Proteome Analysis of Daily Urine Samples of Pregnant Rats Unveils Developmental Processes of Fetus as Well as Physiological Changes in Mother Rats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Rat Caging
2.2. Urine Sample Collection
2.3. Preparation of Urine Samples
2.4. Proteome Analysis of Urine Samples
2.5. Data Processing and Bioinformatic Analysis
3. Results and Discussion
3.1. The Changes in Urine Protein in Pregnancy Rats and Control Rats
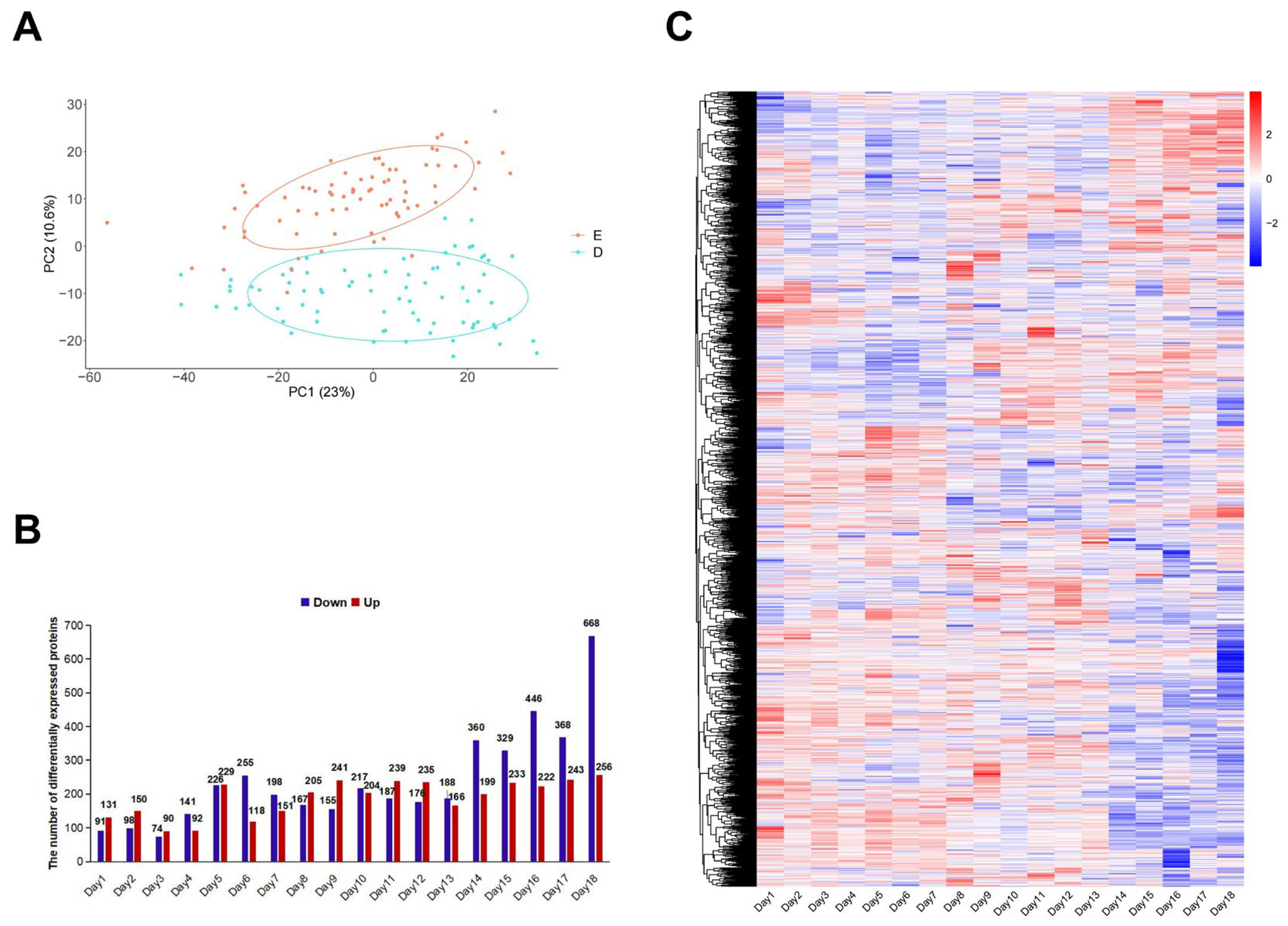
3.2. The Changes Occurring Throughout the Entire Gestation Period in Rats
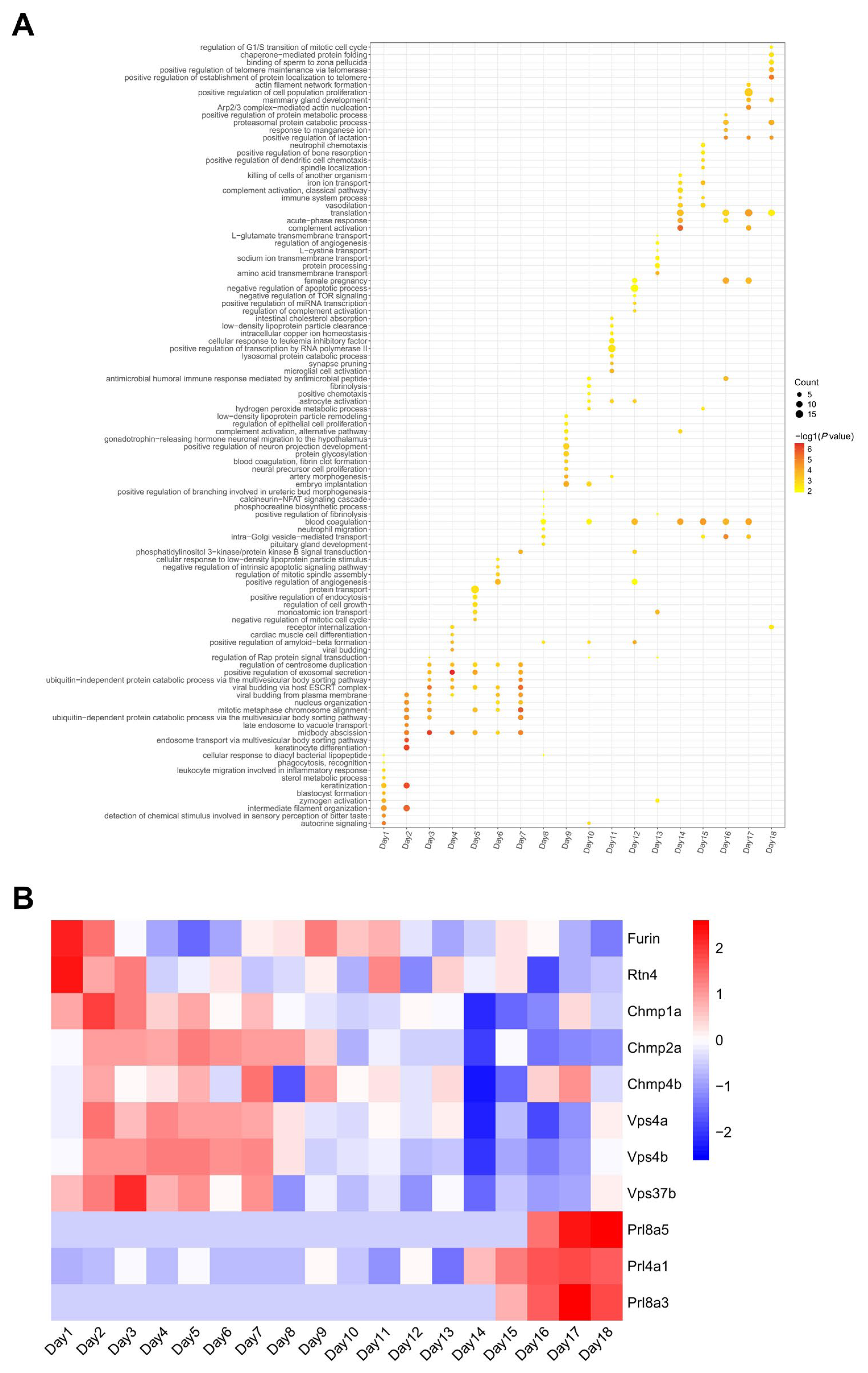
3.3. Dynamic Changes in Embryonic Development Throughout the Whole Pregnancy
3.3.1. Embryonic Development
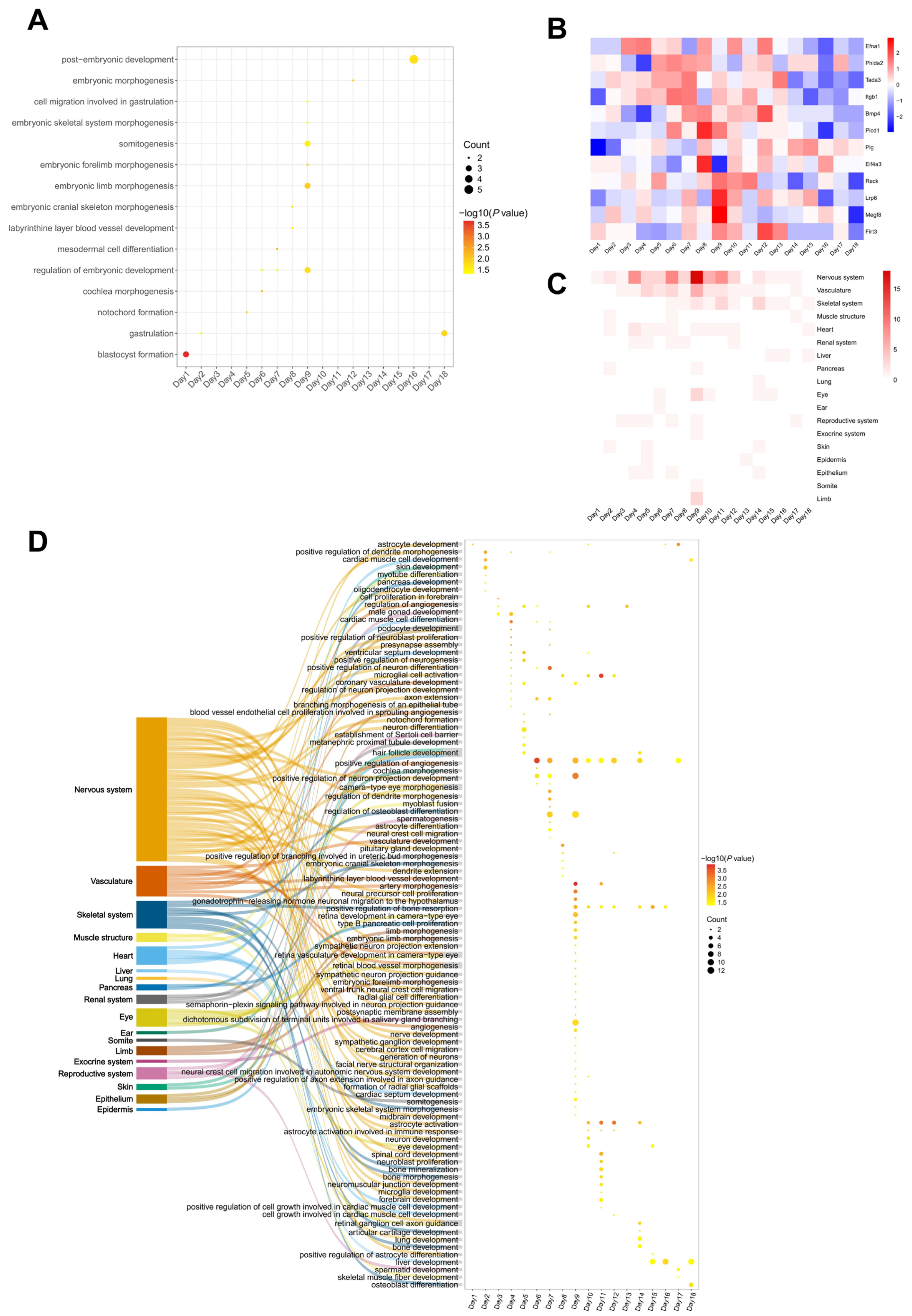
3.3.2. Organ Development of Embryo
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, L.; Rasmussen, M.H.; Piening, B.; Shen, X.; Chen, S.; Röst, H.; Snyder, J.K.; Tibshirani, R.; Skotte, L.; Lee, N.C.; et al. Metabolic Dynamics and Prediction of Gestational Age and Time to Delivery in Pregnant Women. Cell 2020, 181, 1680–1692.e15. [Google Scholar] [CrossRef]
- Soma-Pillay, P.; Nelson-Piercy, C.; Tolppanen, H.; Mebazaa, A. Physiological changes in pregnancy. Cardiovasc. J. Afr. 2016, 27, 89–94. [Google Scholar] [CrossRef]
- Dang, Y.; Zhu, L.; Yuan, P.; Liu, Q.; Guo, Q.; Chen, X.; Gao, S.; Liu, X.; Ji, S.; Yuan, Y.; et al. Functional profiling of stage-specific proteome and translational transition across human pre-implantation embryo development at a single-cell resolution. Cell Discov. 2023, 9, 10. [Google Scholar] [CrossRef]
- Rossant, J.; Tam, P.P.L. Early human embryonic development: Blastocyst formation to gastrulation. Dev. Cell 2022, 57, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Graupner, O.; Enzensberger, C. Prediction of Adverse Pregnancy Outcome Related to Placental Dysfunction Using the sFlt-1/PlGF Ratio: A Narrative Review. Geburtshilfe Frauenheilkd. 2021, 81, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Triunfo, S.; Lanzone, A. Impact of maternal under nutrition on obstetric outcomes. J. Endocrinol. Investig. 2014, 38, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Feldt-Rasmussen, U.; Mathiesen, E.R. Endocrine disorders in pregnancy: Physiological and hormonal aspects of pregnancy. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 875–884. [Google Scholar] [CrossRef]
