Genome-Wide Discovery and Characterization of the Auxin Response Factor (ARF) Gene Family in Avicennia marina That Regulates Phytohormone Levels and Responds to Salt and Auxin Treatments
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Conditions
2.2. Phytohormone (IAA and ABA) Content Determination
2.3. Identification and Physicochemical Aspects of ARF Members in A. marina
2.4. Phylogenetic Analysis, Conserved Motifs, and Gene Structure Assessment
2.5. Putative Promoter Region Analysis
2.6. Proposed miRNA Targeting ARFs and Evaluation of Their Functions
2.7. Gene Ontology and Enrichment Analysis
2.8. Collinearity, Ka/Ks Ratios, and Protein Similarities of AmARFs
2.9. Prediction of Protein–Protein Interaction, 3D Structures, and Chromosomal Localization
2.10. Analysis of ARF Genes Using Quantitative Real-Time PCR (qRT-PCR)
2.11. Data Statistics and Analysis
3. Results
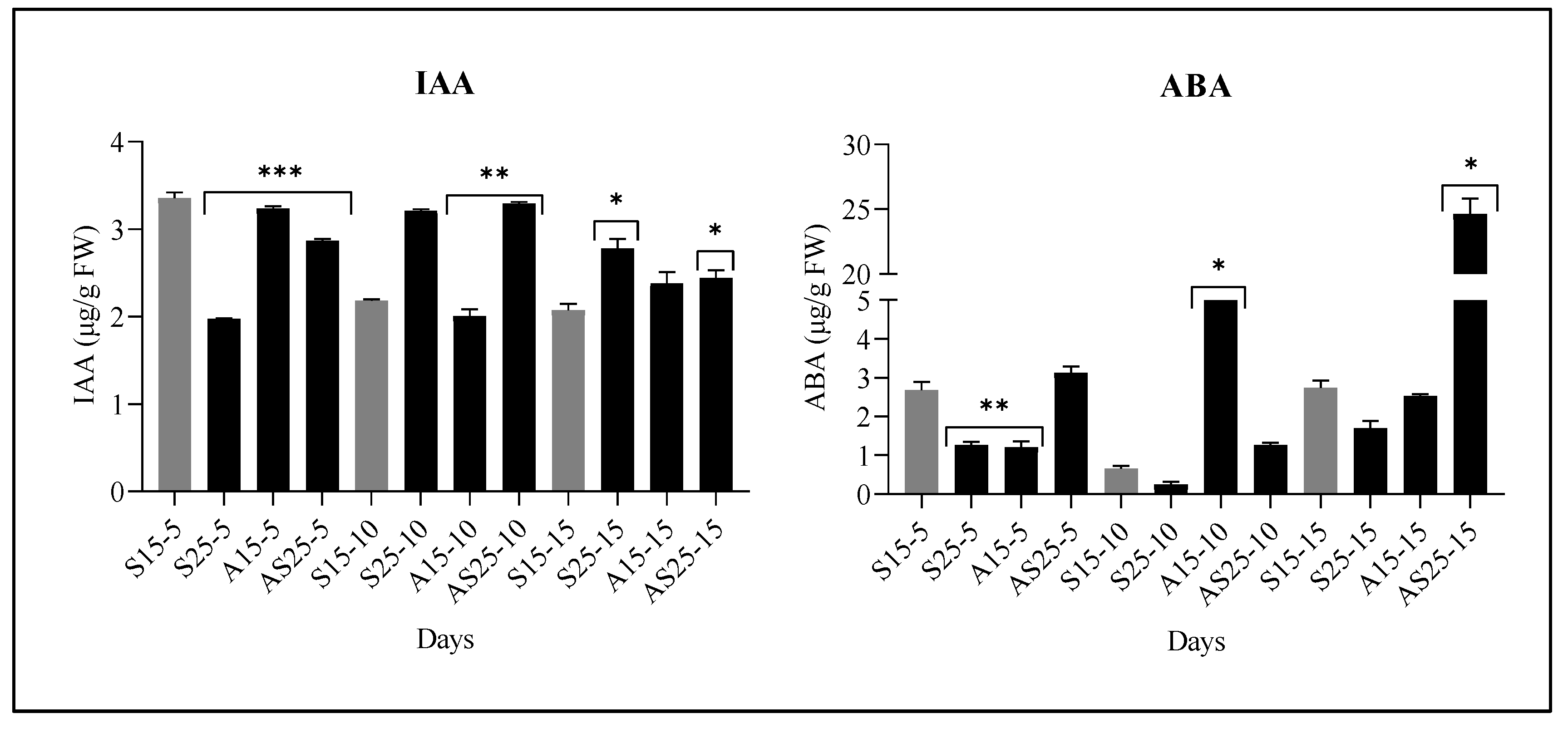
3.1. Changes in IAA and ABA Levels Under Salt and IAA Stress
3.2. Identification and Comprehensive Characterization of ARF Members in the Avicennia marina Genome
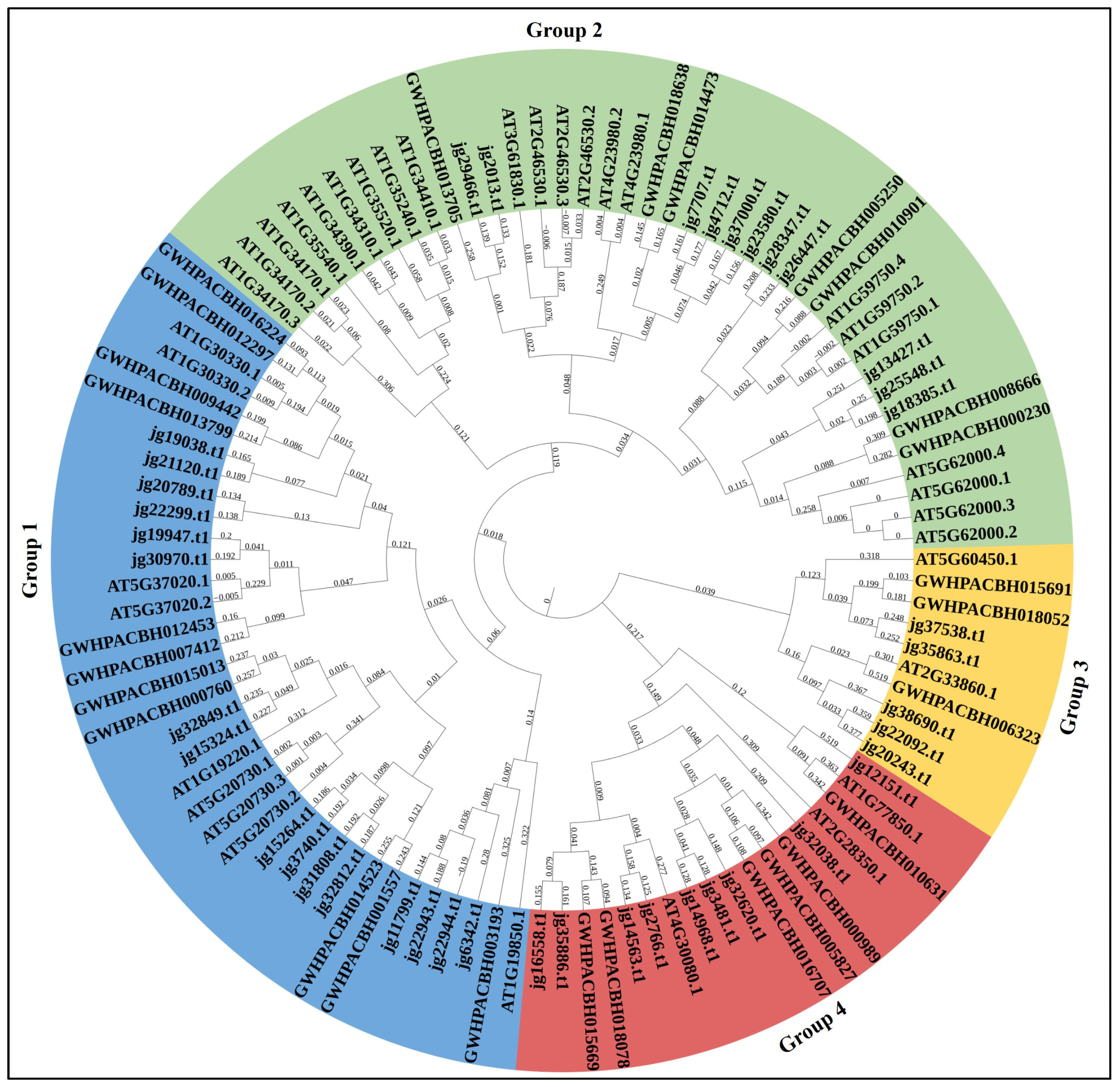
3.3. Phylogenetic Relationship of ARF Proteins
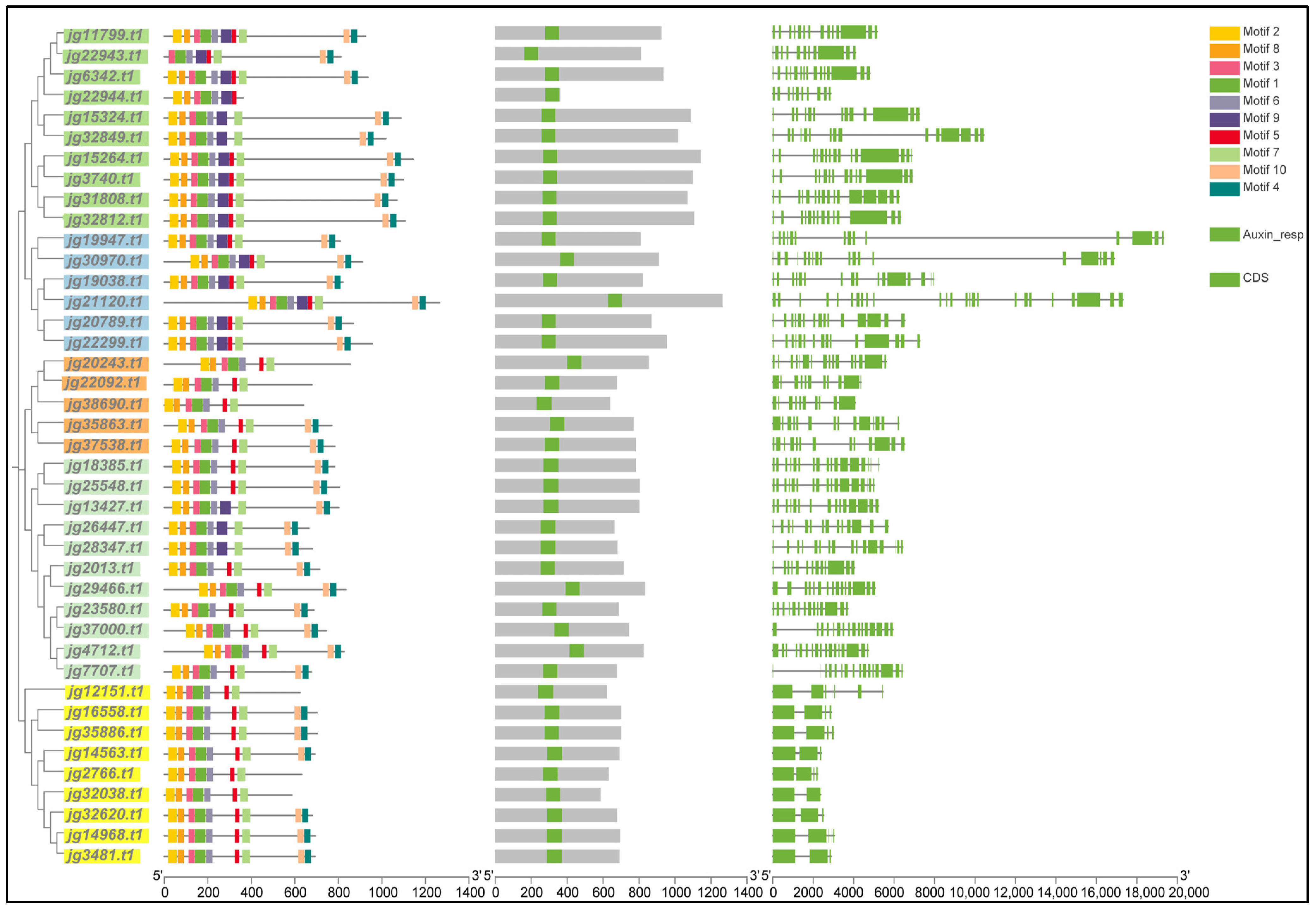
3.4. Motif Compositions, Conserved Domains, and Gene Structures of AmARF
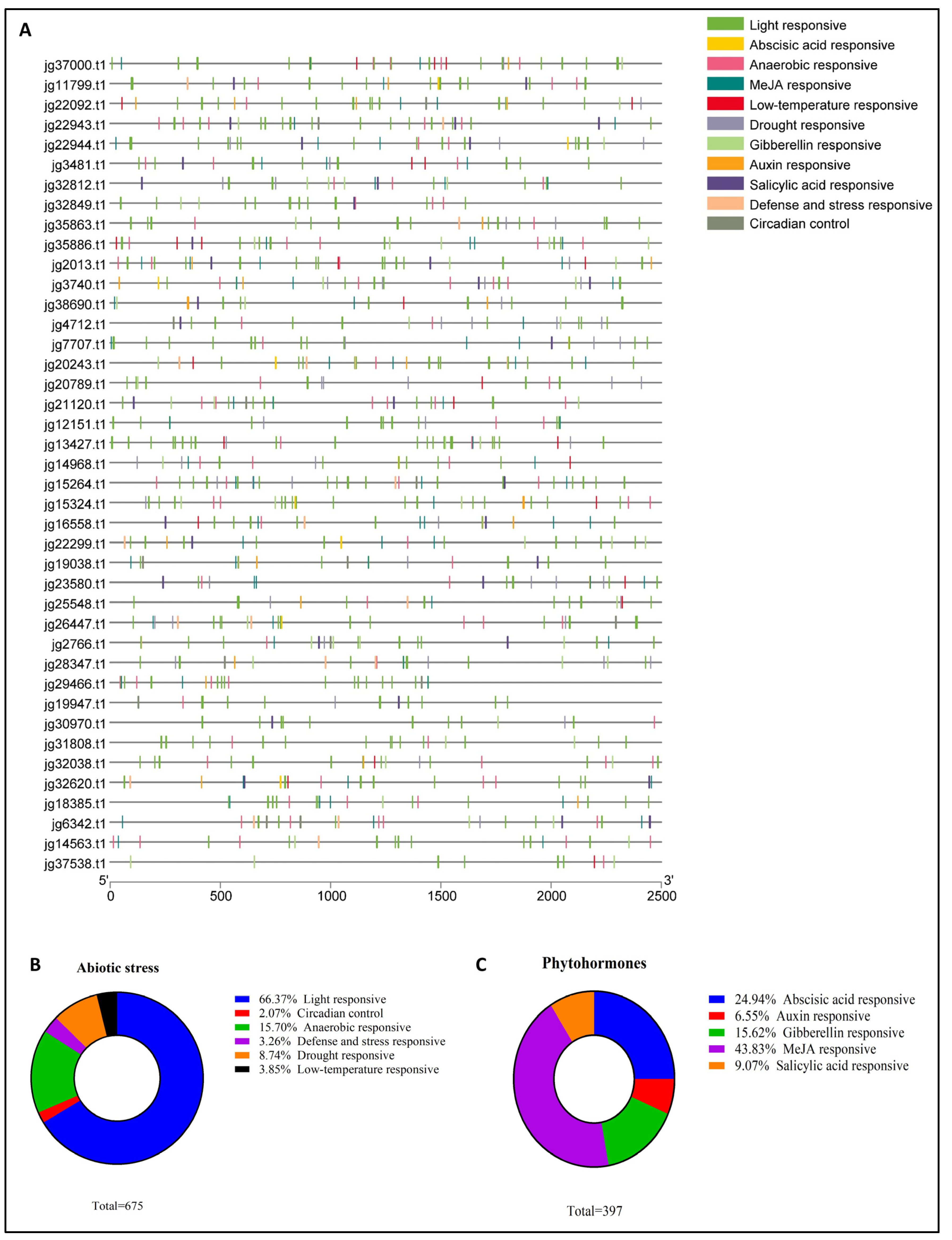
3.5. Examination of Cis-Regulatory Elements Within the Promoter Region of the AmARF Gene Family
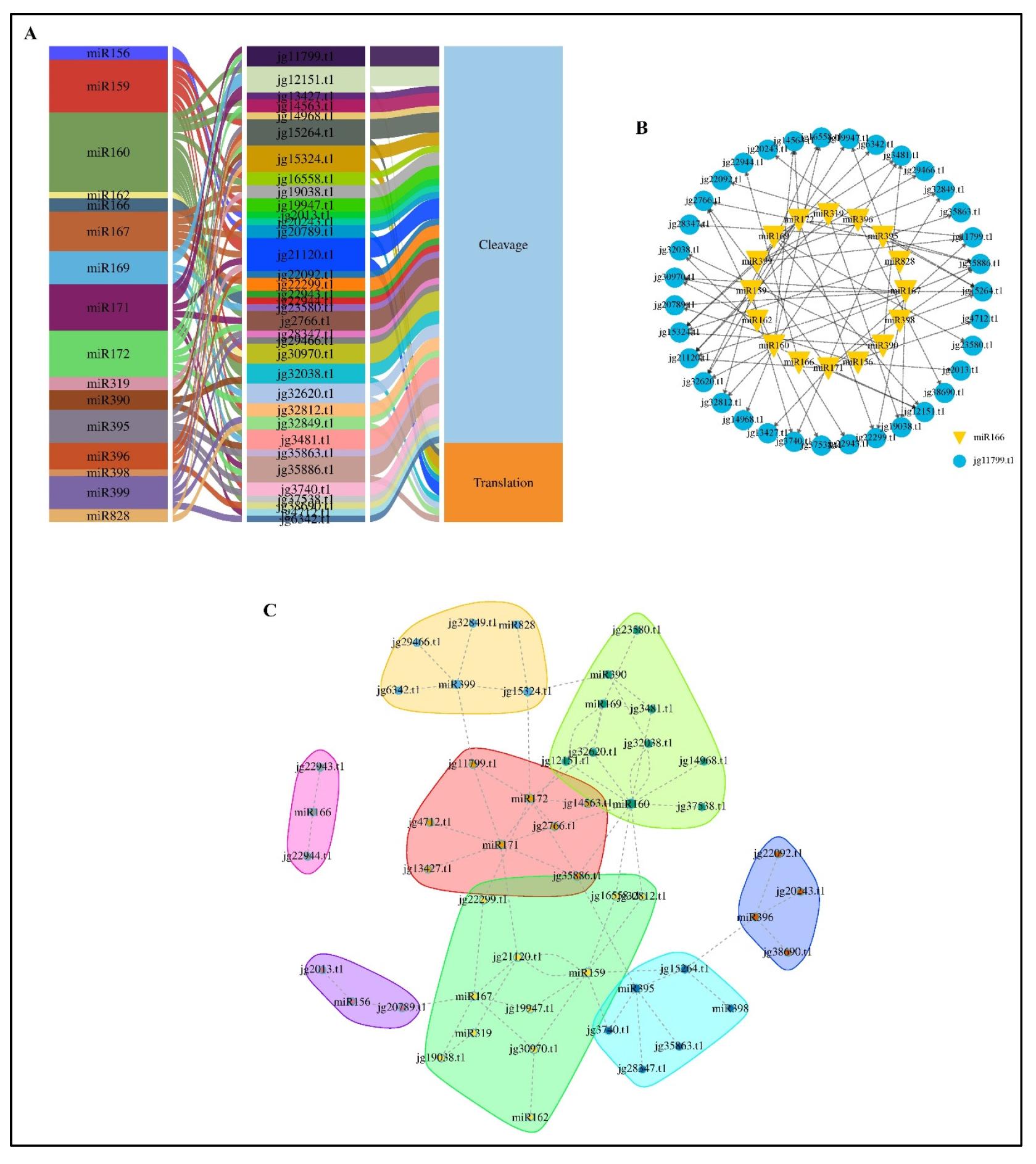
3.6. Identification of miRNAs That Target AmARF Genes
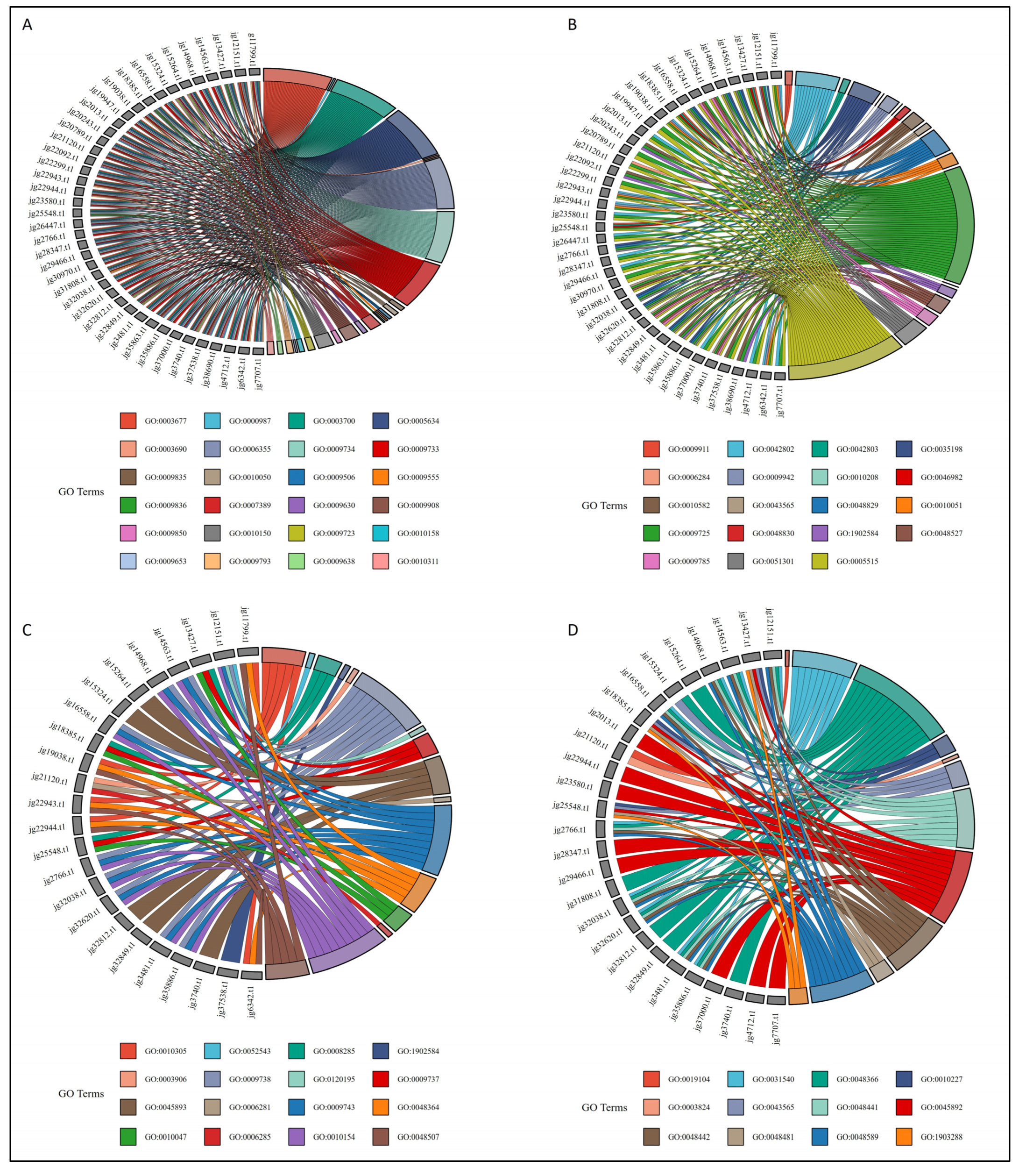
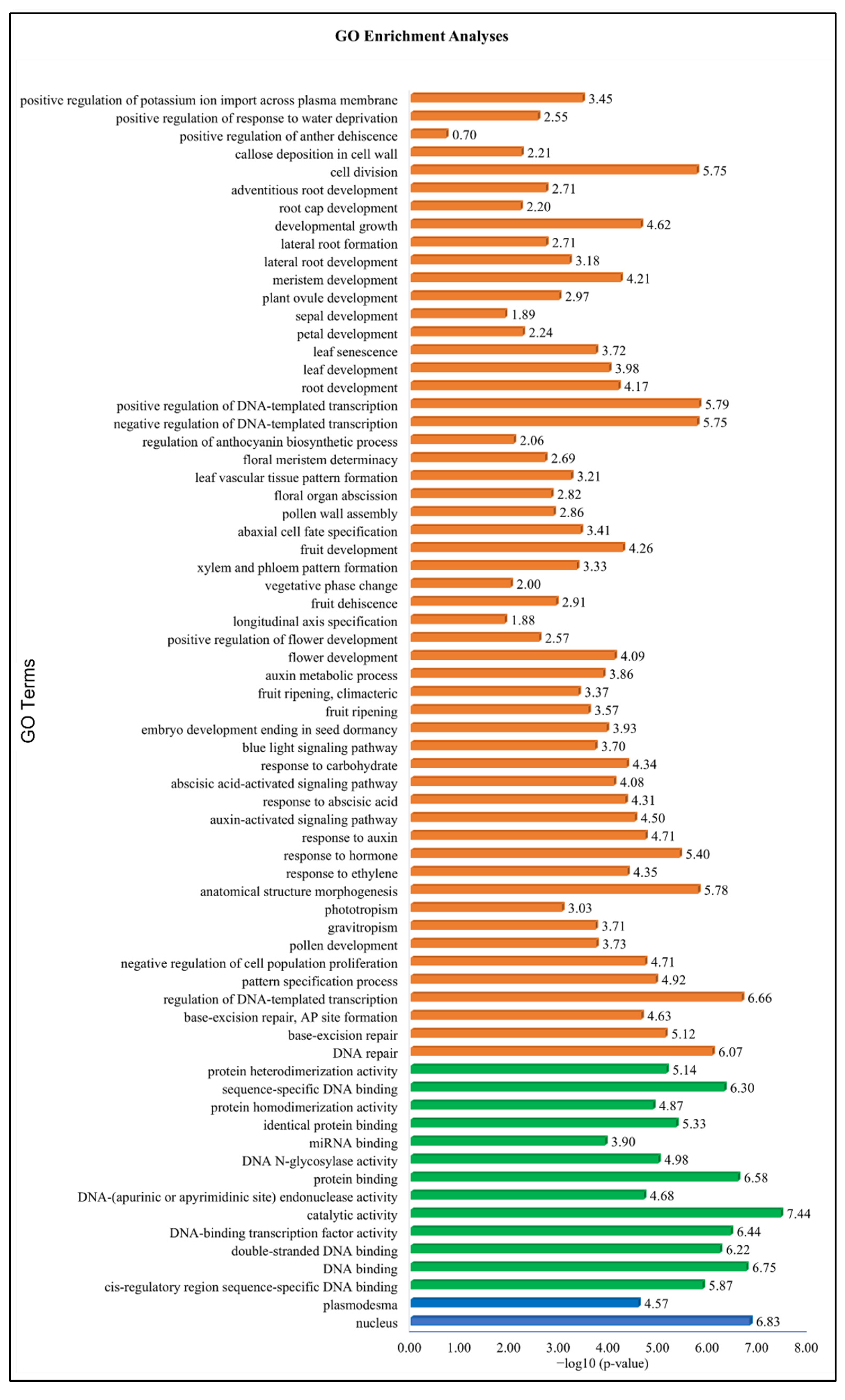
3.7. Gene Ontology Enrichment Analysis
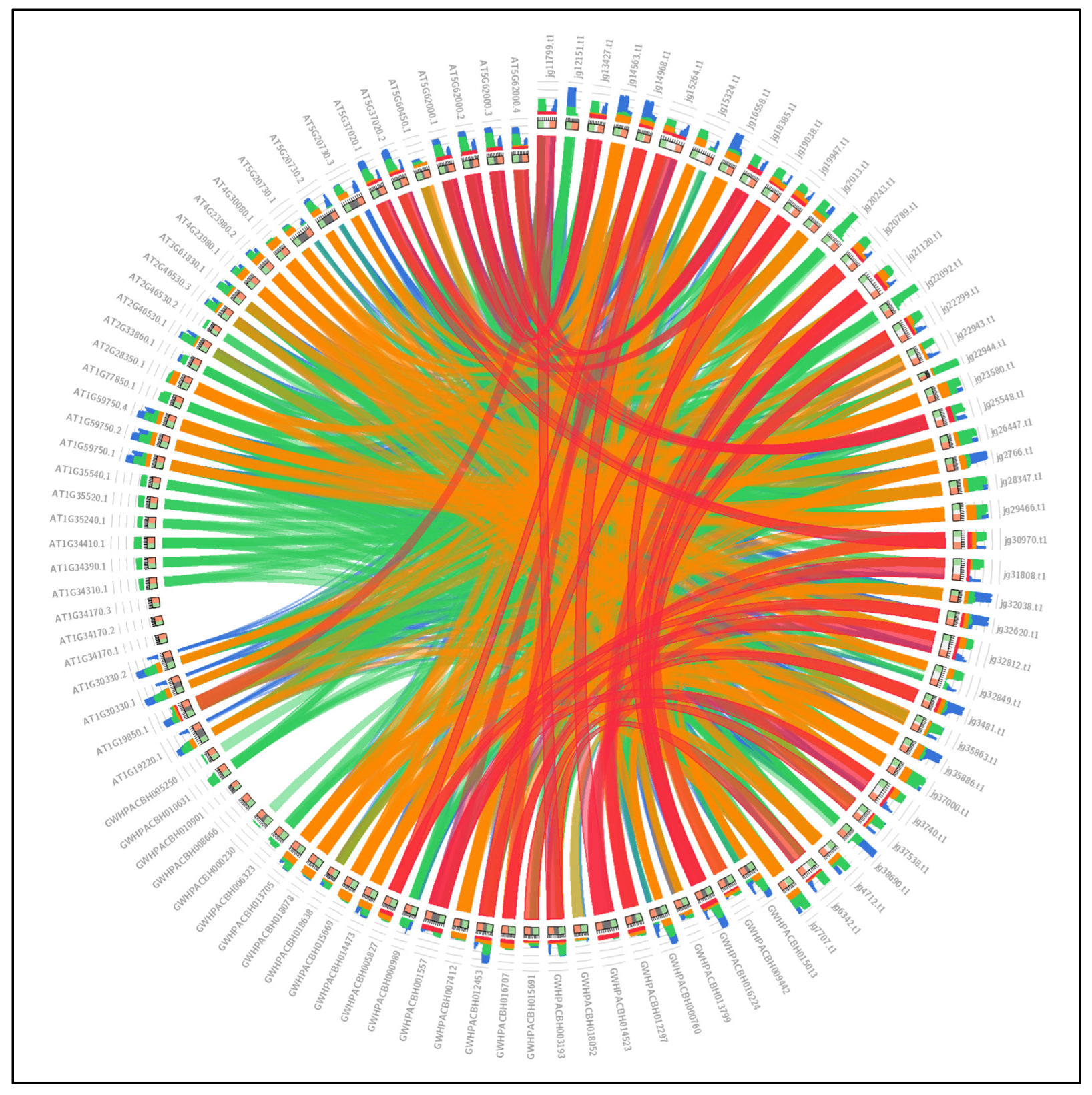
3.8. ARFs Collinearity Analysis
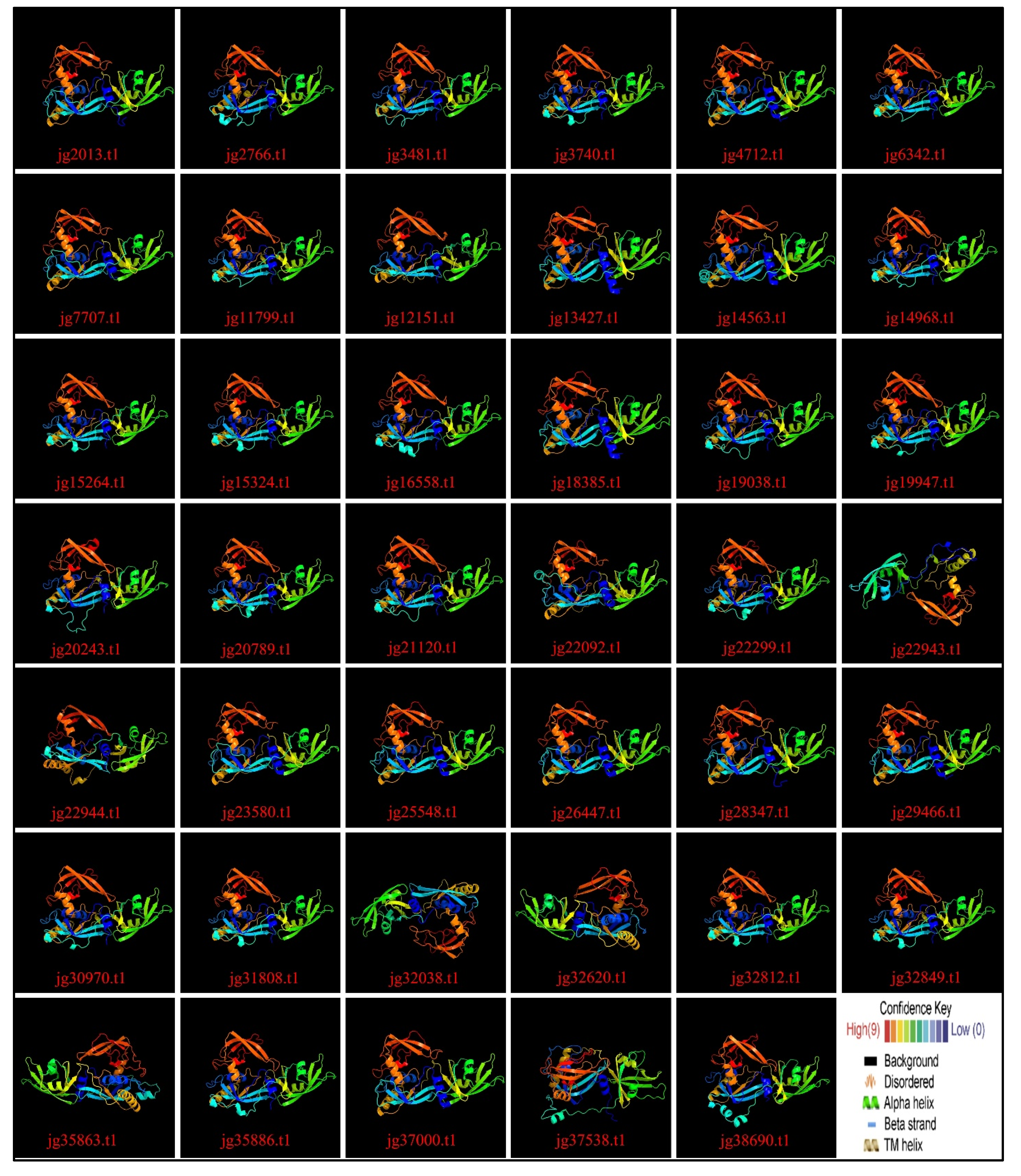
3.9. Variation Across the ARF Family in Terms of 3D and Secondary Structure
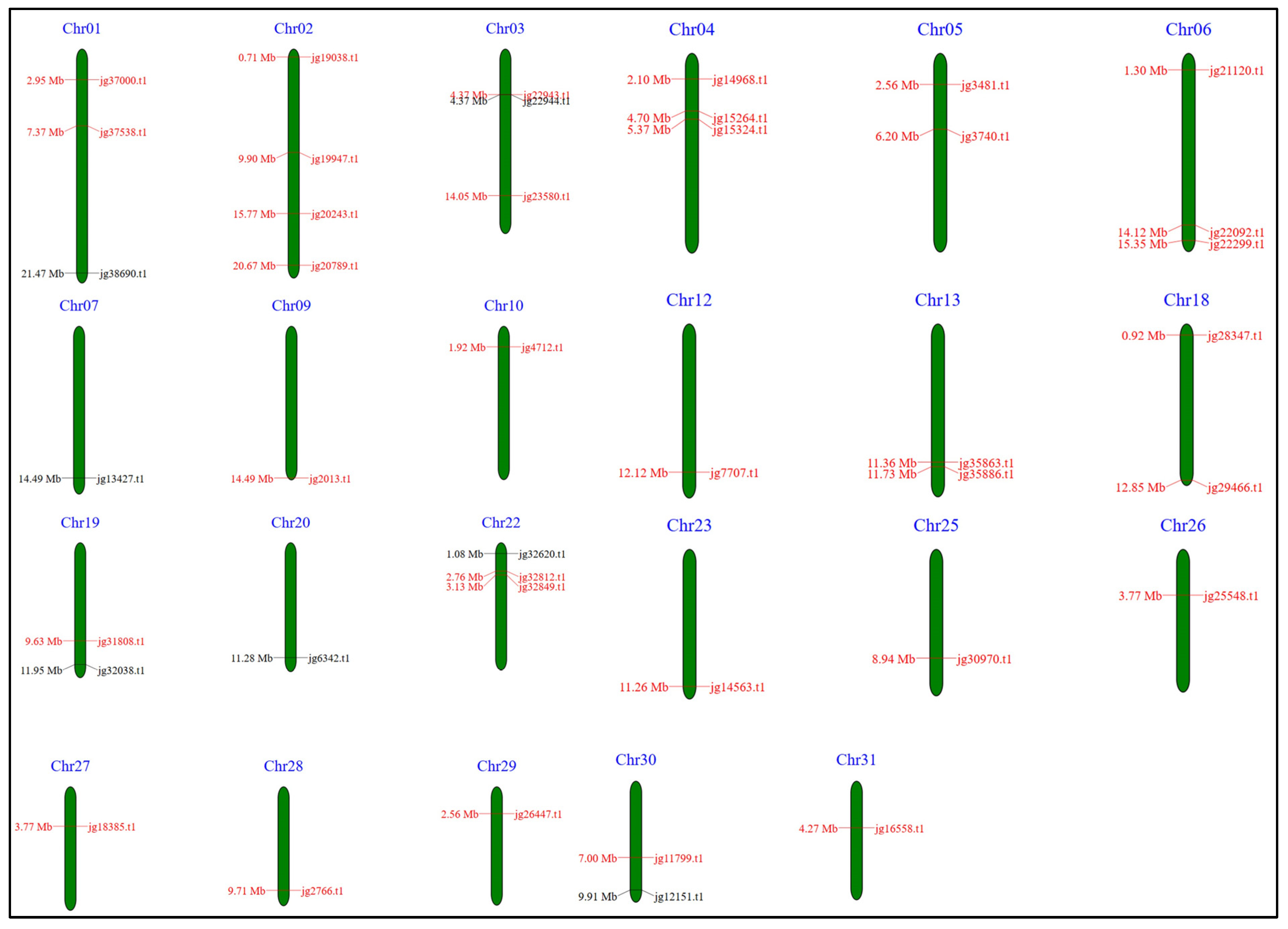
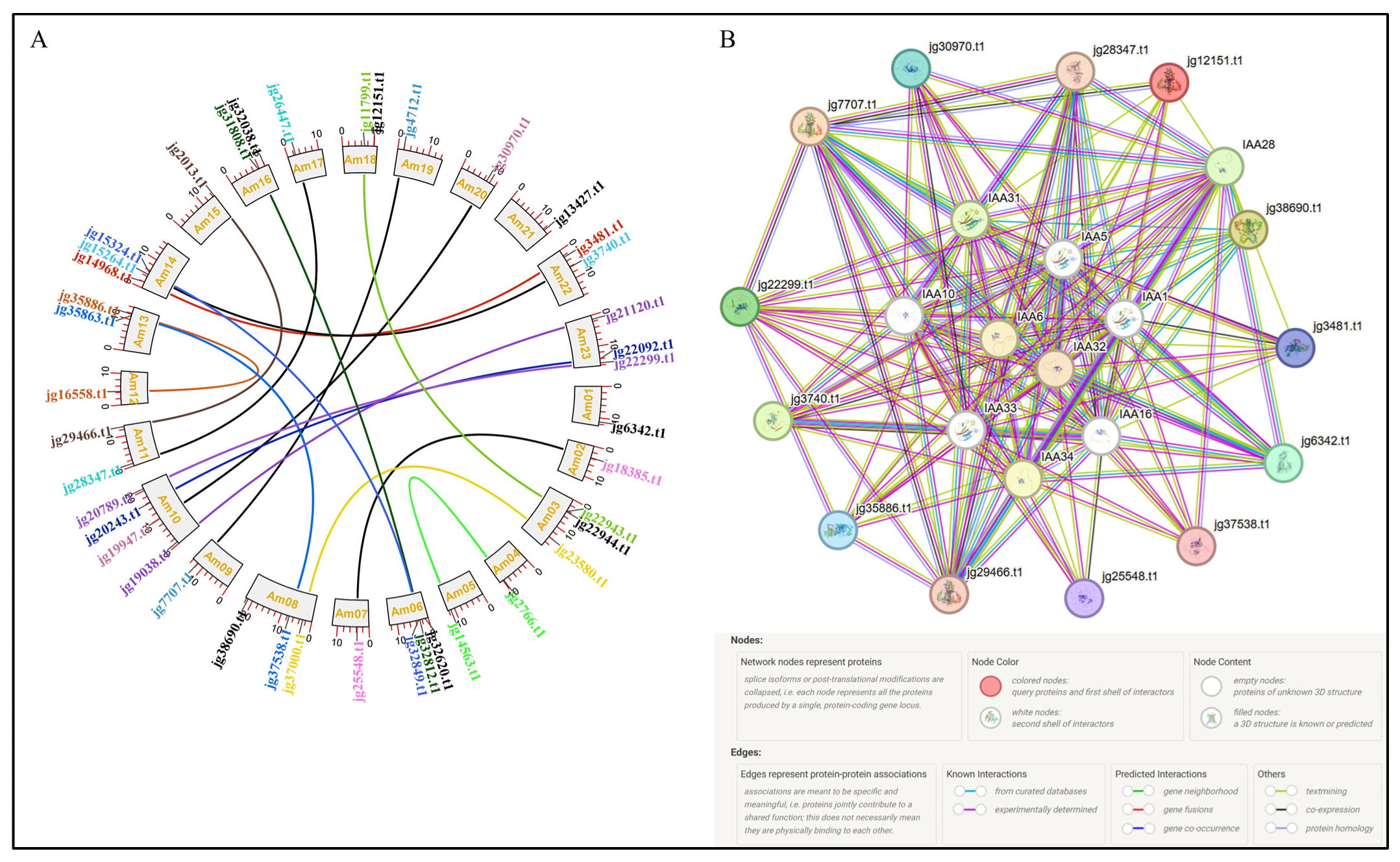
3.10. Chromosomal Localization, Duplication Analysis, and Protein–Protein Interaction of the ARF Genes in A. marina
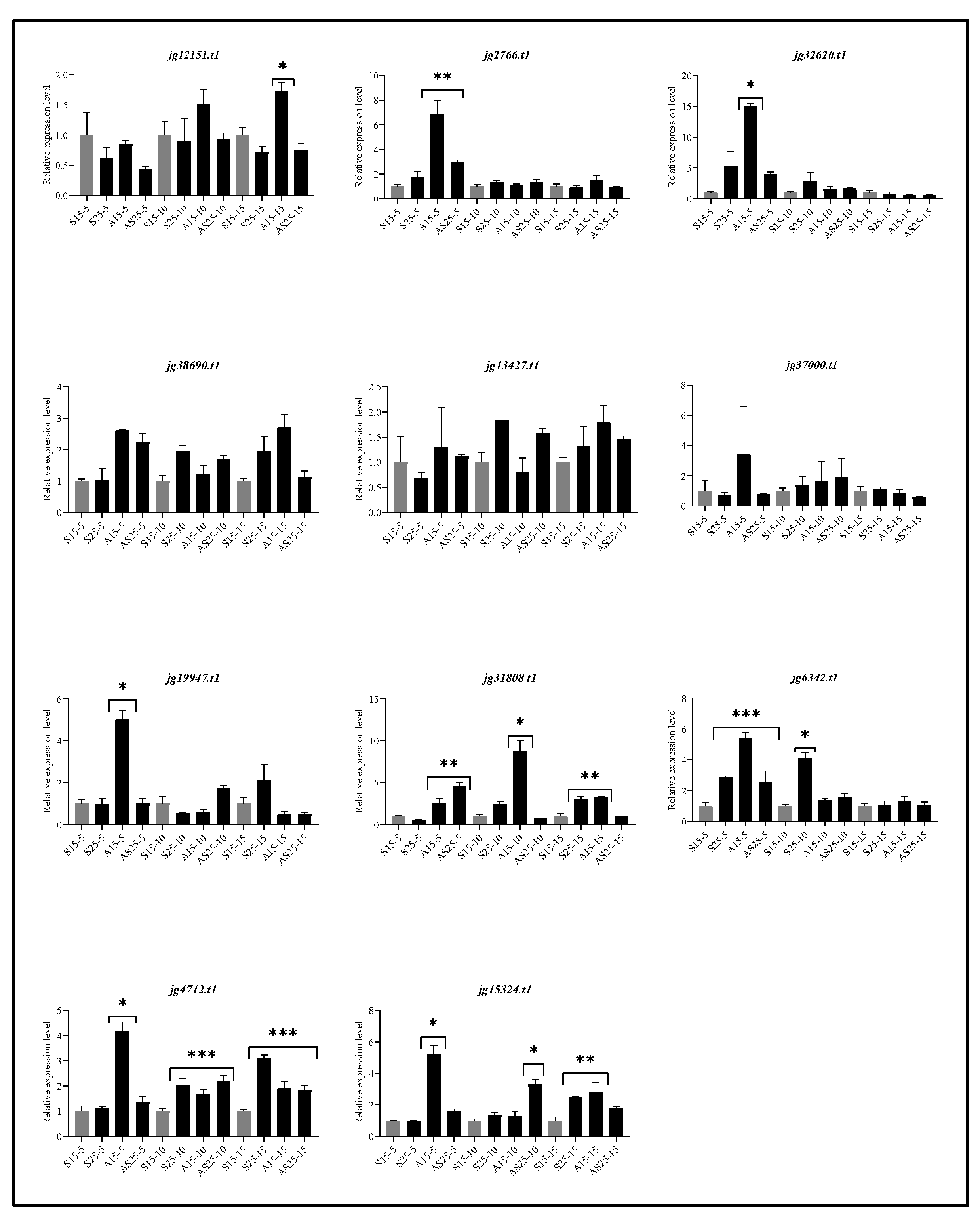
3.11. Expression Analysis of ARF Genes in A. marina Leaves Under Salt and Auxin Stresses.
4. Discussion
5. Limitations and Future Directions
