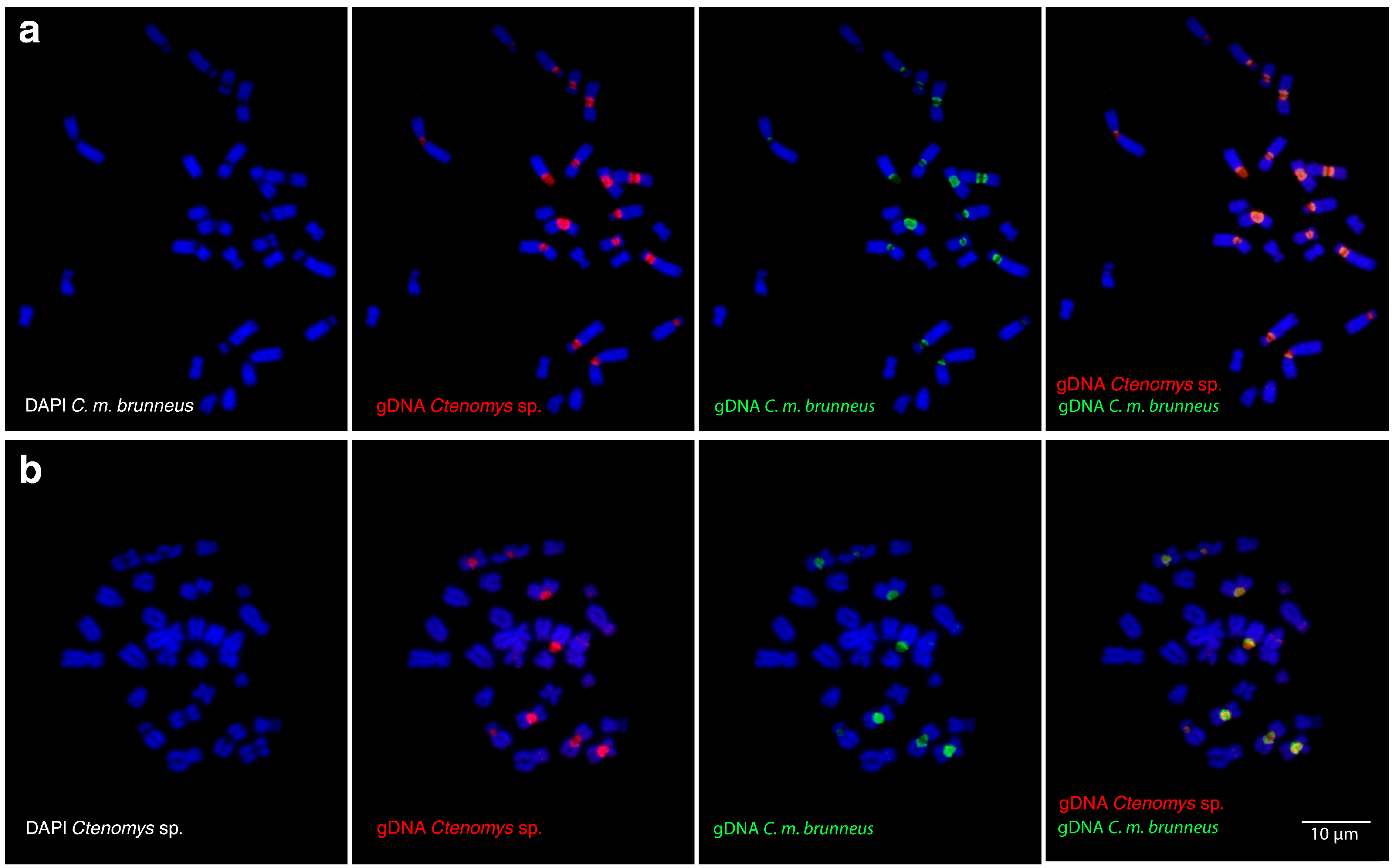
3.1. Cytogenetic Characterization of Repetitive Sequences
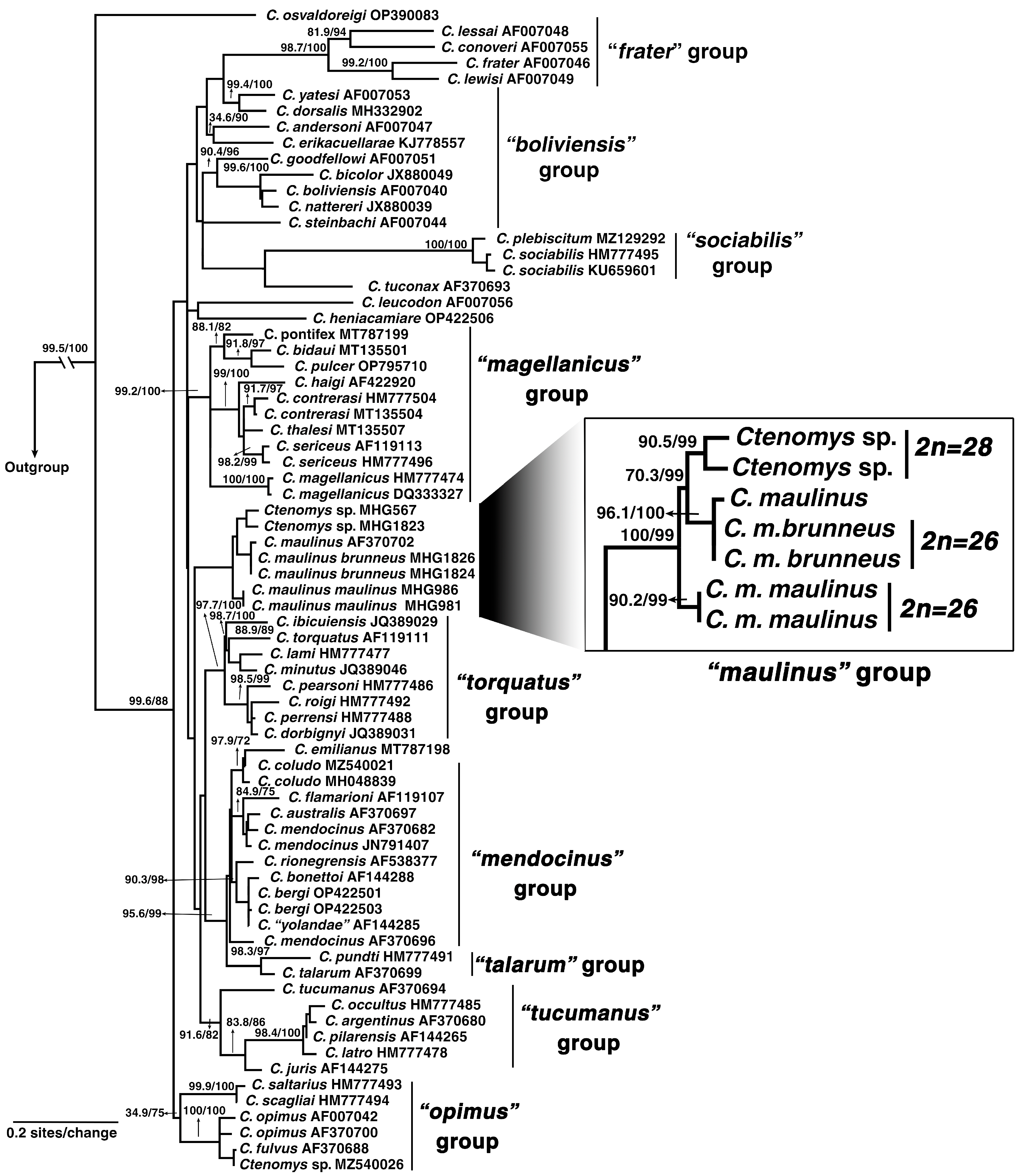
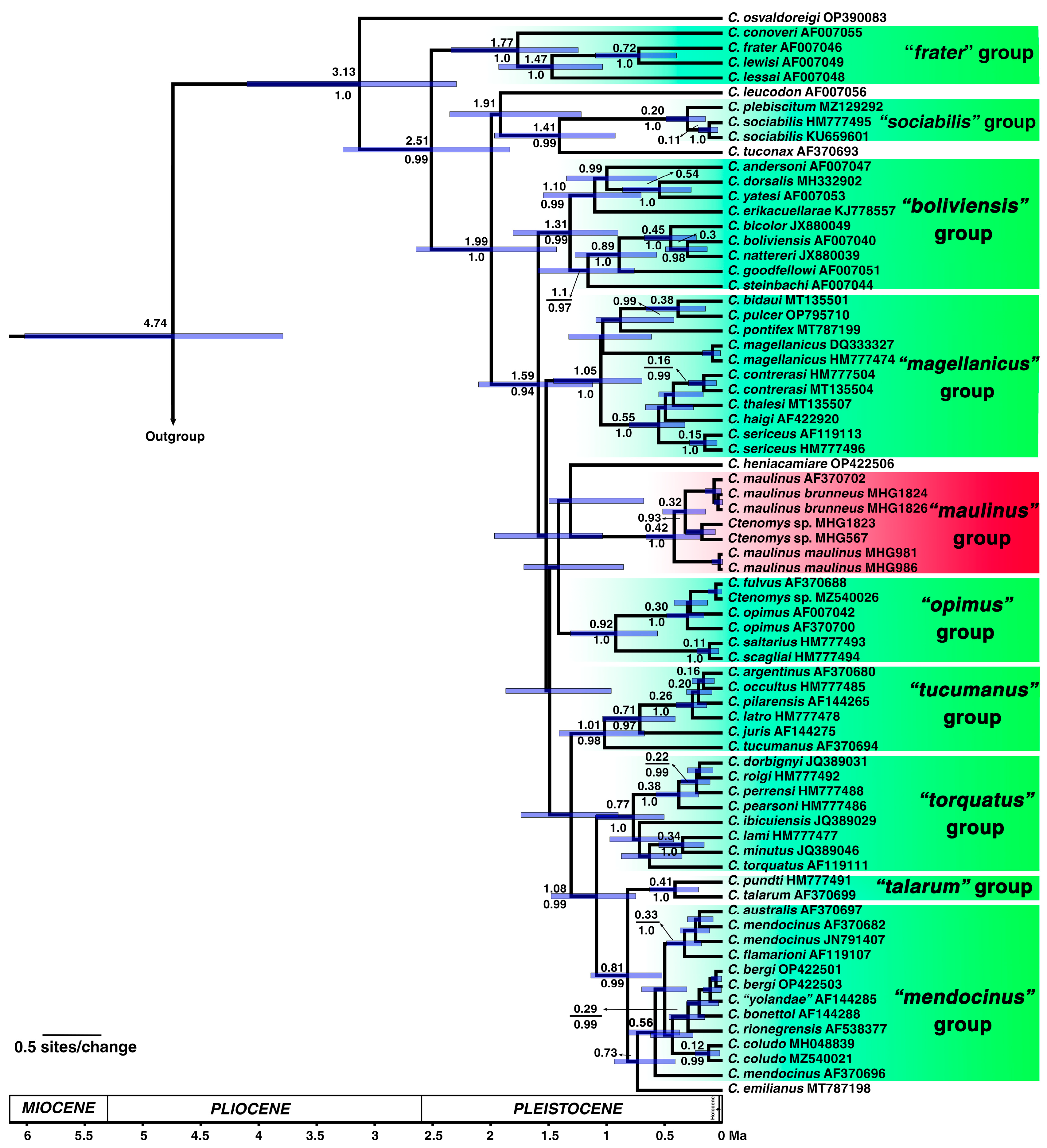
3.2. Phylogeny and Divergence Times in Ctenomys
4. Discussion
4.1. Phylogenetic Context and Karyotypic Evolution in the maulinus Group
4.2. Repetitive DNA Dynamics in Parapatric Ctenomys
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Altemose, N. A classical revival: Human satellite DNAs enter the genomics era. Semin. Cell Dev. Biol. 2022, 128, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Craig-Holmes, A.P.; Shaw, M.W. Polymorphism of human constitutive heterochromatin. Science 1971, 174, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Gadgil, R.; Barthelemy, J.; Lewis, T.; Leffak, M. Replication stalling and DNA microsatellite instability. Biophys. Chem. 2017, 225, 38–48. [Google Scholar] [CrossRef]
- Kim, J.C.; Mirkin, S.M. The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 2013, 23, 280–288. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Palacios-Gimenez, O.M. Repetitive DNAs: The ‘invisible’ regulators of insect adaptation and speciation. Curr. Opin. Insect Sci. 2025, 67, 101295. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Mehrotra, S.; Goel, S.; Sharma, S.; Raina, S.N.; Rajpal, V.R. Sequence analysis of KpnI repeat sequences to revisit the phylogeny of the Genus Carthamus L. Appl. Biochem. Biotechnol. 2013, 169, 1109–1125. [Google Scholar] [CrossRef]
- Silva, D.M.; Pansonato-Alves, J.C.; Utsunomia, R.; Daniel, S.N.; Hashimoto, D.T.; Oliveira, C.; Porto-Foresti, F.; Foresti, F. Chromosomal organization of repetitive DNA sequences in Astyanax bockmanni (Teleostei, Characiformes): Dispersive location, association and co-localization in the genome. Genetica 2013, 141, 329–336. [Google Scholar] [CrossRef]
- Adega, F.; Guedes-Pinto, H.; Chaves, R. Satellite DNA in the karyotype evolution of domestic animals—clinical considerations. Cytogenet. Genome Res. 2009, 126, 12–20. [Google Scholar] [CrossRef]
