Construction and Performance Evaluation of an Astaxanthin–Chitosan/Chitooligosaccharide Hydrogel System for Ex Vivo Culture of Murine Spermatogonial Stem Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Isolation and Purification of Spermatogonia
2.3. Preparation of the Hydrogel Layer
2.3.1. AST + CO Hydrogel (COnAx, 0 ≤ n ≤ 3, 0 ≤ x ≤ 2)
2.3.2. AST + COS Hydrogel (COS nAx, 0 ≤ n ≤ 3, 0 ≤ x ≤ 2)
2.4. Three-Dimensional Hydrogel Cultures of Mouse Spermatogonial Stem Cells
2.5. Assessment of Mouse Spermatogonial Stem Cell Proliferation
2.5.1. EdU-Based Proliferation Assay
2.5.2. CCK-8 Viability Assay
2.6. Scanning Electron Microscopy (SEM)
2.7. Alkaline Phosphatase (AP) Staining
2.8. Indirect Immunofluorescence Microscopy
2.9. Sequencing Analysis
2.10. Statistical Analyses of Experimental Data
3. Results
3.1. Hydrogel Fabrication and Its Impact on Cellular Viability
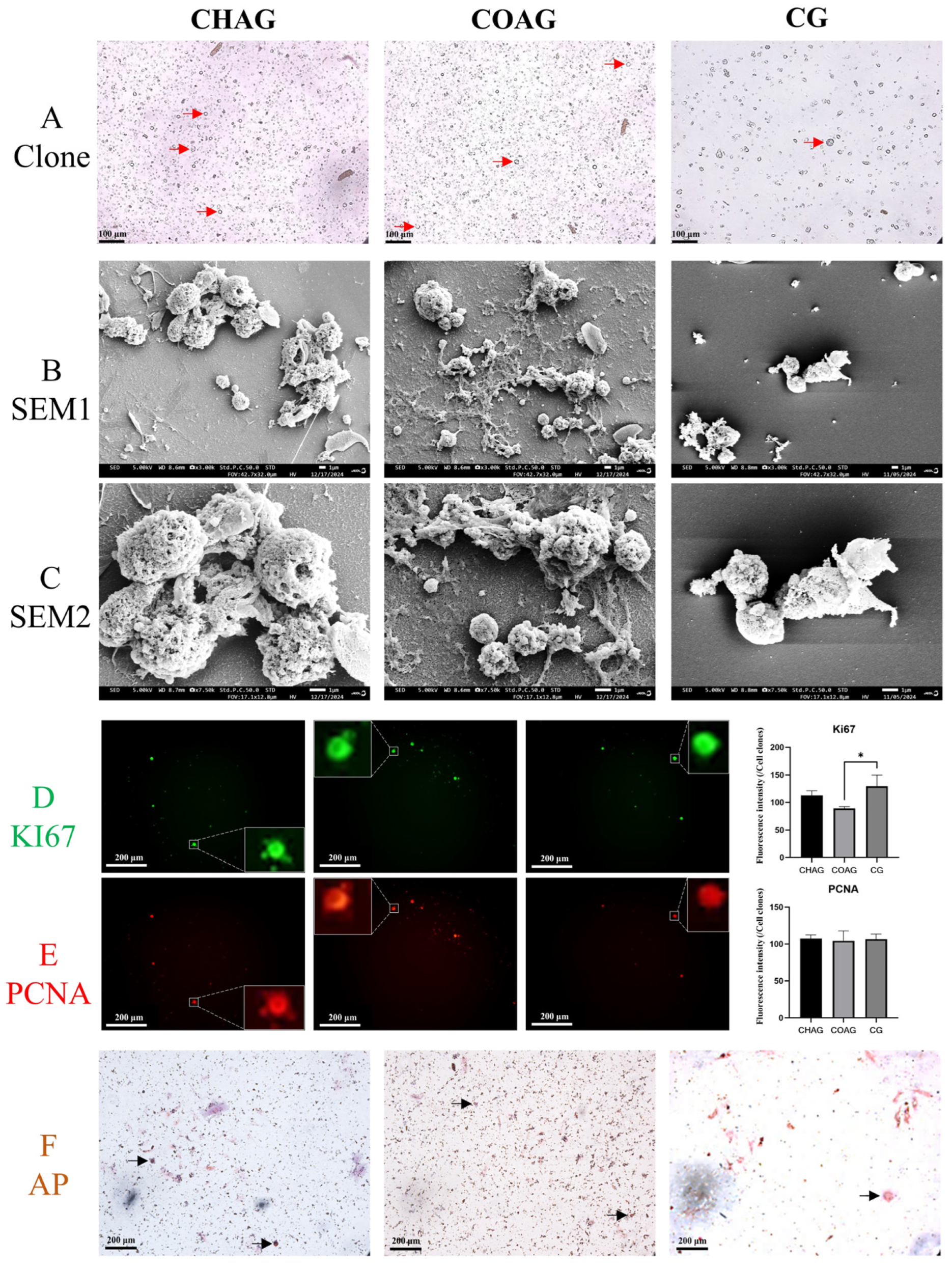
3.2. Isolation, Culture, and Phenotypic Characterization of SSCs
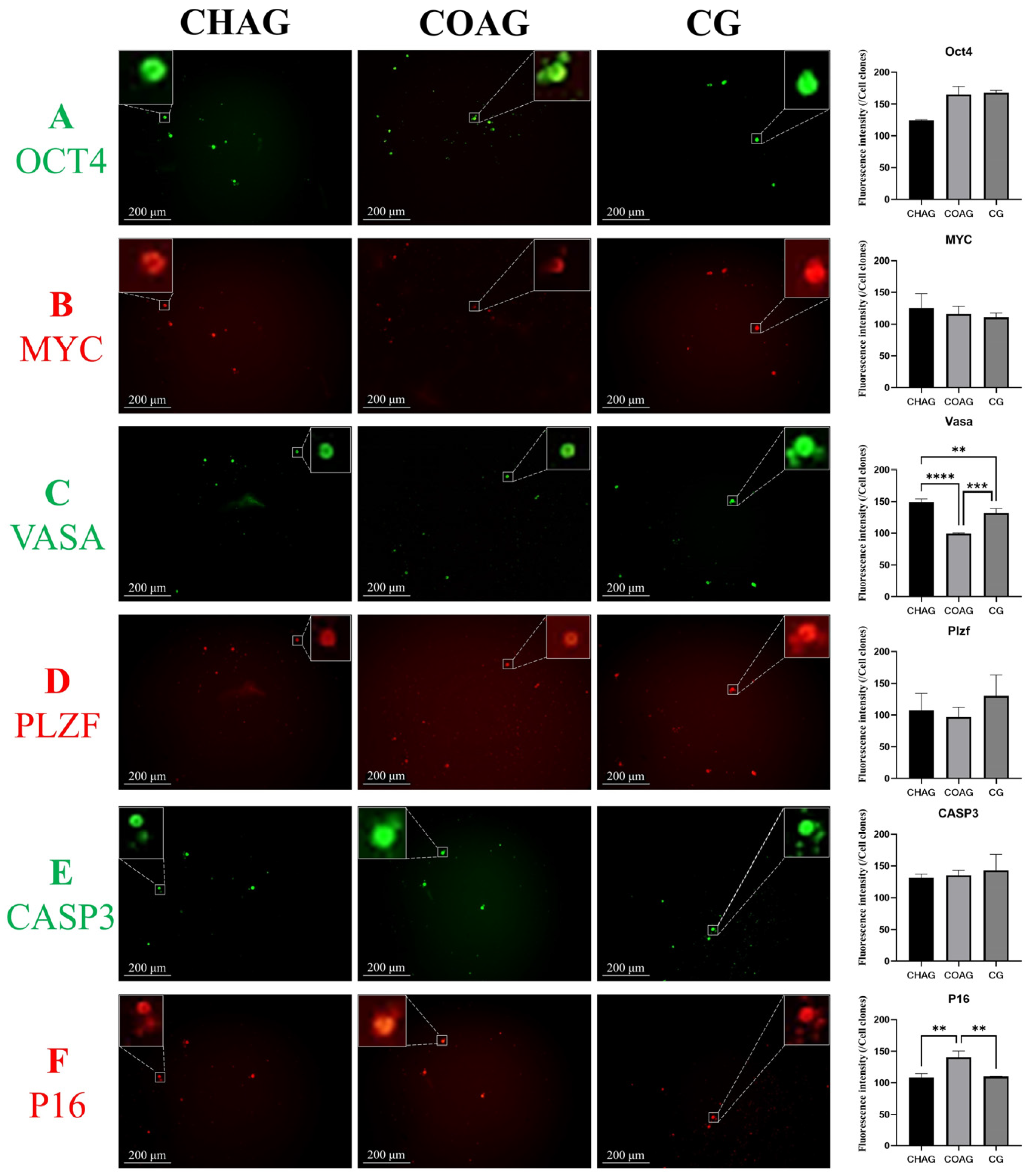
3.3. Immunofluorescence Assessment of SSC Identity, Stemness Maintenance, and Senescence Markers
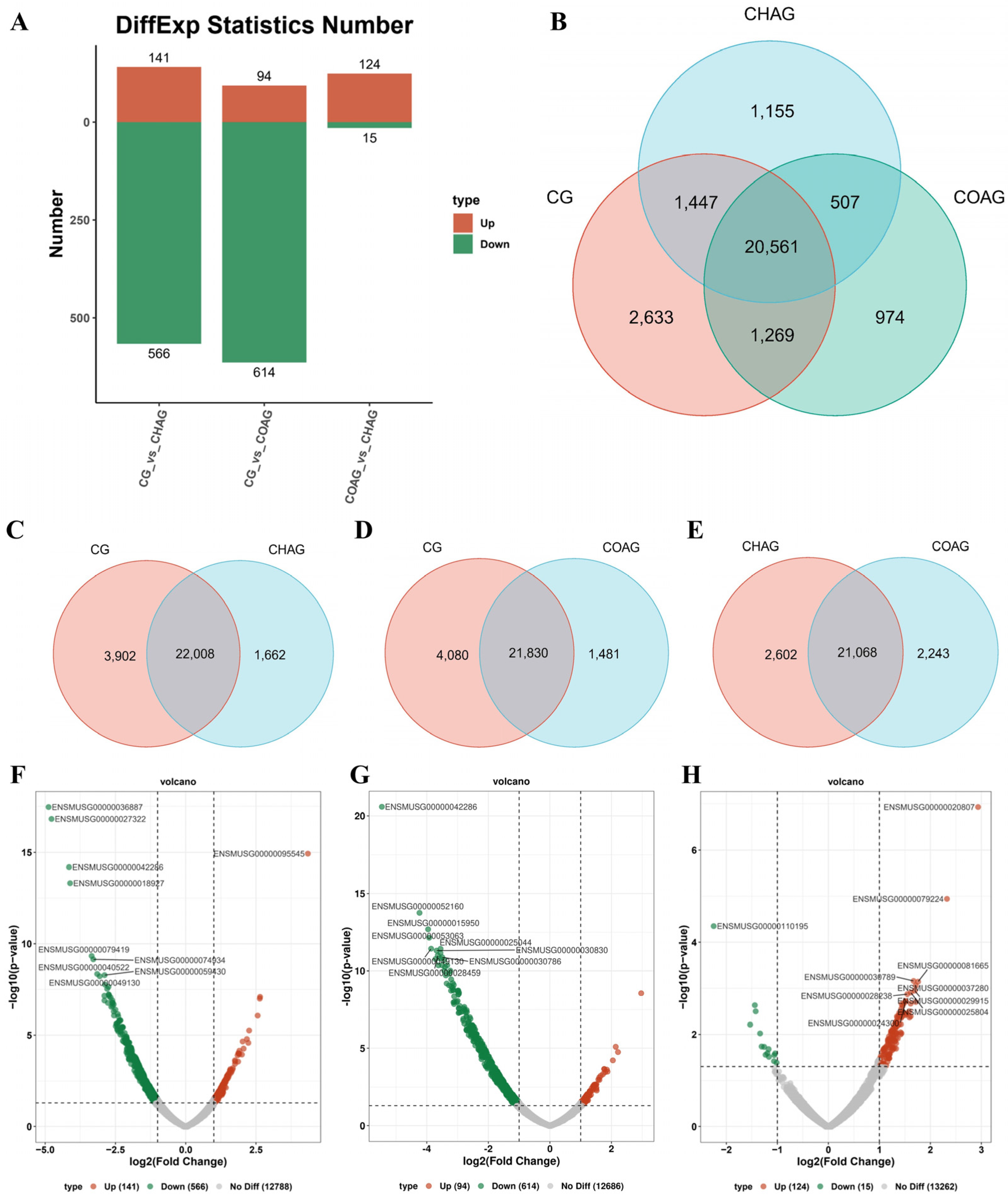
3.4. Comparative mRNA Profiling of SSCs Cultured Under Distinct Hydrogel Systems
3.5. Pathway-Centric Clustering Analysis of Differentially Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP | Alkaline phosphatase |
| AST | Astaxanthin |
| CCK8 | Cell Counting Kit-8 |
| CO | Chitosan |
| COS | Chitosan oligosaccharide |
| DMEM/F12 | Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 |
| EDU | 5-ethynyl-2′-deoxyuridine |
| GO | Gene Ontology |
| GFRα1 | GDNF family receptor alpha 1 |
| KI67 | The cell proliferation antigen Ki-67 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MYC | Myelocytomatosis oncogene |
| OCT4 | Octamer binding transcription factor 4 |
| PCNA | Proliferating Cell Nuclear Antigen |
| PBS | Phosphate-buffered saline |
| P16 | Cyclin-Dependent Kinase Inhibitor 2A |
| P21 | Cyclin-Dependent Kinase Inhibitor 1A |
| PCR | Polymerase Chain Reaction |
| PLZF | Promyelocytic Leukemia Zinc Finger |
| Sirt1 | The silent information regulator sirtuin 1 |
| SSCs | Spermatogonial stem cells |
| TERT | Telomerase reverse transcriptase |
| VASA | DEAD-box helicase 4 |
References
- Wu, J.; Kang, K.; Liu, S.; Ma, Y.; Yu, M.; Zhao, X. Recent Progress of In vitro 3D Culture of Male Germ Stem Cells. J. Funct. Biomater. 2023, 14, 543. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, S.; Li, X.; Zou, K. Progress in in vitro culture and gene editing of porcine spermatogonial stem cells. Zool. Res. 2019, 40, 343–348. [Google Scholar] [CrossRef]
- Du, X.; Wu, S.; Wei, Y.; Yu, X.; Ma, F.; Zhai, Y.; Yang, D.; Zhang, M.; Liu, W.; Zhu, H.; et al. PAX7 promotes CD49f-positive dairy goat spermatogonial stem cells’ self-renewal. J. Cell. Physiol. 2021, 236, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Ibtisham, F.; Zhao, Y.; Wu, J.; Nawab, A.; Mei, X.; Li, G.; An, L. The optimized condition for the isolation and in vitro propagation of mouse spermatogonial stem cells. Biol. Futura 2019, 70, 79–87. [Google Scholar] [CrossRef]
- Liu, S.; Wu, J.; Zhao, X.; Yu, M.; Taniguchi, M.; Bao, H.; Kang, K. Recent Progress of Induced Spermatogenesis In vitro. Int. J. Mol. Sci. 2024, 25, 8524. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Ren, Y.; Ren, F.; Li, Y.; Feng, T.; Wang, Z.; Du, Y.; Zhang, L.; Hu, J. Recent advances in isolation, identification, and culture of mammalian spermatogonial stem cells. Asian J. Androl. 2022, 24, 5–14. [Google Scholar] [CrossRef]