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, D.; Ding, Q.; Guo, Z.; Xu, C.; Liang, P.; Zhao, Z.; Song, S.; Zheng, H.L. The Genome of a Mangrove Plant, Avicennia marina, Provides Insights into Adaptation to Coastal Intertidal Habitats. Planta 2022, 256, 6. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Xu, C.; Zhang, L.; Li, J.; Jiang, L.; Ma, D.; Guo, Z.; Wang, Q.; Wang, X.; Zheng, H. Trehalose along with ABA Promotes the Salt Tolerance of Avicennia marina by Regulating Na+ Transport. Plant J. 2024, 119, 2349–2362. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Murugesan, A.K.; Govindan, G.; Gopalakrishnan, A.; Kumar, R.; Duraisamy, P.; Balaji, R.; Tanuja; Shyamli, P.S.; Parida, A.K.; et al. A Reference-Grade Genome Identifies Salt-Tolerance Genes from the Salt-Secreting Mangrove Species Avicennia marina. Commun. Biol. 2021, 4, 851. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Voβ, U.; Farcot, E.; Bennett, M.J.; Bishopp, A. Systems Biology Approaches to Understand the Role of Auxin in Root Growth and Development. Physiol. Plant. 2014, 151, 73–82. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis and Its Role in Plant Development. Annu. Rev. Plant Biol. 2011, 61, 49–64. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Xu, X.; Wang, R.; Liu, Y.; Huang, S.; Wei, H.; Wei, Z. Molecular Mechanisms of Diverse Auxin Responses During Plant Growth and Development. Int. J. Mol. Sci. 2022, 23, 12495. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.; Yu, J.; Huang, S.; Zhang, Y.; Wei, H.; Wei, Z. Genome-Wide Identification and Characterization of Auxin Response Factor (ARF) Gene Family Involved in Wood Formation and Response to Exogenous Hormone Treatment in Populus trichocarpa. Int. J. Mol. Sci. 2023, 24, 740. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-Box Protein TIR1 Is an Auxin Receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Mockaitis, K.; Estelle, M. Auxin Receptors and Plant Development: A New Signaling Paradigm. Annu. Rev. Cell Dev. Biol. 2008, 24, 55–80. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin Response Factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Li, S.; Xie, Z.; Hu, C.; Zhang, J. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Woodward, A.; Bartel, B. A Receptor for Auxin. Plant Cell 2005, 17, 2425–2429. [Google Scholar] [CrossRef]