- Paco, A.; Adega, F.; Mestrovic, N.; Plohl, M.; Chaves, R. The puzzling character of repetitive DNA in Phodopus genomes (Cricetidae, Rodentia). Chromosome Res. 2015, 23, 427–440. [Google Scholar] [CrossRef]
- Liao, X.; Zhu, W.; Zhou, J.; Li, H.; Xu, X.; Zhang, B.; Gao, X. Repetitive DNA sequence detection and its role in the human genome. Commun. Biol. 2023, 6, 954. [Google Scholar] [CrossRef]
- Shapiro, J.A.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. 2005, 80, 227–250. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef]
- Bladen, J.; Phadnis, N. Genome evolution: A story of species and satellites. Curr. Biol. 2022, 32, R736–R738. [Google Scholar] [CrossRef]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef] [PubMed]
- D’Elía, G.; Teta, P.; Lessa, E.P. A short overview of the systematics of Ctenomys: Species limits and phylogenetic relationships. In Tuco-Tucos: An Evolutionary Approach to the Diversity of a Neotropical Subterranean Rodent; Freitas, T.R.O.d., Gonçalves, G.L., Maestri, R., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 17–41. [Google Scholar]
- Teta, P.; Jayat, J.P.; Alvarado-Larios, R.; Ojeda, A.A.; Cuello, P.; D’Elía, G. An appraisal of the species richness of the Ctenomys mendocinus species group (Rodentia: Ctenomyidae), with the description of two new species from the Andean slopes of west-central Argentina. Vertebr. Zool. 2023, 73, 451–474. [Google Scholar] [CrossRef]
- Gallardo, M.H.; Bickham, J.W.; Kausel, G.; Kohler, N.; Honeycutt, R.L. Gradual and quantum genome size shifts in the hystricognath rodents. J. Evol. Biol. 2003, 16, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Ruedas, L.A.; Cook, J.A.; Yates, T.L.; Bickham, J.W. Conservative genome size and rapid chromosomal evolution in the South American tuco-tucos (Rodentia: Ctenomyidae). Genome 1993, 36, 449–458. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Cook, J.A.; Lessa, E.P.; Rossi, M.S. Recurrent amplifications and deletions of satellite DNA accompanied chromosomal diversification in south American tuco-tucos (genus Ctenomys, Rodentia: Octodontidae): A phylogenetic approach. Mol. Biol. Evol. 2001, 18, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Caraballo, D.A. Reassessing the causal connection between satDNA dynamics and chromosomal evolution in Ctenomys (Rodentia, Ctenomyidae): Unveiling the overlooked importance of the Y chromosome. Contrib. Zool. 2023, 92, 533–546. [Google Scholar] [CrossRef]
