Genome-Wide Analysis of the LEA Gene Family in Pineapple (Ananas comosus L.) Reveals Its Potential Roles in Cold Stress Response and Reproductive Development
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of AcLEA Family in Pineapple
2.2. Phylogenetic Analysis of AcLEAs
2.3. Gene Structure, Conserved Motif, and Cis-Regulatory Element Analysis of AcLEAs
2.4. Three-Dimensional Structural Prediction of AcLEAs
2.5. Chromosomal Distribution and Gene Duplication Analysis of AcLEAs
2.6. Transcription Factor Binding Network Prediction
2.7. Plant Materials, Growth Conditions, and Cold Treatment
2.8. RNA Extraction and qRT-PCR Analysis
3. Results
3.1. Genome-Wide Identification and Basic Features of the AcLEA Gene Family
3.2. Phylogenetic Analyses of the AcLEA Genes
3.3. Gene Structure, Conserved Motif, and Domain Analysis of AcLEAs
3.4. Protein Model Prediction of AcLEAs
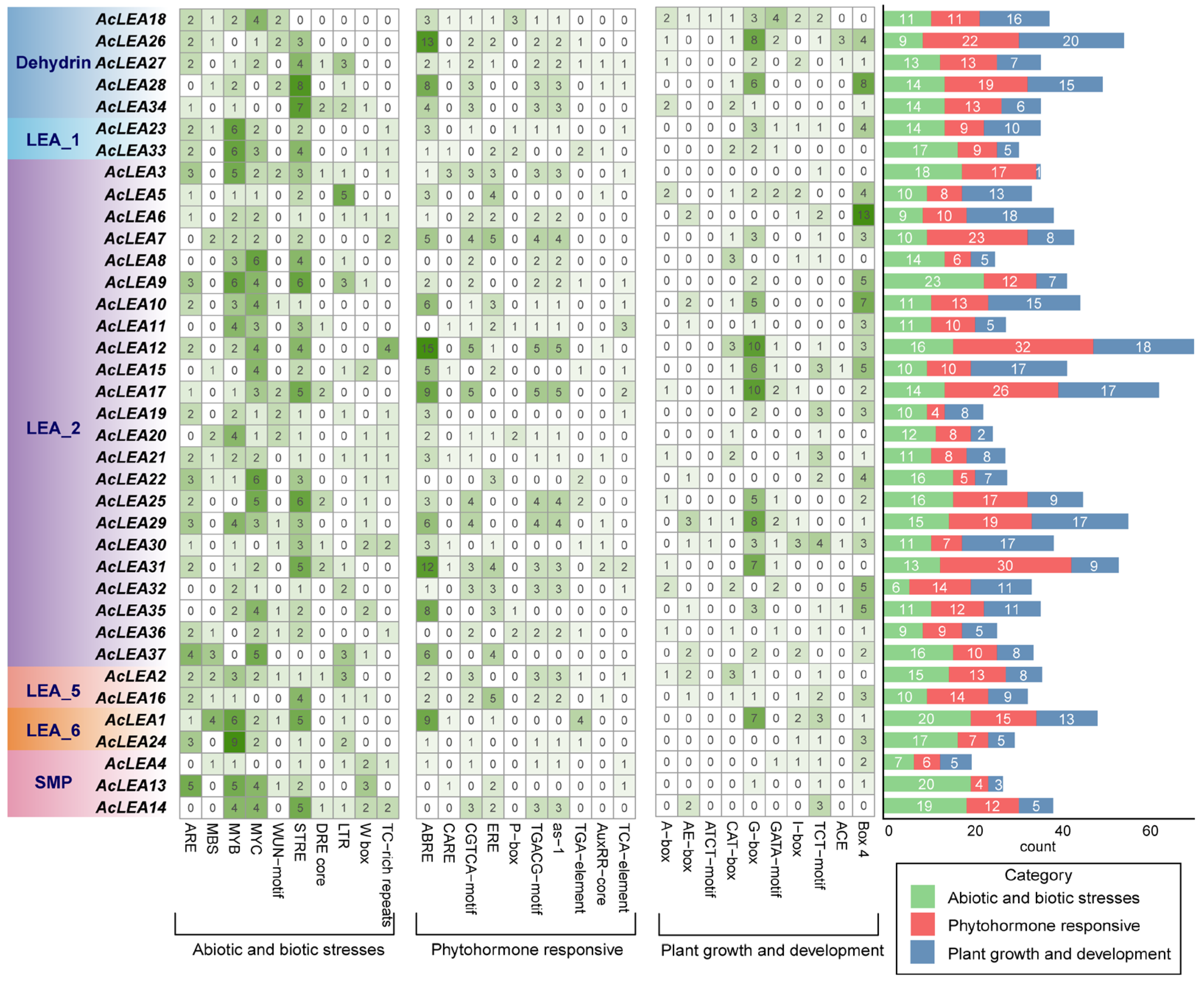
3.5. Cis-Regulatory Elements Identification in AcLEA Promoters
3.6. Chromosomal Distribution, Collinearity and Evolution Analysis of AcLEAs
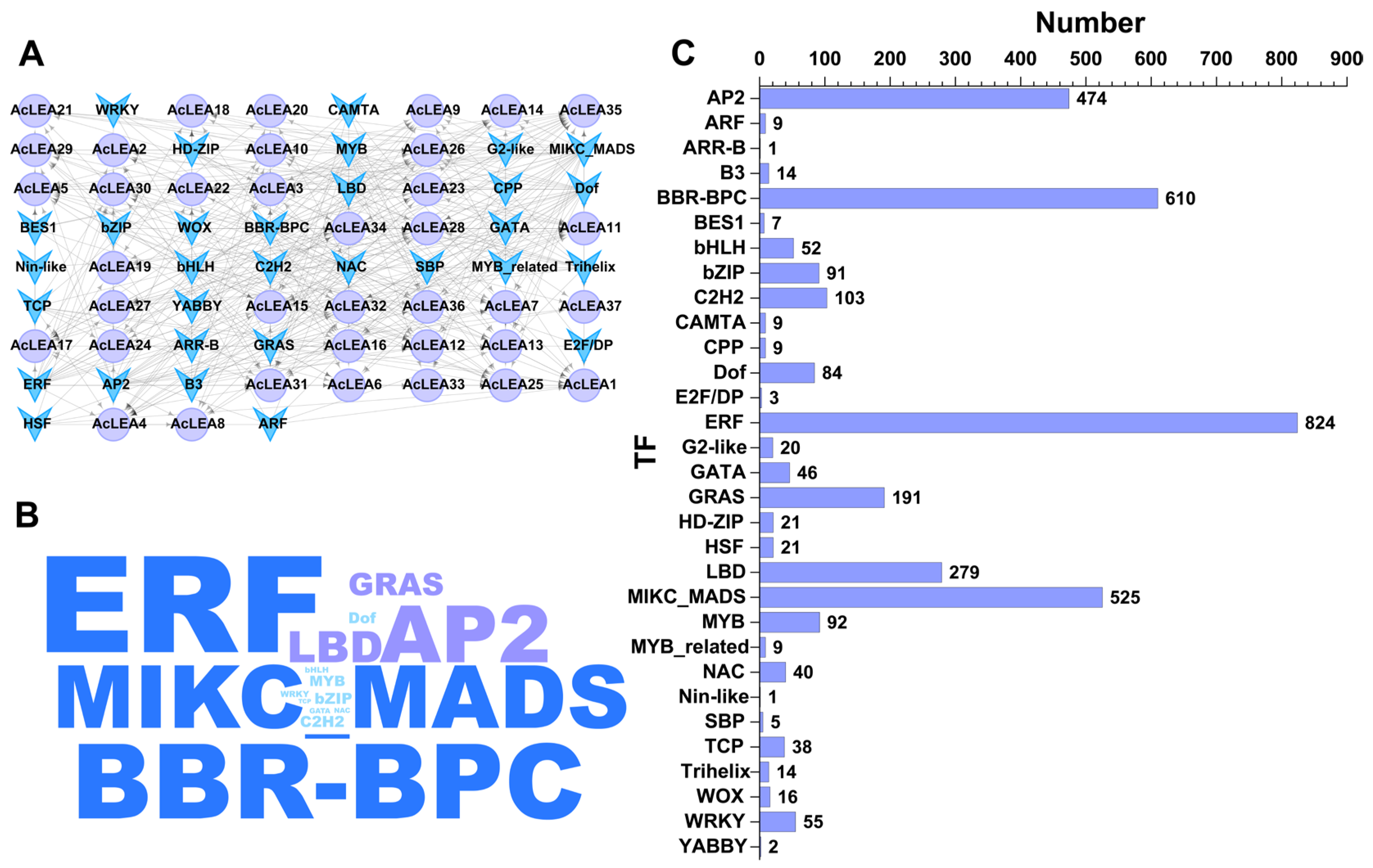
3.7. Transcription Factor Regulatory Network of AcLEA Genes
3.8. miRNA-Mediated Regulatory Mechanisms of AcLEA Genes
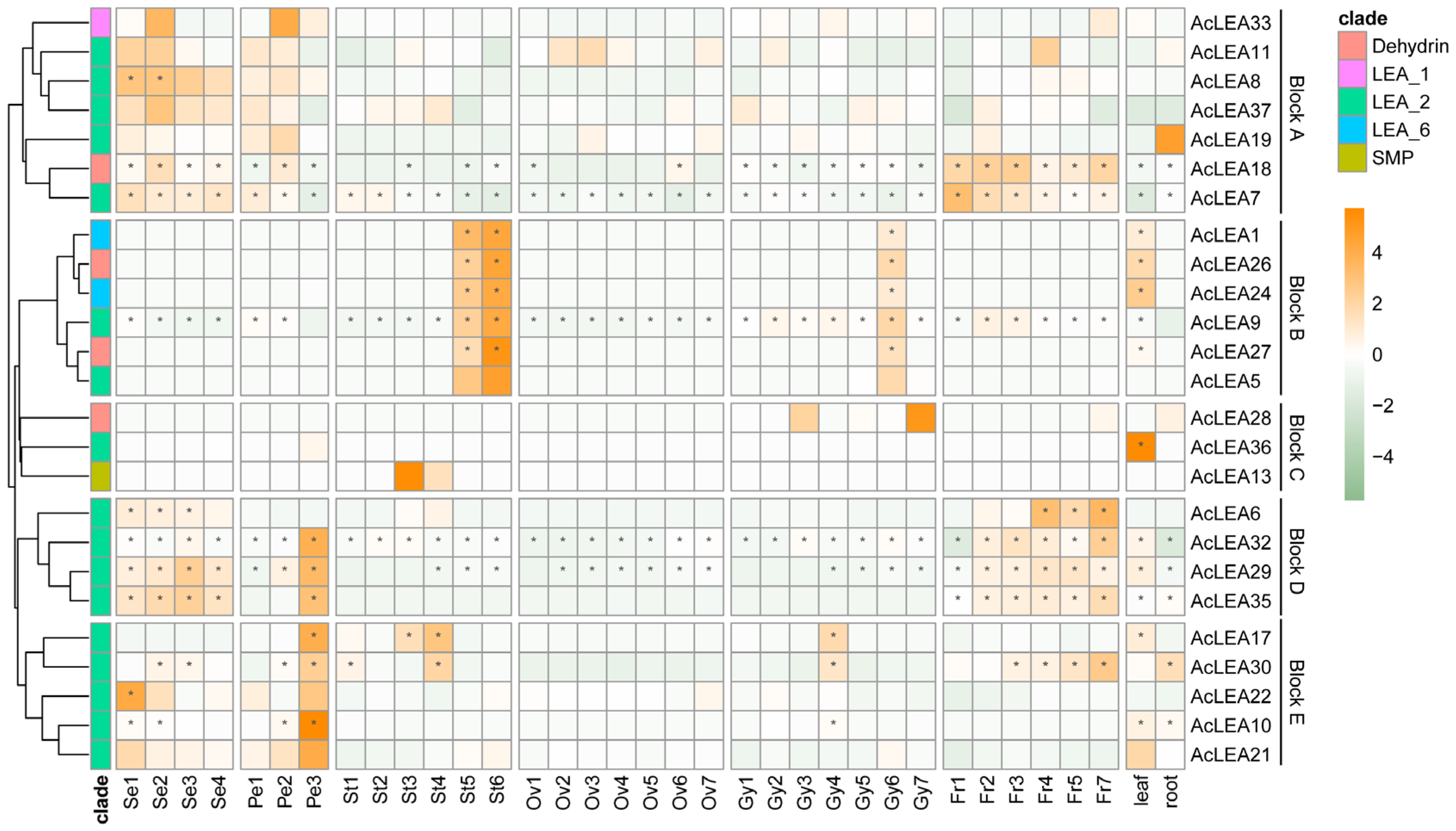
3.9. Expression Profile of AcLEAs in Different Tissues