- Shah, D.D.; Raghani, N.R.; Chorawala, M.R.; Singh, S.; Prajapati, B.G. Harnessing three-dimensional (3D) cell culture models for pulmonary infections: State of the art and future directions. Naunyn Schmiedebergs Arch. Pharmacol. 2023, 396, 2861–2880. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lei, X.; He, F.; He, J.; Liu, Y.; Ye, Y.; Deng, X.; Duan, E.; Yin, D. Silk fibroin/chitosan scaffold with tunable properties and low inflammatory response assists the differentiation of bone marrow mesenchymal stem cells. Int. J. Biol. Macromol. 2017, 105, 584–597. [Google Scholar] [CrossRef]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, J.; Park, S.; Kim, Y.; Yoon, J.; Park, H.S.; Hyun, J.; Joung, Y.K.; Lee, T.I.; Bhang, S.H. Subaqueous 3D stem cell spheroid levitation culture using anti-gravity bioreactor based on sound wave superposition. Biomater. Res. 2023, 27, 51. [Google Scholar] [CrossRef]
- Kim, J.A.; Hong, S.; Rhee, W.J. Microfluidic three-dimensional cell culture of stem cells for high-throughput analysis. World J. Stem Cells 2019, 11, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Y Baena, A.R.; Casasco, A.; Monti, M. Hypes and Hopes of Stem Cell Therapies in Dentistry: A Review. Stem Cell Rev. Rep. 2022, 18, 1294–1308. [Google Scholar] [CrossRef]
- Kehler, J.; Tolkunova, E.; Koschorz, B.; Pesce, M.; Gentile, L.; Boiani, M.; Lomelí, H.; Nagy, A.; Mclaughlin, K.J.; Schöler, H.R.; et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004, 5, 1078–1083. [Google Scholar] [CrossRef]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Kanatsu-Shinohara, M.; Tanaka, T.; Ogonuki, N.; Ogura, A.; Morimoto, H.; Cheng, P.F.; Eisenman, R.N.; Trumpp, A.; Shinohara, T. Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 2016, 30, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Bai, Y.; Li, S.; Wei, H.; Zhang, X.; Li, L.; Tian, X.C.; Jiang, Q.; Wang, C.; Qin, L.; et al. Characteristics of spermatogonial stem cells derived from neonatal porcine testis. Andrologia 2015, 47, 765–778. [Google Scholar] [CrossRef]
- Meng, X.; Lindahl, M.; Hyvönen, M.E.; Parvinen, M.; de Rooij, D.G.; Hess, M.W.; Raatikainen-Ahokas, A.; Sainio, K.; Rauvala, H.; Lakso, M.; et al. Regulation of Cell Fate Decision of Undifferentiated Spermatogonia by GDNF. Science 2000, 287, 1489–1493. [Google Scholar] [CrossRef]
- Kolesnichenko, M.; Vogt, P.K. Understanding PLZF. Cell Cycle 2014, 10, 771–775. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jung, H.J.; Yoon, M.J. VASA (DDX4) is a Putative Marker for Spermatogonia, Spermatocytes and Round Spermatids in Stallions. Reprod. Domest. Anim. 2015, 50, 1032–1038. [Google Scholar] [CrossRef]
- Hermann, B.P.; Sukhwani, M.; Lin, C.; Sheng, Y.; Tomko, J.; Rodriguez, M.; Shuttleworth, J.J.; Mcfarland, D.; Hobbs, R.M.; Pandolfi, P.P.; et al. Characterization, Cryopreservation, and Ablation of Spermatogonial Stem Cells in Adult Rhesus Macaques. Stem Cells 2007, 25, 2330–2338. [Google Scholar] [CrossRef]
- Ma, F.; Zhou, Z.; Li, N.; Zheng, L.; Wu, C.; Niu, B.; Tang, F.; He, X.; Li, G.; Hua, J. Lin28a promotes self-renewal and proliferation of dairy goat spermatogonial stem cells (SSCs) through regulation of mTOR and PI3K/AKT. Sci. Rep. 2016, 6, 38805. [Google Scholar] [CrossRef]
- Azizi, H.; Ghasemi Hamidabadi, H.; Skutella, T. Differential Proliferation Effects after Short-Term Cultivation of Mouse Spermatogonial Stem Cells on Different Feeder Layers. Cell J. 2019, 21, 186–193. [Google Scholar] [CrossRef]
- Mai, F.Y.; He, P.; Ye, J.Z.; Xu, L.H.; Ouyang, D.Y.; Li, C.G.; Zeng, Q.Z.; Zeng, C.Y.; Zhang, C.C.; He, X.H.; et al. Caspase-3-mediated GSDME activation contributes to cisplatin- and doxorubicin-induced secondary necrosis in mouse macrophages. Cell Prolif. 2019, 52, e12663. [Google Scholar] [CrossRef]