- Gao, Y. Urine—An untapped goldmine for biomarker discovery? Sci. China Life Sci. 2013, 56, 1145–1146. [Google Scholar] [CrossRef]
- Syngelaki, A.; Hammami, A.; Bower, S.; Zidere, V.; Akolekar, R.; Nicolaides, K.H. Diagnosis of fetal non-chromosomal abnormalities on routine ultrasound examination at 11–13 weeks’ gestation. Ultrasound Obstet. Gynecol. 2019, 54, 468–476. [Google Scholar] [CrossRef]
- Zimmerli, L.U.; Schiffer, E.; Zürbig, P.; Good, D.M.; Kellmann, M.; Mouls, L.; Pitt, A.R.; Coon, J.J.; Schmieder, R.E.; Peter, K.H.; et al. Urinary Proteomic Biomarkers in Coronary Artery Disease. Mol. Cell. Proteom. 2008, 7, 290–298. [Google Scholar] [CrossRef]
- Chang, Q.; Chen, Y.; Yin, J.; Wang, T.; Dai, Y.; Wu, Z.; Guo, Y.; Wang, L.; Zhao, Y.; Yuan, H.; et al. Comprehensive Urinary Proteome Profiling Analysis Identifies Diagnosis and Relapse Surveillance Biomarkers for Bladder Cancer. J. Proteome Res. 2024, 23, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, J.; Wei, J.; Zhao, Y.; Gao, Y. Urinary biomarker discovery in gliomas using mass spectrometry-based clinical proteomics. Chin. Neurosurg. J. 2020, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Huan, Y.; Gao, Y. Urinary proteome profiling for children with autism using data-independent acquisition proteomics. Transl. Pediatr. 2021, 10, 1765–1778. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Liu, W.; Ding, X.; Liang, S.; Zheng, Y.; Zhu, X.; Quan, S.; Yi, X.; Xiang, N.; Du, J.; et al. Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19. Cell Rep. 2022, 38, 110271. [Google Scholar] [CrossRef]
- Zhan, S.; Zhou, X.; Fu, J. Noninvasive Urinary Biomarkers for Obesity-Related Metabolic Diseases: Diagnostic Applications and Future Directions. Biomolecules 2025, 15, 633. [Google Scholar] [CrossRef]
- Chebotareva, N.; Vinogradov, A.; McDonnell, V.; Zakharova, N.V.; Indeykina, M.I.; Moiseev, S.; Nikolaev, E.N.; Kononikhin, A.S. Urinary Protein and Peptide Markers in Chronic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 12123. [Google Scholar] [CrossRef]
- Reyes-Thomas, J.; Blanco, I.; Putterman, C. Urinary biomarkers in lupus nephritis. Clin. Rev. Allergy Immunol. 2011, 40, 138–150. [Google Scholar] [CrossRef]
- An, M.; Gao, Y. Urinary Biomarkers of Brain Diseases. Urin. Biomark. Brain Dis. 2015, 13, 345–354. [Google Scholar] [CrossRef]