- Buschiazzo, L.M.; Caraballo, D.A.; Labaroni, C.A.; Teta, P.; Rossi, M.S.; Bidau, C.J.; Lanzone, C. Comprehensive cytogenetic analysis of the most chromosomally variable mammalian genus from South America: Ctenomys (Rodentia: Caviomorpha: Ctenomyidae). Mamm. Biol. 2022, 102, 1963–1979. [Google Scholar] [CrossRef]
- Gallardo, M.H. Karyotypic evolution in Ctenomys (Rodentia, Ctenomyidae). J. Mammal. 1991, 72, 11–21. [Google Scholar] [CrossRef]
- Ortells, M.O. Phylogenetic analysis of G-banded karyotypes among the South American subterranean rodents of the genus Ctenomys (Caviomorpha: Octodontidae), with special reference to chromosomal evolution and speciation. Biol. J. Linn. Soc. Lond. 1995, 54, 43–70. [Google Scholar] [CrossRef]
- Gallardo, M.H. Las especies chilenas de Ctenomys. I. Estabilidad cariotípica. Arch. Biol. Y Med. Exp. 1979, 12, 71–82. [Google Scholar]
- Aguilar, G. Relaciones Sistemáticas entre Citotipos del Género Ctenomys (Rodentia, Ctenomyidae), en Chile Central; Universidad Austral de Chile: Valdivia, Chile, 1988. [Google Scholar]
- Ramírez, P.A. Morfometría Craneal en Tres Formas del Género Ctenomys (Blainville, 1826) de la Zona Centro Sur de Chile; Universidad Austral de Chile: Valdivia, Chile, 2017. [Google Scholar]
- Suárez-Villota, E.Y.; Pansonato-Alves, J.C.; Foresti, F.; Gallardo, M.H. Homomorphic sex chromosomes and the intriguing Y chromosome of Ctenomys rodent species (Rodentia, Ctenomyidae). Cytogenet. Genome Res. 2014, 143, 232–240. [Google Scholar] [CrossRef] [PubMed]
- She, C.; Liu, J.; Diao, Y.; Hu, Z.; Song, Y. The Distribution of Repetitive DNAs Along Chromosomes in Plants Revealed by Self-genomic in situ Hybridization. J. Genet. Genom. 2007, 34, 437–448. [Google Scholar] [CrossRef]