- Yan, J.; Chen, S.; Yi, Z.; Zhao, R.; Zhu, J.; Ding, S.; Wu, J. The role of p21 in cellular senescence and aging-related diseases. Mol. Cells 2024, 47, 100113. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Monavari, M.; Homaeigohar, S.; Medhekar, R.; Nawaz, Q.; Monavari, M.; Zheng, K.; Boccaccini, A.R. A 3D-Printed Wound-Healing Material Composed of Alginate Dialdehyde–Gelatin Incorporating Astaxanthin and Borate Bioactive Glass Microparticles. ACS Appl. Mater. Interfaces 2023, 15, 50626–50637. [Google Scholar] [CrossRef] [PubMed]
- Afzali, A.; Amidi, F.; Koruji, M.; Nazari, H.; Gilani, M.A.S.; Sanjbad, A.S. Astaxanthin Relieves Busulfan-Induced Oxidative Apoptosis in Cultured Human Spermatogonial Stem Cells by Activating the Nrf-2/HO-1 pathway. Reprod. Sci. 2022, 29, 374–394. [Google Scholar] [CrossRef]
- Choi, B.Y.; Chalisserry, E.P.; Kim, M.H.; Kang, H.W.; Choi, I.; Nam, S.Y. The Influence of Astaxanthin on the Proliferation of Adipose-derived Mesenchymal Stem Cells in Gelatin-Methacryloyl (GelMA) Hydrogels. Materials 2019, 12, 2416. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Chao, H.; Chern, E.; Hsu, S. Chitosan 3D cell culture system promotes naïve-like features of human induced pluripotent stem cells: A novel tool to sustain pluripotency and facilitate differentiation. Biomaterials 2021, 268, 120575. [Google Scholar] [CrossRef]
- Ding, B.; Gao, H.; Song, J.; Li, Y.; Zhang, L.; Cao, X.; Xu, M.; Cai, J. Tough and Cell-Compatible Chitosan Physical Hydrogels for Mouse Bone Mesenchymal Stem Cells in vitro. ACS Appl. Mater. Interfaces 2016, 8, 19739–19746. [Google Scholar] [CrossRef]
- Moeinzadeh, A.; Ashtari, B.; Garcia, H.; Koruji, M.; Velazquez, C.A.; Bagher, Z.; Barati, M.; Shabani, R.; Davachi, S.M. The Effect of Chitosan/Alginate/Graphene Oxide Nanocomposites on Proliferation of Mouse Spermatogonial Stem Cells. J. Funct. Biomater. 2023, 14, 556. [Google Scholar] [CrossRef]
- Kamal, M.; Sindi, R.A.; El Azzazi, F.E.; Kishk, W.H.; Khalil, H.A.; Abdel Khalek, A.M.; Ayoub, M.A.; Tufarelli, V.; Abd El Hack, M.E. Sexual behaviour response, testicular development and semen quality of New Zealand white rabbit bucks as influenced by dietary chitosan. Reprod. Domest. Anim. 2023, 58, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Sun, G.; Yao, W.; Zhu, Y.; Wang, R.; Cai, L.; Liu, K.; Zhang, Q.; Liu, X.W.; Wan, Q. 3-Aminodeoxypyranoses in Glycosylation: Diversity-Oriented Synthesis and Assembly in Oligosaccharides. Angew. Chem.-Int. Edit. 2017, 56, 5227–5231. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ye, H.; Zhu, F.; Hu, C.; Zheng, Y. The role of Chito-oligosaccharide in regulating ovarian germ stem cells function and restoring ovarian function in chemotherapy mice. Reprod. Biol. Endocrinol. 2021, 19, 14. [Google Scholar] [CrossRef]
- Zheng, K.; Hong, W.; Ye, H.; Zhou, Z.; Ling, S.; Li, Y.; Dai, Y.; Zhong, Z.; Yang, Z.; Zheng, Y. Chito-oligosaccharides and macrophages have synergistic effects on improving ovarian stem cells function by regulating inflammatory factors. J. Ovarian Res. 2023, 16, 76. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.; et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Jamaluddin, M.F.B.; Ghosh, A.; Ingle, A.; Mohammed, R.; Ali, A.; Bahrami, M.; Kaiko, G.; Gibb, Z.; Filipe, E.C.; Cox, T.R.; et al. Bovine and human endometrium-derived hydrogels support organoid culture from healthy and cancerous tissues. Proc. Natl. Acad. Sci. USA 2022, 119, e2208040119. [Google Scholar] [CrossRef]