- Quint, M.; Gray, W.M. Auxin Signaling Marcel. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Mei, M.; Ai, W.; Liu, L.; Xu, X.; Lu, X. Genome-Wide Identification of the Auxin Response Factor (ARF) Gene Family in Magnolia sieboldii and Functional Analysis of MsARF5. Front. Plant Sci. 2022, 13, 958816. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The Roles of Auxin Response Factor Domains in Auxin-Responsive Transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhang, J.; Ha, X.; Ma, H. Genome-Wide Identification and Expression Analysis of the Auxin-Response Factor (ARF) Gene Family in Medicago sativa under Abiotic Stress. BMC Genom. 2023, 24, 498. [Google Scholar] [CrossRef]
- Su, L.; Xu, M.; Zhang, J.; Wang, Y.; Lei, Y.; Li, Q. Genome-Wide Identification of Auxin Response Factor (ARF) Family in Kiwifruit (Actinidia chinensis) and Analysis of Their Inducible Involvements in Abiotic Stresses. Physiol. Mol. Biol. Plants 2021, 27, 1261–1276. [Google Scholar] [CrossRef]
- Hu, W.L.; Luo, C.; Xia, L.M.; Liang, R.Z.; Zhu, J.W.; Li, Y.Z.; Zhang, Y.L.; Lan, M.Y.; Liu, Y.; Nong, S.F.; et al. Genome-Wide Identification of the Mango Auxin Response Factor Family and the Ectopic Expression of Two ARF (MiARF18A) Genes Confers Early Flowering and Increases Silique Number in Transgenic Arabidopsis. Sci. Hortic. 2025, 342, 114017. [Google Scholar] [CrossRef]
- Li, S.; Ouyang, W.; Hou, X.; Xie, L.; Hu, C.; Zhang, J.Z. Genome-Wide Identification, Isolation and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Sweet Orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 119. [Google Scholar] [CrossRef]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-Wide Identification of the ARF (Auxin Response Factor) Gene Family in Peach and Their Expression Analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef]
- Ljung, K. Auxin Metabolism and Homeostasis During Plant Development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Kalve, S.; Sizani, B.L.; Markakis, M.N.; Helsmoortel, C.; Vandeweyer, G.; Laukens, K.; Sommen, M.; Naulaerts, S.; Vissenberg, K.; Prinsen, E.; et al. Osmotic Stress Inhibits Leaf Growth of Arabidopsis thaliana by Enhancing ARF-Mediated Auxin Responses. New Phytol. 2020, 226, 1766–1780. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Khurana, J.P. Transcript Profiling Reveals Diverse Roles of Auxin-Responsive Genes During Reproductive Development and Abiotic Stress in Rice. FEBS J. 2009, 276, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-Wide Analysis of Auxin Response Factor (ARF) Gene Family from Tomato and Analysis of Their Role in Flower and Fruit Development. Mol. Genet. Genom. 2011, 285, 245–260. [Google Scholar] [CrossRef]