- Pita, M.; Fernández, J.L.; Gosálvez, J. Whole-comparative genomic hybridization and “cell code” estimation: An application for assessment of cellular chimerism. Eur. J. Med. Res. 2007, 12, 206–211. [Google Scholar]
- Pita, M.; Luis Fernández, J.; Gosálvez, J. Whole-comparative genomic hybridization (W-CGH): 1. The quick overview of repetitive DNA sequences on a genome. Chromosome Res. 2003, 11, 673–679. [Google Scholar] [CrossRef]
- Verma, R.S.; Babu, A. Human Chromosomes: Principles and Techniques, 2nd ed.; McGraw-Hill: New York, NY, USA, 1995. [Google Scholar]
- Sharma, R.; Sharad, S.; Minhas, G.; Sharma, D.R.; Bhatia, K.; Sharma, N.K. Chapter 12—DNA, RNA isolation, primer designing, sequence submission, and phylogenetic analysis. In Basic Biotechniques for Bioprocess and Bioentrepreneurship; Bhatt, A.K., Bhatia, R.K., Bhalla, T.C., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 197–206. [Google Scholar]
- Gosalvez, J.; Crespo, F.; Vega-Pla, J.L.; López-Fernández, C.; Cortés-Gutiérrez, E.I.; Devila-Rodriguez, M.I.; Mezzanotte, R. Shared Y chromosome repetitive DNA sequences in stallion and donkey as visualized using whole-genomic comparative hybridization. Eur. J. Histochem. 2010, 54, e2. [Google Scholar] [CrossRef][Green Version]
- Giardini, M.C.; Milla, F.H.; Conte, C.A.; Lanzavecchia, S.B.; Nieves, M. Exploring the molecular differentiation of sex chromosomes in Anastrepha fraterculus sp. 1 using comparative genomic hybridization (CGH). Mol. Biol. Rep. 2025, 52, 250. [Google Scholar] [CrossRef]
- Pita, M.; Zabal-aguirre, M.; Arroyo, F.; Gosálvez, J.; López-Fernández, C.; De La Torre, J. Arcyptera fusca and Arcyptera tornosi repetitive DNA families: Whole-comparative genomic hybridization (W-CGH) as a novel approach to the study of satellite DNA libraries. J. Evol. Biol. 2008, 21, 352–361. [Google Scholar] [CrossRef]
- Suárez-Villota, E.Y.; Vargas, R.A.; Marchant, C.L.; Torres, J.E.; Köhler, N.; Núñez, J.J.; de la Fuente, R.; Page, J.; Gallardo, M.H. Distribution of repetitive DNAs and the hybrid origin of the red vizcacha rat (Octodontidae). Genome 2012, 55, 105–117. [Google Scholar] [CrossRef]