- Willemse, J.; van Tienderen, G.; van Hengel, E.; Schurink, I.; van der Ven, D.; Kan, Y.; de Ruiter, P.; Rosmark, O.; Westergren-Thorsson, G.G.; Schneeberger, K.; et al. Hydrogels derived from decellularized liver tissue support the growth and differentiation of cholangiocyte organoids. Biomaterials 2022, 284, 121473. [Google Scholar] [CrossRef]
- Dou, X.Q.; Feng, C.L. Amino Acids and Peptide-Based Supramolecular Hydrogels for Three-Dimensional Cell Culture. Adv. Mater. 2017, 29, 1604062. [Google Scholar] [CrossRef]
- Shi, X.; Janmey, P.A. Large Polyacrylamide Hydrogels for Large-Batch Cell Culture and Mechanobiological Studies. Macromol. Biosci. 2023, 23, 2300042. [Google Scholar] [CrossRef]
- Liu, Q.; Dai, W.; Gao, Y.; Dong, L.; Jia, H.; Li, S.; Guo, L.; Fan, Y.; Zhang, X. The synergistic regulation of chondrogenesis by collagen-based hydrogels and cell co-culture. Acta Biomater. 2022, 154, 194–211. [Google Scholar] [CrossRef]
- Jury, M.; Matthiesen, I.; Rasti Boroojeni, F.; Ludwig, S.L.; Civitelli, L.; Winkler, T.E.; Selegård, R.; Herland, A.; Aili, D. Bioorthogonally Cross-Linked Hyaluronan–Laminin Hydrogels for 3D Neuronal Cell Culture and Biofabrication. Adv. Healthc. Mater. 2022, 11, 2102097. [Google Scholar] [CrossRef]
- Xu, X.; Feng, Q.; Ma, X.; Deng, Y.; Zhang, K.; Ooi, H.S.; Yang, B.; Zhang, Z.; Feng, B.; Bian, L. Dynamic gelatin-based hydrogels promote the proliferation and self-renewal of embryonic stem cells in long-term 3D culture. Biomaterials 2022, 289, 121802. [Google Scholar] [CrossRef]
- Lou, J.; Mooney, D.J. Chemical strategies to engineer hydrogels for cell culture. Nat. Rev. Chem. 2022, 6, 726–744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, C. Enhanced proliferation and differentiation of mesenchymal stem cells by astaxanthin-encapsulated polymeric micelles. PLoS ONE 2019, 14, e0216755. [Google Scholar] [CrossRef]
- Naeemi, S.; Eidi, A.; Khanbabaee, R.; Sadri-Ardekani, H.; Kajbafzadeh, A. Differentiation and proliferation of spermatogonial stem cells using a three-dimensional decellularized testicular scaffold: A new method to study the testicular microenvironment in vitro. Int. Urol. Nephrol. 2021, 53, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, J.; Luo, J.; Cui, Y.; Chen, J.; Zeng, B.; Deng, Z.; Shao, L. “Double-sided protector” Janus hydrogels for skin and mucosal wound repair: Applications, mechanisms, and prospects. J. Nanobiotechnology 2025, 23, 387. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Z.; Liu, S.; Gao, F.; Zhang, J.; Peng, Z.; Wang, L.; Pan, X. Astaxanthin improves the development of the follicles and oocytes through alleviating oxidative stress induced by BPA in cultured follicles. Sci. Rep. 2022, 12, 7853. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kang, X.F.; Shang, X.J. Astaxanthin in male reproduction: Advances in studies. Zhonghua Nan Ke Xue 2016, 22, 938–943. [Google Scholar]
- Uslu, B.; Biltekin, B.; Denir, S.; Ozbas-Turan, S.; Arbak, S.; Akbuga, J.; Bilir, A. Effects of different forms of chitosan on intercellular junctions of mouse fibroblasts in vitro. Biotech. Histochem. 2016, 91, 20–29. [Google Scholar] [CrossRef]
- Boonthum, C.; Namdee, K.; Khongkow, M.; Temisak, S.; Chatdarong, K.; Sajomsang, W.; Ponglowhapan, S.; Yata, T. Gonadotropin-releasing hormone-modified chitosan as a safe and efficient gene delivery vector for spermatogonia cells. Reprod. Domest. Anim. 2018, 53 (Suppl. S3), 23–28. [Google Scholar] [CrossRef]