- Achard, P.; Genschik, P. Releasing the Brakes of Plant Growth: How GAs Shutdown DELLA Proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef]
- Die, J.V.; Gil, J.; Millan, T. Genome-Wide Identification of the Auxin Response Factor Gene Family in Cicer arietinum. BMC Genom. 2018, 19, 301. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The Interaction between MiR160 and MiR165/166 in the Control of Leaf Development and Drought Tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef]
- Tang, Y.; Du, G.; Xiang, J.; Hu, C.; Li, X.; Wang, W.; Zhu, H.; Qiao, L.; Zhao, C.; Wang, J.; et al. Genome-Wide Identification of Auxin Response Factor (ARF) Gene Family and the MiR160-ARF18-Mediated Response to Salt Stress in Peanut (Arachis hypogaea L.). Genomics 2022, 114, 171–184. [Google Scholar] [CrossRef]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-Regulation of AUXIN RESPONSE FACTORS 6 and 8 by MicroRNA 167 Leads to Floral Development Defects and Female Sterility in Tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef]
- Wu, M.-F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 Controls Patterns of ARF6 and ARF8 Expression, and Regulates Both Female and Male Reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Damodharan, S.; Zhao, D.; Arazi, T. A Common MiRNA160-Based Mechanism Regulates Ovary Patterning, Floral Organ Abscission and Lamina Outgrowth in Tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Hussain, Q.; Zheng, M.; Hänninen, H.; Bhalerao, R.P.; Riaz, M.W.; Sajjad, M.; Zhang, R.; Wu, J. Effect of the Photoperiod on Bud Dormancy in Liriodendron chinense. J. Plant Physiol. 2022, 279, 153835. [Google Scholar] [CrossRef] [PubMed]
- Friis, G.; Vizueta, J.; Smith, E.G.; Nelson, D.R.; Khraiwesh, B.; Qudeimat, E.; Salehi-ashtiani, K.; Ortega, A.; Marshell, A.; Duarte, C.M.; et al. A High-Quality Genome Assembly and Annotation of the Gray mangrove, Avicennia marina. G3 Genes Genomes Genet. 2021, 11, jkaa025. [Google Scholar] [CrossRef] [PubMed]