- Liao, M.; Zheng, J.; Wang, Z.; Wang, Y.; Zhang, J.; Cai, M. Molecular cytogenetic of the Amoy croaker, Argyrosomus amoyensis (Teleostei, Sciaenidae). J. Oceanol. Limnol. 2018, 36, 842–849. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed]
- De Santi, N.A.; Olivares, A.I.; Piñero, P.; Villoldo, J.A.F.; Verzi, D.H. An exceptionally well-preserved fossil rodent of the South American subterranean clade Ctenomys (Rodentia, Ctenomyidae). Phylogeny and adaptive profile. J. Mamm. Evol. 2024, 31, 35. [Google Scholar] [CrossRef]
- Smith, M.F.; Patton, J.L. The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol. J. Linn. Soc. 1993, 50, 149–177. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Verzi, D.H.; Olivares, A.I.; Morgan, C.C. The oldest South American tuco-tuco (Late Pliocene, northwestern Argentina) and the boundaries of the genus Ctenomys (Rodentia, Ctenomyidae). Mamm. Biol. 2010, 75, 243–252. [Google Scholar] [CrossRef]
- Soibelzon, E.; Prevosti, F.J.; Bidegain, J.C.; Rico, Y.; Verzi, D.H.; Tonni, E.P. Correlation of Late Cenozoic sequences of southeastern Buenos Aires province: Biostratigraphy and magnetostratigraphy. Quat. Int. 2009, 210, 51–56. [Google Scholar] [CrossRef]
- Lessa, E.P.; Cook, J.A. The molecular phylogenetics of tuco-tucos (genus Ctenomys, Rodentia: Octodontidae) suggests an early burst of speciation. Mol. Phylogenetics Evol. 1998, 9, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.H.; Cortinas, M.N.; Lessa, E.P. Rapid diversification of South American tuco-tucos (Ctenomys; Rodentia, Ctenomyidae): Contrasting mitochondrial and nuclear intron sequences. J. Mammal. 2005, 86, 170–179. [Google Scholar] [CrossRef]
- Cook, J.A.; Lessa, E.P. Are rates of diversification in subterranean south american tuco-tucos (genus Ctenomys, Rodentia: Octodontidae) unusually high? Evolution 1998, 52, 1521–1527. [Google Scholar] [CrossRef]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res./Rev. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef]
- Rabassa, J.; Coronato, A.; MartÍnez, O. Late Cenozoic glaciations in Patagonia and Tierra del Fuego: An updated review. Biol. J. Linn. Soc. 2011, 103, 316–335. [Google Scholar] [CrossRef]