- Liu, E.; Nisenblat, V.; Farquhar, C.; Fraser, I.; Bossuyt, P.M.; Johnson, N.; Hull, M.L. Urinary biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst. Rev. 2015, 2015, CD012019. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Jones, E.R.; Barr, C.E.; O’Flynn, H.; Whetton, A.D.; Crosbie, E.J. Urinary Biomarkers and Their Potential for the Non-Invasive Detection of Endometrial Cancer. Front. Oncol. 2020, 10, 559016. [Google Scholar] [CrossRef]
- Wu, D.; Ni, J.; Beretov, J.; Cozzi, P.; Willcox, M.; Wasinger, V.; Walsh, B.; Graham, P.; Li, Y. Urinary biomarkers in prostate cancer detection and monitoring progression. Crit. Rev. Oncol./Hematol. 2017, 118, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Gao, Y. Urinary Proteome Changes during Pregnancy in Rats. Biomolecules 2022, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, C.; Wang, J.; Wang, L.; Zhao, Y.; Sun, L.; Sun, W.; Zhu, Y.; Li, J.; Wu, S. BioLadder: A bioinformatic platform primarily focused on proteomic data analysis. iMeta 2024, 3, e215. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Tao, Q.; Du, H.; Zhao, Z.; Dong, Y.; He, S.; Shao, R.; Wang, Y.; Han, W.; Wang, X.; et al. Tengdan Capsule Prevents Hypertensive Kidney Damage in SHR by Inhibiting Periostin-Mediated Renal Fibrosis. Front. Pharmacol. 2021, 12, 638298. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, J.; Hu, X.; Xu, P.; Huang, S.; Cui, S.; Guo, Y.; Yang, H.; Chen, X.; Jiang, C. Yi-shen-hua-shi granules modulate immune and inflammatory damage via the ALG3/PPARγ/NF-κB pathway in the treatment of immunoglobulin a nephropathy. J. Ethnopharmacol. 2024, 319, 117204. [Google Scholar] [CrossRef]
- Ben-Hur, S.; Sernik, S.; Afar, S.; Kolpakova, A.; Politi, Y.; Gal, L.; Florentin, A.; Golani, O.; Sivan, E.; Dezorella, N.; et al. Egg multivesicular bodies elicit an LC3-associated phagocytosis-like pathway to degrade paternal mitochondria after fertilization. Nat. Commun. 2024, 15, 5715. [Google Scholar] [CrossRef]
- Whitlock, K.E. Origin and development of GnRH neurons. Trends Endocrinol. Metab. TEM 2005, 16, 145–151. [Google Scholar] [CrossRef]
- Tomassy, G.S.; Lodato, S.; Trayes-Gibson, Z.; Arlotta, P. Development and regeneration of projection neuron subtypes of the cerebral cortex. Sci. Prog. 2010, 93, 151–169. [Google Scholar] [CrossRef]
- Zhang, M.; Su, L.; Wang, W.; Li, C.; Liang, Q.; Ji, F.; Jiao, J. Endothelial cells regulated by RNF20 orchestrate the proliferation and differentiation of neural precursor cells during embryonic development. Cell Rep. 2022, 40, 111350. [Google Scholar] [CrossRef]