- Tiryaki, I. Biosynthesis, Metabolism and Signaling Pathway of Auxin in Plants. Philipp. Agric. Sci. 2009, 92, 243–253. [Google Scholar]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin Biosynthesis and Storage Forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The Main Auxin Biosynthesis Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Galvan-ampudia, C.S.; Testerink, C. Salt Stress Signals Shape the Plant Root. Curr. Opin. Plant Biol. 2011, 14, 296–302. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Shani, E.; Salehin, M.; Zhang, Y.; Sanchez, S.E.; Wang, R.; Mangado, C.C.; Song, L.; Tal, I.; Ecker, J.R.; Kay, S.A.; et al. Plant Stress Tolerance Requires Auxin-Sensitive Aux/IAA Transcriptional Repressors. Curr. Biol. 2018, 27, 437–444. [Google Scholar] [CrossRef]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-Wide Analysis of the Auxin Response Factors (ARF) Gene Family in Rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wang, L.; Li, W.; Zhao, L.; Huang, X.; Azam, S.M.; Qin, Y. Genome-Wide Identification of Auxin Response Factor (ARF) Genes Family and Its Tissue-Specific Prominent Expression in Pineapple (Ananas comosus). Trop. Plant Biol. 2017, 10, 86–96. [Google Scholar] [CrossRef]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-Wide Identification, Molecular Evolution, and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 336. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF Gene Family in Land Plants: Old Domains, New Tricks. Mol. Biol. Evol. 2012, 30, 45–56. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-Responsive Gene Expression: Genes, Promoters and Regulatory Factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Shen, C.; Yue, R.; Sun, T.; Zhang, L.; Xu, L.; Tie, S.; Wang, H.; Yang, Y. Genome-Wide Identification and Expression Analysis of Auxin Response Factor Gene Family in Medicago truncatula. Front. Plant Sci. 2015, 6, 73. [Google Scholar] [CrossRef]
- Boer, D.R.; Freire-rios, A.; Van Den Berg, W.A.M.; Saaki, T.; Manfield, I.W.; Kepinski, S.; López-Vidrieo, I.; Franco-Zorrilla, J.M.; De Vries, S.C.; Solano, R.; et al. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, N.; Dong, C.; Shang, Q. Genome-Wide Identification and Expression of ARF Gene Family During Adventitious Root Development in Hot Pepper (Capsicum annuum). Hortic. Plant J. 2017, 3, 151–164. [Google Scholar] [CrossRef]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-Wide Identification and Expression Profiling of Auxin Response Factor (ARF) Gene Family in Maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef]