- Saranya, N.; Moorthi, A.; Saravanan, S.; Devi, M.P.; Selvamurugan, N. Chitosan and its derivatives for gene delivery. Int. J. Biol. Macromol. 2011, 48, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Jing, H.; Yu, W.; Lei, F.; Wang, R.; Hu, C.; Li, M.; Lin, T.; Zhou, H.; Wang, F.; et al. A bone matrix-simulating scaffold to alleviate replicative senescence of mesenchymal stem cells during long-term expansion. J. Biomed. Mater. Res. Part A 2020, 108, 1955–1967. [Google Scholar] [CrossRef]
- Tanaka, H.; Hamaya, Y.; Nishiwaki, N.; Ishida, H. A concise synthesis of rhamnan oligosaccharides with alternating α-(1→2)/(1→3)-linkages and repeating α-(1→3)-linkages by iterative α-glycosylation using disaccharide building blocks. Carbohydr. Res. 2018, 455, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Dan, P.; Schlachet, I.; Rouxel, D.; Menu, P.; Sosnik, A. Chitosan ascorbate hydrogel improves water uptake capacity and cell adhesion of electrospun poly(epsilon-caprolactone) membranes. Int. J. Pharm. 2019, 559, 420–426. [Google Scholar] [CrossRef]
- Tashakkorian, H.; Hasantabar, V.; Mostafazadeh, A.; Golpour, M. Transparent chitosan based nanobiocomposite hydrogel: Synthesis, thermophysical characterization, cell adhesion and viability assay. Int. J. Biol. Macromol. 2020, 144, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Wang, Y.; Xie, F.; Wang, S.; Xu, W.; Xu, J.; Sun, C. Carboxymethyl Chitosan and Gelatin Hydrogel Scaffolds Incorporated with Conductive PEDOT Nanoparticles for Improved Neural Stem Cell Proliferation and Neuronal Differentiation. Molecules 2022, 27, 8326. [Google Scholar] [CrossRef]
- Zhao, W.; Li, R.; Xiao, Z.; Yang, F.; Chen, S.; Miao, J.; Ma, G.; Wang, Y.; Chen, Y.; Fan, S. Rhein-chitosan in situ hydrogel promotes wound healing in diabetic mice. Int. J. Biol. Macromol. 2024, 277, 134472. [Google Scholar] [CrossRef]
- Ding, Q.; Liu, W.; Zhang, S.; Sun, S.; Yang, J.; Zhang, L.; Wang, N.; Ma, S.; Chai, G.; Shen, L.; et al. Hydrogel loaded with thiolated chitosan modified taxifolin liposome promotes osteoblast proliferation and regulates Wnt signaling pathway to repair rat skull defects. Carbohydr. Polym. 2024, 336, 122115. [Google Scholar] [CrossRef]
- Liu, S.; Ma, L. Therapeutic effects of chitosan/β-glycerophosphate/collagen hydrogel combined with MSCs on chronic achilles tendon injury via the Akt/GSK-3β pathway. J. Orthop. Surg. Res. 2025, 20, 204. [Google Scholar] [CrossRef]
- Zhai, X.; Yang, X.; Zou, P.; Shao, Y.; Yuan, S.; Abd El Aty, A.M.; Wang, J. Protective Effect of Chitosan Oligosaccharides Against Cyclophosphamide-Induced Immunosuppression and Irradiation Injury in Mice. J. Food Sci. 2018, 83, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.J.; Shie, M.Y.; Hoshiba, T.; Kawazoe, N.; Chen, G.; Chang, H.C. Osteogenic differentiation and immune response of human bone-marrow-derived mesenchymal stem cells on injectable calcium-silicate-based bone grafts. Tissue Eng. Part A 2010, 16, 2343–2354. [Google Scholar] [CrossRef] [PubMed]

| 0.1%AST | 0.2%AST | |
|---|---|---|
| 0.1%CO | 0.1%CO + 0.1%AST (COA) | 0.1%CO + 0.2%AST (COA2) |
| 0.2%CO | 0.2%CO + 0.1%AST (CO2A) | 0.2%CO + 0.2%AST (CO2A2) |
| 0.3%CO | 0.3%CO + 0.1%AST (CO3A) | 0.3%CO + 0.2%AST (CO3A2) |
| 0.1%COS | 0.1%COS + 0.1%AST (COSA) | 0.1%COS + 0.2%AST (COSA2) |
| 0.2%COS | 0.2%COS + 0.1%AST (COS2A) | 0.2%COS + 0.2%AST (COS2A2) |
| 0.3%COS | 0.3%COS + 0.1%AST (COS3A1) | 0.3%COS + 0.2%AST (COS3A2) |
| CHAG Group | COAG Group | CG Group |
|---|---|---|
| 0.3% chitosan with 0.2% astaxanthin loading | 0.2% astaxanthin-loaded 0.2% chitosan oligosaccharide | No gel |
| Pathway ID | Pathway Description | GeneRatio | p-Value | Names of the Genes in Pathway |
|---|---|---|---|---|