- Urasoko, Y.; He, X.J.; Ebata, T.; Kinoshita, Y.; Kobayashi, J.; Mochizuki, M.; Ikeya, M. Changes in blood parameters and coagulation-related gene expression in pregnant rats. J. Am. Assoc. Lab. Anim. Sci. 2009, 48, 272–278. [Google Scholar]
- Chighizola, C.B.; Lonati, P.A.; Trespidi, L.; Meroni, P.L.; Tedesco, F. The Complement System in the Pathophysiology of Pregnancy and in Systemic Autoimmune Rheumatic Diseases During Pregnancy. Front. Immunol. 2020, 11, 2084. [Google Scholar] [CrossRef]
- Hannan, F.M.; Elajnaf, T.; Vandenberg, L.N.; Kennedy, S.H.; Thakker, R.V. Hormonal regulation of mammary gland development and lactation. Nat. Rev. Endocrinol. 2022, 19, 46–61. [Google Scholar] [CrossRef]
- Masso-Welch, P.A.; Darcy, K.M.; Stangle-Castor, N.C.; Ip, M.M. A developmental atlas of rat mammary gland histology. J. Mammary Gland Biol. Neoplasia 2000, 5, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Bessonnard, S.; Mesnard, D.; Constam, D.B. PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation. J. Cell Biol. 2015, 210, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, B.; Xiao, Z.; Chen, B.; Han, J.; Wang, X.; Zhang, J.; Gao, S.; Zhao, Y.; Dai, J. Nogo-66 Regulates Nanog Expression Through Stat3 Pathway in Murine Embryonic Stem Cells. Stem Cells Dev. 2010, 19, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Colf, L.A.; Karren, M.A.; Sandrin, V.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA 2010, 107, 12889–12894. [Google Scholar] [CrossRef]
- Alam, S.M.K.; Konno, T.; Soares, M.J. Identification of target genes for a prolactin family paralog in mouse decidua. Reproduction 2015, 149, 625–632. [Google Scholar] [CrossRef]
- Stelloo, S.; Alejo-Vinogradova, M.T.; van Gelder, C.A.G.H.; Zijlmans, D.W.; van Oostrom, M.J.; Valverde, J.M.; Lamers, L.A.; Rus, T.; Sobrevals Alcaraz, P.; Schäfers, T.; et al. Deciphering lineage specification during early embryogenesis in mouse gastruloids using multilayered proteomics. Cell Stem Cell 2024, 31, 1072–1090.e1078. [Google Scholar] [CrossRef]
- Solnica-Krezel, L.; Sepich, D.S. Gastrulation: Making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 2012, 28, 687–717. [Google Scholar] [CrossRef]
- Sarkar, A.A.; Sabatino, J.A.; Sugrue, K.F.; Zohn, I.E. Abnormal labyrinthine zone in the Hectd1-null placenta. Placenta 2016, 38, 16–23. [Google Scholar] [CrossRef]