- Zouine, M.; Fu, Y.; Chateigner-Boutin, A.-L.; Mila, I.; Frasse, P.; Wang, H.; Audran, C.; Roustan, J.-P.; Bouzayen, M. Characterization of the Tomato ARF Gene Family Uncovers a Multi-Levels Post-Transcriptional Regulation Including Alternative Splicing. PLoS ONE 2014, 9, e84203. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) Are Potential Mediators of Auxin Action in Tomato Response to Biotic and Abiotic Stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Wang, Y.; Li, B.; Li, Y.; Du, W.; Zhang, Y.; Han, Y.; Liu, C.; Fan, S.; Hao, J. Application of Exogenous Auxin and Gibberellin Regulates the Bolting of Lettuce (Lactuca sativa L.). Open Life Sci. 2022, 17, 438–446. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-Related Gene Families in Abiotic Stress Response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Song, S.; Hao, L.; Zhao, P.; Xu, Y.; Zhong, N.; Zhang, H.; Liu, N. Genome-Wide Identification, Expression Profiling and Evolutionary Analysis of Auxin Response Factor Gene Family in Potato (Solanum tuberosum Group Phureja). Sci. Rep. 2019, 9, 1755. [Google Scholar] [CrossRef]
| Sequence ID | AA 1 | CDS 2 | MW 3 | pI 4 | II 5 | AI 6 | GRAVY 7 | SL 8 |
|---|---|---|---|---|---|---|---|---|
| jg11799.t1 | 923 | 2769 | 102,030.6 | 5.3 | 56.34 | 73.92 | −0.432 | nucleus |
| jg12151.t1 | 621 | 1863 | 67,860.82 | 6.74 | 40.79 | 82.29 | −0.241 | nucleus |
| jg13427.t1 | 801 | 2403 | 89,332.21 | 6.18 | 61.23 | 64.87 | −0.663 | nucleus |
| jg14563.t1 | 691 | 2073 | 76,248.19 | 6.56 | 44.43 | 74.79 | −0.385 | nucleus |
| jg14968.t1 | 693 | 2079 | 75,977.67 | 7.21 | 49.44 | 69.06 | −0.41 | nucleus |
| jg15264.t1 | 1142 | 3426 | 124,981.77 | 6.16 | 63.77 | 75.9 | −0.505 | nucleus |
| jg15324.t1 | 1086 | 3258 | 120,456.08 | 6.32 | 61.65 | 75.81 | −0.566 | nucleus |
| jg16558.t1 | 700 | 2100 | 77,331.65 | 6.39 | 51.84 | 75.91 | −0.362 | nucleus |
| jg18385.t1 | 782 | 2346 | 87,712.9 | 6.52 | 62.6 | 68.57 | −0.623 | nucleus |
| jg19038.t1 | 819 | 2457 | 91,062.47 | 6.32 | 58.67 | 72.53 | −0.451 | nucleus |
| jg19947.t1 | 808 | 2424 | 90,706.42 | 6.26 | 58.04 | 72.86 | −0.422 | nucleus |
| jg2013.t1 | 713 | 2139 | 79,216.69 | 6.51 | 53.32 | 74.1 | −0.426 | nucleus |
| jg20243.t1 | 854 | 2562 | 93,784.01 | 8.61 | 48.58 | 71.57 | −0.355 | nucleus |