| mmu04810 | Regulation of actin cytoskeleton | 26/370 | 9.94 × 10−7 | Actg2, Myh11, Itgal, Itgam, Itga4, Fn1, Itgb2, Itga6, Pdgfb, Itgax, Vav1, Cxcr4, Pik3cd, Lpar4, Fgf10, Myh10, Diaph3, Acta2, Itga2, Itga1, Iqgap2, Nckap1l, Itgb5, Scin, Vav3, Iqgap3 |
| mmu04510 | Focal adhesion | 22/370 | 1.10 × 10−5 | Actg2, Col6a3, Ccnd1, Itga4, Parvg, Igf1, Fn1, Lama5, Itga6, Pdgfb, Pgf, Thbs2, Vav1, Pik3cd, Thbs3, Parvb, Acta2, Itga2, Itga1, Itgb5, Lamc3, Vav3 |
| mmu04110 | Cell cycle | 15/370 | 0.000622 | Ccnd1, Tgfb3, Fbxo5, Sfn, Skp2, Pkmyt1, Bub1b, Aurkb, E2f2, Mcm5, Tgfb2, Cdc20, Ndc80, Cdc25c, Cdc25b |
| mmu04520 | Adherens junction | 10/370 | 0.005086 | Actg2, Ptprb, Was, Ptpn6, Cdh5, Map3k7cl, Acta2, Iqgap2, Cdh1, Iqgap3 |
| mmu04512 | ECM–receptor interaction | 12/370 | 0.000166 | Col6a3, Itga4, Fn1, Lama5, Itga6, Thbs2, Thbs3, Npnt, Itga2, Itga1, Itgb5, Lamc3 |
| mmu04514 | Cell adhesion molecules | 23/370 | 0.002206 | Siglec1, Itgal, H2-T24, Selplg, Itga4, Cdh3, Evi2a, Sdc3, L1cam, Milr1, Vsir, Itgb2, Itga6, Slitrk4, F11r, Cdh5, Ptprc, Cd28, Mpzl2, Cntnap2, Cd80, Cdh1, Spn |
| mmu05205 | Proteoglycans in cancer | 23/370 | 2.40 × 10−6 | Actg2, Ccnd1, Camk2a, Igf1, Plau, Fn1, Mmp2, Thbs2, Ptpn6, Vav1, Pik3ip1, Pik3cd, Mapk13, Hcls1, Gpc3, Acta2, Plaur, Itga2, Iqgap2, Itgb5, Tgfb2, Vav3, Iqgap3 |
| mmu05410 | Hypertrophic cardiomyopathy | 14/370 | 4.17 × 10−5 | Actg2, Des, Itga4, Igf1, Tgfb3, Itga6, Ace, Tmem178, Acta2, Itga2, Itga1, Mylk, Itgb5, Tgfb2 |
| mmu05412 | Arrhythmogenic right ventricular cardiomyopathy | 11/370 | 0.00078 | Actg2, Des, Itga4, Itga6, Tmem178, Jup, Acta2, Itga2, Itga1, Itgb5, Dsp |
| mmu05032 | Morphine addiction | 10/370 | 0.001603 | Pde2a, Pde7b, Cacna1a, Gabra4, Gng2, Arrb2, Gabbr1, Pde3b, Pde1b, Grk3 |
| mmu05206 | MicroRNAs in cancer | 14/370 | 0.003315 | Ccnd1, Hmga2, Plau, Pdgfb, Thbs2, Pik3ip1, Mmp16, Pik3cd, E2f2, Kif23, Notch1, Tgfb2, Cdc25c, Cdc25b |
| mmu05418 | Fluid shear stress and atherosclerosis | 13/370 | 0.004012 | Actg2, Ncf1, Milr1, Pdgfb, Mmp2, Trpv4, Pik3cd, Cdh5, Mapk13, Map3k7cl, Acta2, Ncf2, Il1a |
| mmu05219 | Bladder cancer | 6/370 | 0.005041 | Ccnd1, Mmp2, Thbs2, E2f2, Upk3b, Cdh1 |
| mmu04380 | Osteoclast differentiation | 23/370 | 5.81 × 10−10 | Ncf1, Csf1r, Milr1, Sirpa, Tnfrsf11a, Fcgr2b, Pira2, Lcp2, Lilrb4a, Blnk, Pik3cd, Fcgr3, Mapk13, Tyrobp, Btk, Map3k7cl, Spi1, Tnfrsf11b, Tgfb2, Ncf2, Il1a, Pira12, Ppp3r2 |
| mmu04062 | Chemokine signaling pathway | 21/370 | 8.37 × 10−6 | Ccl6, Ncf1, Ccl9, Was, Dock2, Pf4, Cxcl1, Pik3cg, Cxcl2, Hck, Plcb2, Vav1, Cxcr4, Gng2, Arrb2, Prex1, Pik3cd, Lyn, Rasgrp2, Vav3, Grk3 |
| mmu04670 | Leukocyte transendothelial migration | 17/370 | 9.86 × 10−6 | Actg2, Ncf1, Itgal, Itga4, Milr1, Itgb2, Mmp2, Vav1, Cxcr4, F11r, Rapgef3, Pik3cd, Cdh5, Mapk13, Acta2, Ncf2, Vav3 |
| mmu04611 | Platelet activation | 16/370 | 3.52 × 10−5 | Actg2, Fcer1g, Pik3cg, Plcb2, Lcp2, Tbxas1, Pik3cd, Mapk13, Btk, Prkg2, Fermt3, Lyn, Acta2, Itga2, Apbb1ip, Rasgrp2 |
| mmu04662 | B cell receptor signaling pathway | 17/370 | 0.001055 | Cd72, Inpp5d, Milr1, Fcgr2b, Pira2, Ptpn6, Vav1, Lilrb4a, Blnk, Pik3cd, Pik3ap1, Fcgr3, Btk, Lyn, Pira12, Vav3, Ppp3r2 |
| mmu04914 | Progesterone-mediated oocyte maturation | 10/370 | 0.002033 | Igf1, Pgr, Pik3cd, Pkmyt1, Mapk13, Pde3b, Rps6ka1, Kif22, Cdc25c, Cdc25b |
| Pathway ID | Pathway Description | GeneRatio | p-Value | Names of the Genes in Pathway |
|---|---|---|---|---|
| mmu04510 | Focal adhesion | 24/375 | 1.18 × 10−6 | Actg2, Itga4, Ccnd1, Vav1, Parvb, Igf1, Col6a3, Prkcb, Pdgfb, Rac2, Itga6, Fn1, Spp1, Thbs2, Pgf, Rasgef1b, Vav3, Pik3cd, Plxdc1, Lama5, Vwf, Itga2, Efs, Thbs3 |