- Simmons, D.G.; Natale, D.R.; Begay, V.; Hughes, M.; Leutz, A.; Cross, J.C. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 2008, 135, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Zhang, D.; Wang, W.; Ding, Y.; Wang, Y.; Gu, S.; Shang, Y.; Gan, J.; Jiang, L.; Meng, F.; et al. Cross-Species Insights into Trophoblast Invasion During Placentation Governed by Immune-Featured Trophoblast Cells. Adv. Sci. 2024, 11, e2407221. [Google Scholar] [CrossRef] [PubMed]
- McDole, K.; Guignard, L.; Amat, F.; Berger, A.; Malandain, G.; Royer, L.A.; Turaga, S.C.; Branson, K.; Keller, P.J. In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level. Cell 2018, 175, 859–876.E33. [Google Scholar] [CrossRef] [PubMed]
- de Haan, S.; He, J.; Corbat, A.A.; Belicova, L.; Ratz, M.; Vinsland, E.; Frisén, J.; Kelley, M.W.; Andersson, E.R. Ectoderm barcoding reveals neural and cochlear compartmentalization. Science 2025, 388, 60–68. [Google Scholar] [CrossRef]
- Granström, G.; Jacobsson, C.; Magnusson, B.C. Enzyme histochemical analysis of craniofacial malformations induced by retinoids. Scand. J. Plast. Reconstr. Surg. Hand Surg. 1991, 25, 133–141. [Google Scholar] [CrossRef]
- Simonoska, R.; Stenberg, A.; Masironi, B.; Sahlin, L.; Hultcrantz, M. Estrogen receptors in the inner ear during different stages of pregnancy and development in the rat. Acta Oto-Laryngol. 2009, 129, 1175–1181. [Google Scholar] [CrossRef]
- Morriss-Kay, G. A journey in the world of craniofacial development: From 1968 to the future. J. Anat. 2024, 245, 816–828. [Google Scholar] [CrossRef]
- Power, S.C.; Lancman, J.; Smith, S.M. Retinoic acid is essential for Shh/Hoxd signaling during rat limb outgrowth but not for limb initiation. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1999, 216, 469–480. [Google Scholar] [CrossRef]
- Roelink, H.; Augsburger, A.; Heemskerk, J.; Korzh, V.; Norlin, S.; Ruiz i Altaba, A.; Tanabe, Y.; Placzek, M.; Edlund, T.; Jessell, T.M. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 1994, 76, 761–775. [Google Scholar] [CrossRef]
- Deries, M.; Thorsteinsdóttir, S. Axial and limb muscle development: Dialogue with the neighbourhood. Cell. Mol. Life Sci. CMLS 2016, 73, 4415–4431. [Google Scholar] [CrossRef]