| jg20789.t1 | 868 | 2604 | 95,306.8 | 6.46 | 61.51 | 78.73 | −0.301 | nucleus |
| jg21120.t1 | 1264 | 3792 | 139,295.27 | 7 | 57.41 | 77.23 | −0.391 | nucleus |
| jg22092.t1 | 676 | 2028 | 74,134.34 | 8.24 | 55.2 | 68.93 | −0.412 | nucleus |
| jg22299.t1 | 954 | 2862 | 104,921.35 | 6.72 | 63.82 | 75.41 | −0.401 | nucleus |
| jg22943.t1 | 810 | 2430 | 89,317.38 | 5.53 | 48.96 | 77.38 | −0.393 | nucleus |
| jg22944.t1 | 361 | 1083 | 40,412.34 | 9.39 | 54 | 77.84 | −0.356 | nucleus |
| jg23580.t1 | 685 | 2055 | 76,736.46 | 7.36 | 55.45 | 70.85 | −0.558 | nucleus |
| jg25548.t1 | 803 | 2409 | 89,606.21 | 6.23 | 58.22 | 65.69 | −0.584 | nucleus |
| jg26447.t1 | 663 | 1989 | 75,569.74 | 6.59 | 59.67 | 75.84 | −0.555 | nucleus |
| jg2766.t1 | 631 | 1893 | 69,408.41 | 6.26 | 43.55 | 75.69 | −0.369 | nucleus |
| jg28347.t1 | 680 | 2040 | 76,174.1 | 5.76 | 58.08 | 74.41 | −0.429 | nucleus |
| jg29466.t1 | 833 | 2499 | 93,139.23 | 6.7 | 52.73 | 79.06 | −0.328 | nucleus |
| jg30970.t1 | 910 | 2730 | 102,132.19 | 6.39 | 59.29 | 70.92 | −0.469 | nucleus |
| jg31808.t1 | 1068 | 3204 | 117,372.04 | 6.15 | 66.7 | 72.41 | −0.544 | nucleus |
| jg32038.t1 | 586 | 1758 | 64,740.11 | 8.92 | 50.45 | 68.05 | −0.452 | nucleus |
| jg32620.t1 | 678 | 2034 | 73,752.22 | 6.71 | 46.51 | 71.19 | −0.346 | nucleus |
| jg32812.t1 | 1105 | 3315 | 121,212.97 | 6.15 | 63.25 | 73.87 | −0.498 | nucleus |
| jg32849.t1 | 1016 | 3048 | 112,319.7 | 6.36 | 66.61 | 70.94 | −0.598 | nucleus |
| jg3481.t1 | 691 | 2073 | 75,413.34 | 7.25 | 49.72 | 73.95 | −0.313 | nucleus |
| jg35863.t1 | 769 | 2307 | 85,467.36 | 6.31 | 56.8 | 73.25 | −0.482 | nucleus |
| jg35886.t1 | 700 | 2100 | 76,452.86 | 7.9 | 42.97 | 75.33 | −0.314 | nucleus |
| jg37000.t1 | 744 | 2232 | 83,240.88 | 6.11 | 58.31 | 72.57 | −0.491 | nucleus |
| jg3740.t1 | 1097 | 3291 | 120,835.52 | 6.12 | 60.44 | 75.63 | −0.497 | nucleus |
| jg37538.t1 | 783 | 2349 | 86,909.13 | 5.72 | 53.41 | 71.7 | −0.466 | nucleus |
| jg38690.t1 | 639 | 1917 | 70,890.23 | 6.89 | 60.48 | 75.23 | −0.395 | nucleus |
| jg4712.t1 | 825 | 2475 | 92,646.74 | 8.67 | 56.23 | 75.02 | −0.451 | nucleus |
| jg6342.t1 | 935 | 2805 | 103,811.84 | 5.94 | 47.51 | 74.35 | −0.411 | nucleus |
| jg7707.t1 | 675 | 2025 | 75,633.85 | 7.1 | 60.03 | 73.47 | −0.482 | nucleus |
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| jg11799.t1 | jg22943.t1 | 0.110 | 0.510 | 0.215 |