- Stanton-Yonge, A.; Griffith, W.A.; Cembrano, J.; St. Julien, R.; Iturrieta, P. Tectonic role of margin-parallel and margin-transverse faults during oblique subduction in the Southern Volcanic Zone of the Andes: Insights from boundary element modeling. Tectonics 2016, 35, 1990–2013. [Google Scholar] [CrossRef]
- Barrientos, S.E.; Acevedo-Aránguiz, P.S. Seismological aspects of the 1988–1989 Lonquimay (Chile) volcanic eruption. J. Volcanol. Geotherm. Res. 1992, 53, 73–87. [Google Scholar] [CrossRef]
- Hsu, J.L.; Crawford, J.C.; Tammone, M.N.; Ramakrishnan, U.; Lacey, E.A.; Hadly, E.A. Genomic data reveal a loss of diversity in two species of tuco-tucos (genus Ctenomys) following a volcanic eruption. Sci. Rep. 2017, 7, 16227. [Google Scholar] [CrossRef]
- Gallardo, M.H.; Köhler, N.; Araneda, C. Bottleneck effects in local populations of fossorial Ctenomys (Rodentia, Ctenomyidae) affected by vulcanism. Heredity 1995, 74, 638–646. [Google Scholar] [CrossRef][Green Version]
- Gallardo, M.H.; Köhler, N. Demographic changes and genetic losses in populations of a subterranean rodent (Ctenomys maulinus brunneus) affected by a natural catastrophe. Z. Für Säugetierkunde 1994, 59, 358–365. [Google Scholar]
- Mackintosh, A.; Vila, R.; Martin, S.H.; Setter, D.; Lohse, K. Do chromosome rearrangements fix by genetic drift or natural selection? Insights from Brenthis butterflies. Mol. Ecol. 2024, 33, e17146. [Google Scholar] [CrossRef]
- Templeton, A.R. The reality and importance of founder speciation in evolution. BioEssays 2008, 30, 470–479. [Google Scholar] [CrossRef]
- Brown, R.E.; Freudenreich, C.H. Structure-forming repeats and their impact on genome stability. Curr. Opin. Genet. Dev. 2021, 67, 41–51. [Google Scholar] [CrossRef]
- Rico-Porras, J.M.; Mora, P.; Palomeque, T.; Montiel, E.E.; Cabral-de-Mello, D.C.; Lorite, P. Heterochromatin is not the only place for satDNAs: The high diversity of satDNAs in the euchromatin of the Beetle Chrysolina americana (Coleoptera, Chrysomelidae). Genes. 2024, 15, 395. [Google Scholar] [CrossRef] [PubMed]