- Naruse-Nakajima, C.; Asano, M.; Iwakura, Y. Involvement of EphA2 in the formation of the tail notochord via interaction with ephrinA1. Mech. Dev. 2021, 102, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Mohibi, S.; Gurumurthy, C.B.; Nag, A.; Wang, J.; Mirza, S.; Mian, Y.; Quinn, M.; Katafiasz, B.; Eudy, J.; Pandey, S.; et al. Mammalian Alteration/Deficiency in Activation 3 (Ada3) Is Essential for Embryonic Development and Cell Cycle Progression. J. Biol. Chem. 2012, 287, 29442–29456. [Google Scholar] [CrossRef] [PubMed]
- Tunster, S.J.; Creeth, H.D.J.; John, R.M. The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources. Dev. Biol. 2016, 409, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Shiraki, N.; Yamazoe, T.; Qin, Z.; Ohgomori, K.; Mochitate, K.; Kume, K.; Kume, S. Efficient Differentiation of Embryonic Stem Cells into Hepatic Cells In Vitro Using a Feeder-Free Basement Membrane Substratum. PLoS ONE 2011, 6, e24228. [Google Scholar] [CrossRef]
- Richter, A.; Valdimarsdottir, L.; Hrafnkelsdottir, H.E.; Runarsson, J.F.; Omarsdottir, A.R.; Oostwaard, D.W.-v.; Mummery, C.; Valdimarsdottir, G. BMP4 Promotes EMT and Mesodermal Commitment in Human Embryonic Stem Cells via SLUG and MSX2. Stem Cells 2014, 32, 636–648. [Google Scholar] [CrossRef]
- Tsaytler, P.; Liu, J.; Blaess, G.; Schifferl, D.; Veenvliet, J.V.; Wittler, L.; Timmermann, B.; Herrmann, B.G.; Koch, F. BMP4 triggers regulatory circuits specifying the cardiac mesoderm lineage. Development 2023, 150, dev201450. [Google Scholar] [CrossRef]
- Mahany, E.B.; Han, X.; Borges, B.C.; da Silveira Cruz-Machado, S.; Allen, S.J.; Garcia-Galiano, D.; Hoenerhoff, M.J.; Bellefontaine, N.H.; Elias, C.F. Obesity and High-Fat Diet Induce Distinct Changes in Placental Gene Expression and Pregnancy Outcome. Endocrinology 2018, 159, 1718–1733. [Google Scholar] [CrossRef]
- Nakamura, Y.; Hamada, Y.; Fujiwara, T.; Enomoto, H.; Hiroe, T.; Tanaka, S.; Nose, M.; Nakahara, M.; Yoshida, N.; Takenawa, T.; et al. Phospholipase C-δ1 and -δ3 Are Essential in the Trophoblast for Placental Development. Mol. Cell. Biol. 2023, 25, 10979–10988. [Google Scholar] [CrossRef]
- Miller, E.E.; Kobayashi, G.S.; Musso, C.M.; Allen, M.; Ishiy, F.A.A.; de Caires, L.C.; Goulart, E.; Griesi-Oliveira, K.; Zechi-Ceide, R.M.; Richieri-Costa, A.; et al. EIF4A3 deficient human iPSCs and mouse models demonstrate neural crest defects that underlie Richieri-Costa-Pereira syndrome. Hum. Mol. Genet. 2017, 26, 2177–2191. [Google Scholar] [CrossRef]
- Parsons, K.J.; Albertson, R.C. Roles for Bmp4 and CaM1 in Shaping the Jaw: Evo-Devo and Beyond. Annu. Rev. Genet. 2009, 43, 369–388. [Google Scholar] [CrossRef]