| mmu04810 | Regulation of actin cytoskeleton | 25/375 | 4.19 × 10−6 | Itgal, Itgam, Itgax, Itgb2, Actg2, Itga4, Myh11, Vav1, Nckap1l, Cxcr4, Pdgfb, Rac2, Itga6, Lpar4, Fn1, Rasgef1b, Vav3, Pik3cd, Myh14, Myh10, Scin, Itga2, Fgf10, Efs, Iqgap3 |
| mmu04010 | MAPK signaling pathway | 26/375 | 0.000126 | Csf1r, Cacna1a, Igf1, Prkcb, Ptpn7, Pdgfb, Rac2, Arrb2, Rps6ka1, Pgf, Tgfb3, Tnf, Rasgef1b, Plxdc1, Cd14, Map3k7cl, Cacng8, Tgfb2, Arrb1, Tmem178, Hspa1b, Map3k8, Fgf10, Cdc25b, Rasgrp2, Mapk13 |
| mmu04512 | ECM–receptor interaction | 11/375 | 0.000736 | Itga4, Col6a3, Cd36, Itga6, Fn1, Spp1, Thbs2, Lama5, Vwf, Itga2, Thbs3 |
| mmu05205 | Proteoglycans in cancer | 26/375 | 5.86 × 10−8 | Actg2, Ptpn6, Plau, Ccnd1, Vav1, Igf1, Prkcb, Hcls1, Camk2a, Rac2, Fn1, Thbs2, Tnf, Rasgef1b, Vav3, Pik3cd), Mmp2, Plxdc1, Plcg2, Ank3, Tgfb2, Itga2, Gpc3, Iqgap3, Mapk13, Pik3ip1 |
| mmu05206 | MicroRNAs in cancer | 19/375 | 1.37 × 10−5 | Plau, Ccnd1, Prkcb, Pdgfb, Hmga2, Thbs2, Rasgef1b, Mmp16, Pik3cd, Plxdc1, Plcg2, Tgfb2, Notch1, Brca1, Ccne2, Kif23, Cdc25b, Bcl2l11, Pik3ip1 |
| mmu05417 | Lipid and atherosclerosis | 22/375 | 6.95 × 10−5 | Ncf1, Abcg1, Vav1, Cd36, Camk2a, Ncf2, Casp1, Rac2, Ncf4, Lyn, Tnf, Cxcl1, Pycard, Vav3, Pik3cd, Cd14, Map3k7cl, Lbp, Hspa1b, Cxcl2, Bcl2l14, Mapk13 |
| mmu05134 | Legionellosis | 10/375 | 0.000129 | Itgb2, Casp1, Eef1a2, Naip2, Tnf, Cxcl1, Pycard, Cd14, Hspa1b, Cxcl2 |
mmu05200 | Pathways in cancer | 39/375 | 0.000182 | Csf1r, Spi1, Ccnd1, Igf1, Prkcb, Gng2, Camk2a, Cxcr4, Pdgfb, Rac2, Il2rg, Itga6, Lpar4, Fn1, Csf2ra, Il7r, Csf2rb2, Pgf, Tgfb3, Rasgef1b, Pik3cd, Mmp2, Plxdc1, Plcg2, Lama5, Tgfb2, Jag1, Notch1, Itga2, Ccne2, Skp2, Rad51, Csf2rb, Fgf10, Jup, Rasgrp2, Bcl2l14, Bcl2l11, Gli2 |
| mmu05410 | Hypertrophic cardiomyopathy | 13/375 | 0.000191 | Actg2, Itga4, Igf1, Des, Itga6, Ace, Tgfb3, Tnf, Cacng8, Tgfb2, Itga2, Tmem178, Mybpc3 |
| mmu04933 | AGE-RAGE signaling pathway in diabetic complications | 12/375 | 0.000275 | Ccnd1, Prkcb, Rac2, Fn1, Tgfb3, Tnf, Pik3cd, Mmp2, Plcg2, Tgfb2, Bcl2l14, Mapk13 |
| mmu05214 | Glioma | 10/375 | 0.000739 | Ccnd1, Igf1, Prkcb, Camk2a, Pdgfb, Rasgef1b, Camk1d, Pik3cd, Plcg2, Bcl2l14 |
| mmu04380 | Osteoclast differentiation | 34/375 | 1.85 × 10−19 | Ncf1, Csf1r, Lilrb4a, Tnfrsf11a, Sirpa, Tyrobp, Fcgr3, Spi1, Pira2, Pira12, Milr1, Lcp2, Fcgr2b, Pirb, Lilrb4b, Pira1, Ncf2, Blnk, Btk, Rac2, Ncf4, Gm49339, Syk, Tnfrsf11b, Tnf, Fcgr4, Pik3cd, Plcg2, Map3k7cl, Tgfb2, Fosb, Socs3, Tec, Mapk13 |
| mmu04062 | Chemokine signaling pathway | 28/375 | 5.87 × 10−10 | Ncf1, Ccl9, Was, Dock2, Pik3cg, Vav1, Prkcb, Gng2, Cxcr4, Rac2, Ccr1, Arrb2, Prex1, Hck, Lyn, Cxcl1, Rasgef1b, Vav3, Pik3cd, Plcg2, Pik3r5, Grk3, Arrb1, Tec, Cxcl2, Efs, Rasgrp2, Pf4 |
| mmu04670 | Leukocyte transendothelial migration | 23/375 | 1.01 × 10−9 | Ncf1, Itgal, Itgb2, Actg2, Itga4, Milr1, Vav1, Prkcb, Ncf2, Cxcr4, Rac2, Ncf4, Cdh5, Vav3, Pik3cd, Mmp2, Plcg2, F11r, Rapgef3, Tec, Cldn4, Efs, Mapk13 |
| mmu04662 | B cell receptor signaling pathway | 26/375 | 2.23 × 10−8 | Cd72, Inpp5d, Lilrb4a, Ptpn6, Fcgr3, Pira2, Pira12, Milr1, Vav1, Fcgr2b, Prkcb, Pirb, Lilrb4b, Pira1, Pik3ap1, Blnk, Btk, Rac2, Gm49339, Syk, Lyn, Rasgef1b, Vav3, Pik3cd, Plcg2, Tec |
| mmu04611 | Platelet activation | 18/375 | 2.59 × 10−6 | Fcer1g, Actg2, Pik3cg, Fermt3, Lcp2, Btk, Tbxas1, Syk, Lyn, Pik3cd, Plcg2, Apbb1ip, Pik3r5, Vwf, Itga2, Tec, Rasgrp2, Mapk13 |
| mmu04625 | C-type lectin receptor signaling pathway | 13/375 | 0.000228 | Clec7a, Fcer1g, Clec4n, Lsp1, Casp1, Clec4d, Syk, Tnf, Pycard, Pik3cd, Plcg2, Egr3, Mapk13 |
| Pathway ID | Pathway Description | GeneRatio | p-Value | Names of the Genes in Pathway |
|---|---|---|---|---|