- Šatović-Vukšić, E.; Plohl, M. Satellite DNAs—From localized to highly dispersed genome components. Genes. 2023, 14, 742. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Adega, F.; Meštrović, N.; Plohl, M.; Chaves, R. Evolutionary story of a satellite DNA from Phodopus sungorus (Rodentia, Cricetidae). Genome Biol. Evol. 2014, 6, 2944–2955. [Google Scholar] [CrossRef]
- Corach, D. Repetitive DNA sequence homologies and amplifications in South American cricetid rodents. Genetica 1990, 82, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.H.; Corvelo, A.; Hayashi, C.Y. Rapid molecular diversification and homogenization of clustered major ampullate silk genes in Argiope garden spiders. PLoS Genet. 2022, 18, e1010537. [Google Scholar] [CrossRef]
- Feliciello, I.; Pezer, Ž.; Sermek, A.; Bruvo Mađarić, B.; Ljubić, S.; Ugarković, Đ. Satellite DNA-mediated gene expression regulation: Physiological and evolutionary implication. In Satellite DNAs in Physiology and Evolution; Ugarković, Ð., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 145–167. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas, R.A.; Haro, R.E.; Di-Nizo, C.B.; Suárez-Villota, E.Y. Repetitive DNA Dynamics, Phylogenetic Relationships and Divergence Times in Andean Ctenomys (Rodentia: Ctenomyidae). Biology 2025, 14, 1776. https://doi.org/10.3390/biology14121776
Vargas RA, Haro RE, Di-Nizo CB, Suárez-Villota EY. Repetitive DNA Dynamics, Phylogenetic Relationships and Divergence Times in Andean Ctenomys (Rodentia: Ctenomyidae). Biology. 2025; 14(12):1776. https://doi.org/10.3390/biology14121776
Chicago/Turabian StyleVargas, Rodrigo A., Ronie E. Haro, Camilla Bruno Di-Nizo, and Elkin Y. Suárez-Villota. 2025. "Repetitive DNA Dynamics, Phylogenetic Relationships and Divergence Times in Andean Ctenomys (Rodentia: Ctenomyidae)" Biology 14, no. 12: 1776. https://doi.org/10.3390/biology14121776
APA StyleVargas, R. A., Haro, R. E., Di-Nizo, C. B., & Suárez-Villota, E. Y. (2025). Repetitive DNA Dynamics, Phylogenetic Relationships and Divergence Times in Andean Ctenomys (Rodentia: Ctenomyidae). Biology, 14(12), 1776. https://doi.org/10.3390/biology14121776