| jg15324.t1 | jg32849.t1 | 0.163 | 0.615 | 0.265 |
| jg15264.t1 | jg3740.t1 | 0.111 | 0.539 | 0.207 |
| jg31808.t1 | jg32812.t1 | 0.125 | 0.543 | 0.230 |
| jg19947.t1 | jg30970.t1 | 0.096 | 0.417 | 0.230 |
| jg19038.t1 | jg21120.t1 | 0.081 | 0.417 | 0.194 |
| jg20789.t1 | jg22299.t1 | 0.111 | 0.455 | 0.243 |
| jg20243.t1 | jg22092.t1 | 0.160 | 0.592 | 0.271 |
| jg35863.t1 | jg37538.t1 | 0.146 | 0.581 | 0.252 |
| jg18385.t1 | jg25548.t1 | 0.138 | 0.658 | 0.210 |
| jg26447.t1 | jg28347.t1 | 0.179 | 0.594 | 0.301 |
| jg2013.t1 | jg29466.t1 | 0.131 | 0.457 | 0.286 |
| jg23580.t1 | jg37000.t1 | 0.141 | 0.512 | 0.275 |
| jg4712.t1 | jg7707.t1 | 0.144 | 0.530 | 0.272 |
| jg16558.t1 | jg35886.t1 | 0.152 | 0.722 | 0.210 |
| jg14563.t1 | jg2766.t1 | 0.115 | 0.589 | 0.195 |
| jg14968.t1 | jg3481.t1 | 0.128 | 0.530 | 0.242 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, Q.; Hussain, M.A.; Li, Y.; Zhang, Q.; Shang, C.; Abdel-Maksoud, M.A.; Alrokayan, S.; Alamri, A. Genome-Wide Discovery and Characterization of the Auxin Response Factor (ARF) Gene Family in Avicennia marina That Regulates Phytohormone Levels and Responds to Salt and Auxin Treatments. Biology 2025, 14, 1774. https://doi.org/10.3390/biology14121774
Hussain Q, Hussain MA, Li Y, Zhang Q, Shang C, Abdel-Maksoud MA, Alrokayan S, Alamri A. Genome-Wide Discovery and Characterization of the Auxin Response Factor (ARF) Gene Family in Avicennia marina That Regulates Phytohormone Levels and Responds to Salt and Auxin Treatments. Biology. 2025; 14(12):1774. https://doi.org/10.3390/biology14121774
Chicago/Turabian StyleHussain, Quaid, Muhammad Azhar Hussain, Yingying Li, Qi Zhang, Chenjing Shang, Mostafa A. Abdel-Maksoud, Salman Alrokayan, and Abdulaziz Alamri. 2025. "Genome-Wide Discovery and Characterization of the Auxin Response Factor (ARF) Gene Family in Avicennia marina That Regulates Phytohormone Levels and Responds to Salt and Auxin Treatments" Biology 14, no. 12: 1774. https://doi.org/10.3390/biology14121774
APA StyleHussain, Q., Hussain, M. A., Li, Y., Zhang, Q., Shang, C., Abdel-Maksoud, M. A., Alrokayan, S., & Alamri, A. (2025). Genome-Wide Discovery and Characterization of the Auxin Response Factor (ARF) Gene Family in Avicennia marina That Regulates Phytohormone Levels and Responds to Salt and Auxin Treatments. Biology, 14(12), 1774. https://doi.org/10.3390/biology14121774