| mmu04145 | Phagosome | 13/81 | 5.50 × 10−7 | Atp6v0d2, Mrc1, Itgb2, Clec7a, Cd36, Marco, Msr1, Fcgr3, Ncf4, Rac2, Ctss, Ncf2, Ncf1 |
| mmu04810 | Regulation of actin cytoskeleton | 7/81 | 0.003389 | Itgax, Itgb2, Itgam, Rac2, Nckap1l, Gm49368, Vav1 |
| mmu04060 | Cytokine–cytokine receptor interaction | 8/81 | 0.004224 | Ccr1, Il1a, Csf2rb2, Il2rg, Il7r, Il1rn, Tnf, Tnfrsf11a |
| mmu05140 | Leishmaniasis | 10/81 | 1.55 × 10−6 | Itgb2, Prkcb, Il1a, Fcgr3, Ncf4, Ptpn6, Ncf2, Ncf1, Tnf, Eef1a2 |
| mmu05152 | Tuberculosis | 11/81 | 3.85 × 10−5 | Itgax, Atp6v0d2, Mrc1, Itgb2, Clec7a, Il1a, Itgam, Fcgr3, Ctss, Fcer1g, Tnf |
| mmu05417 | Lipid and atherosclerosis | 8/81 | 0.000598 | Cd36, Casp1, Ncf4, Rac2, Ncf2, Ncf1, Tnf, Vav1 |
| mmu05134 | Legionellosis | 4/81 | 0.00179 | Itgb2, Casp1, Tnf, Eef1a2 |
| mmu05133 | Pertussis | 4/81 | 0.003546 | Itgb2, Il1a, Casp1, Tnf |
| mmu05415 | Diabetic cardiomyopathy | 6/81 | 0.005367 | Prkcb, Cd36, Ncf4, Rac2, Ncf2, Ncf1 |
| mmu04930 | Type II diabetes mellitus | 3/81 | 0.006182 | Hk3, Tnf, Cacna1a |
| mmu05418 | Fluid shear stress and atherosclerosis | 5/81 | 0.007561 | Il1a, Rac2, Ncf2, Ncf1, Tnf |
| mmu04933 | AGE-RAGE signaling pathway in diabetic complications | 4/81 | 0.007944 | Prkcb, Il1a, Rac2, Tnf |
| mmu04380 | Osteoclast differentiation | 16/81 | 6.69 × 10−15 | Pira1, Lilrb4a, Pira12, Il1a, Spi1, Tyrobp, Fcgr3, Ncf4, Lilrb4b, Rac2, Pirb, Gm49339, Ncf2, Ncf1, Tnf, Tnfrsf11a |
| mmu04662 | B cell receptor signaling pathway | 12/81 | 5.50 × 10−8 | Pira1, Prkcb, Lilrb4a, Pira12, Fcgr3, Lilrb4b, Rac2, Inpp5d, Pirb, Gm49339, Ptpn6, Vav1 |
| mmu04650 | Natural killer cell-mediated cytotoxicity | 10/81 | 3.77 × 10−5 | Itgb2, Prkcb, Tyrobp, Fcgr3, Rac2, Lat2, Ptpn6, Fcer1g, Tnf, Vav1 |
| mmu04670 | Leukocyte transendothelial migration | 7/81 | 0.000107 | Itgb2, Prkcb, Ncf4, Rac2, Ncf2, Ncf1, Vav1 |
| mmu04613 | Neutrophil extracellular trap formation | 10/81 | 0.00019 | Itgb2, Prkcb, Clec7a, Casp1, Fcgr3, Ncf4, Rac2, Ncf2, Ncf1, Tlr7 |
| mmu04625 | C-type lectin receptor signaling pathway | 6/81 | 0.000253 | Clec7a, Casp1, Clec4d, Clec4n, Fcer1g, Tnf |
| mmu04666 | Fc gamma R-mediated phagocytosis | 7/81 | 0.001101 | Prkcb, Fcgr3, Ptprc, Rac2, Inpp5d, Ncf1, Vav1 |
| mmu04610 | Complement and coagulation cascades | 4/81 | 0.009357 | Itgax, Itgb2, C3ar1, Itgam |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Liu, S.; Zeng, X.; Li, Y.; Yao, Y.; Wang, J.; Hu, G.; Kang, K. Construction and Performance Evaluation of an Astaxanthin–Chitosan/Chitooligosaccharide Hydrogel System for Ex Vivo Culture of Murine Spermatogonial Stem Cells. Biology 2025, 14, 1664. https://doi.org/10.3390/biology14121664
Wu J, Liu S, Zeng X, Li Y, Yao Y, Wang J, Hu G, Kang K. Construction and Performance Evaluation of an Astaxanthin–Chitosan/Chitooligosaccharide Hydrogel System for Ex Vivo Culture of Murine Spermatogonial Stem Cells. Biology. 2025; 14(12):1664. https://doi.org/10.3390/biology14121664
Chicago/Turabian StyleWu, Jiang, Siqi Liu, Xiaowen Zeng, Yang Li, Yinlin Yao, Jing Wang, Guangdong Hu, and Kai Kang. 2025. "Construction and Performance Evaluation of an Astaxanthin–Chitosan/Chitooligosaccharide Hydrogel System for Ex Vivo Culture of Murine Spermatogonial Stem Cells" Biology 14, no. 12: 1664. https://doi.org/10.3390/biology14121664
APA StyleWu, J., Liu, S., Zeng, X., Li, Y., Yao, Y., Wang, J., Hu, G., & Kang, K. (2025). Construction and Performance Evaluation of an Astaxanthin–Chitosan/Chitooligosaccharide Hydrogel System for Ex Vivo Culture of Murine Spermatogonial Stem Cells. Biology, 14(12), 1664. https://doi.org/10.3390/biology14121664