- Engelhard, C.; Sarsfield, S.; Merte, J.; Wang, Q.; Li, P.; Beppu, H.; Kolodkin, A.L.; Sucov, H.M.; Ginty, D.D. MEGF8 is a modifier of BMP signaling in trigeminal sensory neurons. eLife 2013, 2, e01160. [Google Scholar] [CrossRef]
- Xu, W.; MacDonald, B.T.; Semenov, M.V.; Huang, H.; He, X. Dissecting Molecular Differences between Wnt Coreceptors LRP5 and LRP6. PLoS ONE 2011, 6, e23537. [Google Scholar] [CrossRef]
- Yamamoto, M.; Matsuzaki, T.; Takahashi, R.; Adachi, E.; Maeda, Y.; Yamaguchi, S.; Kitayama, H.; Echizenya, M.; Morioka, Y.; Alexander, D.B.; et al. The transformation suppressor gene Reck is required for postaxial patterning in mouse forelimbs. Biol. Open 2012, 1, 458–466. [Google Scholar] [CrossRef]
- Bottasso-Arias, N.; Leesman, L.; Burra, K.; Snowball, J.; Shah, R.; Mohanakrishnan, M.; Xu, Y.; Sinner, D. BMP4 and Wnt signaling interact to promote mouse tracheal mesenchyme morphogenesis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2022, 322, L224–L242. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.G.; Mai, S.; Chung, K.; Wei, K. Flrt2 and Flrt3 have overlapping and non-overlapping expression during craniofacial development. Gene Expr. Patterns 2009, 9, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Bock, E.; Yavin, Z.; Jørgensen, O.S.; Yavin, E. Nervous system-specific proteins in developing rat cerebral cells in culture. Nerv. Syst.-Specif. Proteins Dev. Rat Cereb. Cells Cult. 1980, 35, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Harik, S.I.; Hall, A.K.; Richey, P.; Andersson, L.; Lundahl, P.; Perry, G. Ontogeny of the erythroid/HepG2-type glucose transporter (GLUT-1) in the rat nervous system. Brain Res. Dev. Brain Res. 1993, 72, 41–49. [Google Scholar] [CrossRef]
- Calderon, D.; Bardot, E.; Dubois, N. Probing early heart development to instruct stem cell differentiation strategies. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2016, 245, 1130–1144. [Google Scholar] [CrossRef]
- Miquerol, L.; Kelly, R.G. Organogenesis of the vertebrate heart. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 17–29. [Google Scholar] [CrossRef]
- Stutt, N.; Song, M.; Wilson, M.D.; Scott, I.C. Cardiac specification during gastrulation—The Yellow Brick Road leading to Tinman. Semin. Cell Dev. Biol. 2022, 127, 46–58. [Google Scholar] [CrossRef]
- Robillard, J.E.; Guillery, E.N.; Segar, J.L.; Merrill, D.C.; Jose, P.A. Influence of renal nerves on renal function during development. Pediatr. Nephrol. 1993, 7, 667–671. [Google Scholar] [CrossRef]
- Serluca, F.C.; Drummond, I.A.; Fishman, M.C. Endothelial signaling in kidney morphogenesis: A role for hemodynamic forces. Curr. Biol. CB 2002, 12, 492–497. [Google Scholar] [CrossRef]
- Duncan, S.A. Mechanisms controlling early development of the liver. Mech. Dev. 2003, 120, 19–33. [Google Scholar] [CrossRef]
- Lemaigre, F.; Zaret, K.S. Liver development update: New embryo models, cell lineage control, and morphogenesis. Curr. Opin. Genet. Dev. 2004, 14, 582–590. [Google Scholar] [CrossRef]
- Diaz-Cuadros, M.; Miettinen, T.P.; Skinner, O.S.; Sheedy, D.; Díaz-García, C.M.; Gapon, S.; Hubaud, A.; Yellen, G.; Manalis, S.R.; Oldham, W.M.; et al. Metabolic regulation of species-specific developmental rates. Nature 2023, 613, 550–557. [Google Scholar] [CrossRef]
- Miao, Y.; Pourquié, O. Cellular and molecular control of vertebrate somitogenesis. Nat. Rev. Mol. Cell Biol. 2024, 25, 517–533. [Google Scholar] [CrossRef]
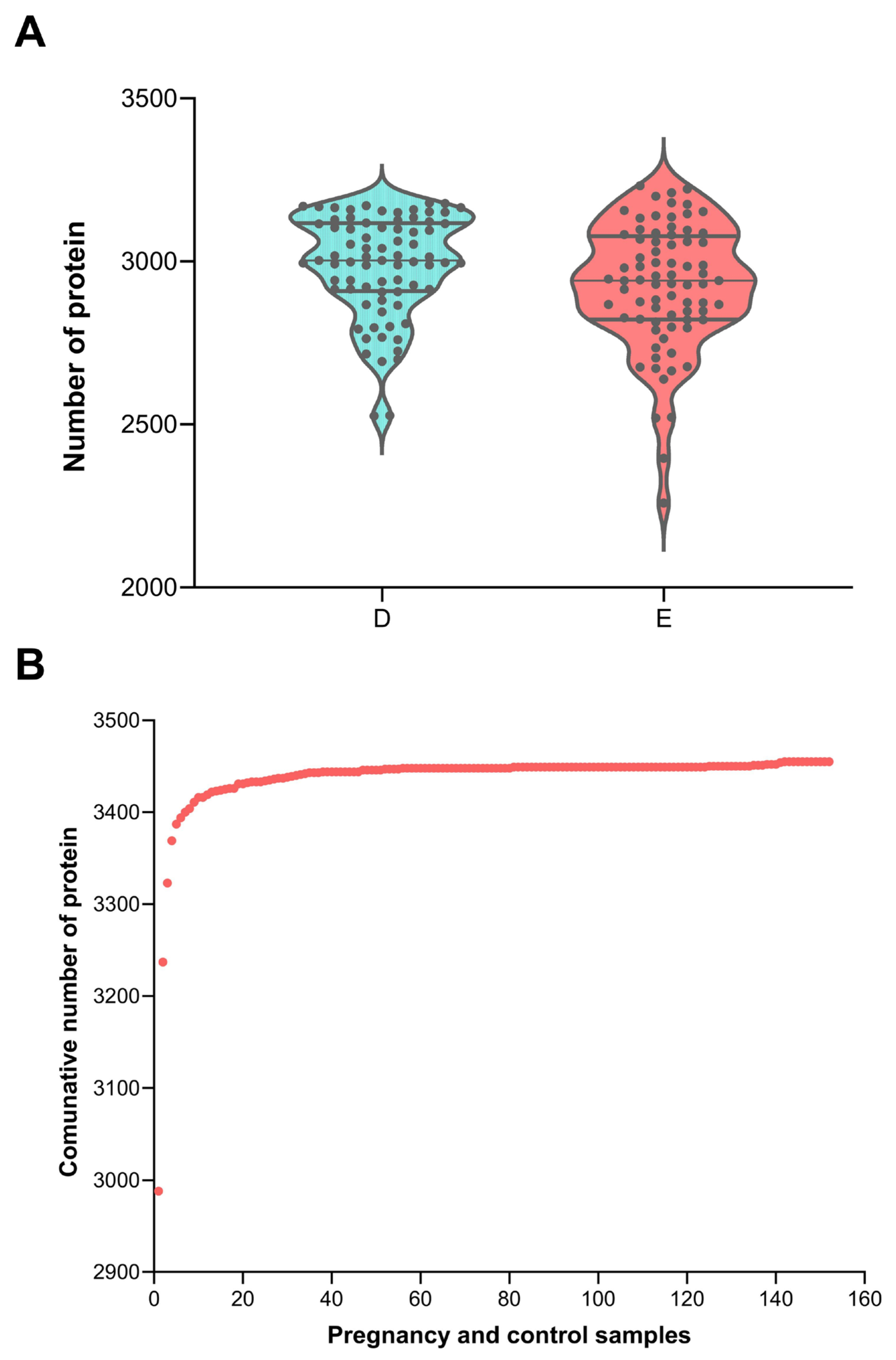
| Group | Number of Samples | Time Point | Number of Identified Proteins |
|---|---|---|---|
| Pregnant group | n = 76 | Day 0–Day 18 | 2924 ± 192 |
| Control group | n = 76 | Day 0–Day 18 | 2987 ± 155 |
| All | n = 152 | Day 0–Day 18 | 2956 ± 177 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Ge, L.; Chen, S.; Sun, L.; Sun, W.; Gao, Y. Proteome Analysis of Daily Urine Samples of Pregnant Rats Unveils Developmental Processes of Fetus as Well as Physiological Changes in Mother Rats. Biology 2025, 14, 1700. https://doi.org/10.3390/biology14121700
Wang H, Ge L, Chen S, Sun L, Sun W, Gao Y. Proteome Analysis of Daily Urine Samples of Pregnant Rats Unveils Developmental Processes of Fetus as Well as Physiological Changes in Mother Rats. Biology. 2025; 14(12):1700. https://doi.org/10.3390/biology14121700
Chicago/Turabian StyleWang, Haitong, Linna Ge, Sijie Chen, Longqin Sun, Wei Sun, and Youhe Gao. 2025. "Proteome Analysis of Daily Urine Samples of Pregnant Rats Unveils Developmental Processes of Fetus as Well as Physiological Changes in Mother Rats" Biology 14, no. 12: 1700. https://doi.org/10.3390/biology14121700
APA StyleWang, H., Ge, L., Chen, S., Sun, L., Sun, W., & Gao, Y. (2025). Proteome Analysis of Daily Urine Samples of Pregnant Rats Unveils Developmental Processes of Fetus as Well as Physiological Changes in Mother Rats. Biology, 14(12), 1700. https://doi.org/10.3390/biology14121700