3.10. Expression Patterns of AcLEA Genes in Response to Clod Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nowicka, B.; Ciura, J.; Szymanska, R.; Kruk, J. Improving photosynthesis, plant productivity and abiotic stress tolerance—Current trends and future perspectives. J. Plant Physiol. 2018, 231, 415–433. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-Induced CBF-PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef]
- Wang, G.; Xu, X.; Gao, Z.; Liu, T.; Li, Y.; Hou, X. Genome-wide identification of LEA gene family and cold response mechanism of BcLEA4-7 and BcLEA4-18 in non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Plant Sci. 2022, 321, 111291. [Google Scholar] [CrossRef]
- Zhao, Y.; Antoniou-Kourounioti, R.L.; Calder, G.; Dean, C.; Howard, M. Temperature-dependent growth contributes to long-term cold sensing. Nature 2020, 583, 825–829. [Google Scholar] [CrossRef]
- Dure, L., 3rd; Pyle, J.B.; Chlan, C.A.; Baker, J.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: XVII. Developmental expression of genes for the principal storage proteins. Plant Mol. Biol. 1983, 2, 199–206. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Colmenero-Flores, J.M.; Garciarrubio, A.; Covarrubias, A.A. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 2000, 275, 5668–5674. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, R.; Furuki, T.; Shimizu, T.; Takezawa, D.; Kikawada, T.; Sakurai, M.; Sugawara, Y. Biochemical and structural characterization of an endoplasmic reticulum-localized late embryogenesis abundant (LEA) protein from the liverwort Marchantia polymorpha. Biochem. Biophys. Res. Commun. 2014, 454, 588–593. [Google Scholar] [CrossRef]
- Hara, M.; Fujinaga, M.; Kuboi, T. Metal binding by citrus dehydrin with histidine-rich domains. J. Exp. Bot. 2005, 56, 2695–2703. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloids Surf. B Biointerfaces 2005, 45, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses. Sci. Rep. 2019, 9, 14123. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar] [CrossRef]
- Pantelic, A.; Stevanovic, S.; Komic, S.M.; Kilibarda, N.; Vidovic, M. In Silico Characterisation of the Late Embryogenesis Abundant (LEA) Protein Families and Their Role in Desiccation Tolerance in Ramonda serbica Panc. Int. J. Mol. Sci. 2022, 23, 3547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, P.; Xu, Y.; Liu, Q.; Yang, Y.; Liu, S. Overexpression of CsLEA11, a Y3SK2-type dehydrin gene from cucumber (Cucumis sativus), enhances tolerance to heat and cold in Escherichia coli. AMB Express 2017, 7, 182. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, J.; Sun, L.; Yang, X.; Li, D. Group 3 LEA Protein, ZmLEA3, Is Involved in Protection from Low Temperature Stress. Front. Plant Sci. 2016, 7, 1011. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Xia, W.; Hou, M.; Ruan, N.; Li, J.; Zhu, J. Cloning and function analysis of a Saussurea involucrata LEA4 gene. Front. Plant Sci. 2022, 13, 957133. [Google Scholar] [CrossRef]
- Shi, H.; He, X.; Zhao, Y.; Lu, S.; Guo, Z. Constitutive expression of a group 3 LEA protein from Medicago falcata (MfLEA3) increases cold and drought tolerance in transgenic tobacco. Plant Cell Rep. 2020, 39, 851–860. [Google Scholar] [CrossRef]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem. Biophys. Res. Commun. 2016, 478, 703–709. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, W.; Li, H.; Zhao, Z.; Zhu, J.; Li, J. The Pyrus sinkiangensis Yu PsLEA4 Gene Enhances the Cold Resistance of Solanum lycopersicum. Plants 2025, 14, 180. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, F.; Ma, M.; Gong, J.; Wang, Q.; Jia, P.; Zheng, G.; Liu, H. Overexpression of AtLEA3-3 confers resistance to cold stress in Escherichia coli and provides enhanced osmotic stress tolerance and ABA sensitivity in Arabidopsis thaliana. Mol. Biol. 2011, 45, 851–862. [Google Scholar] [CrossRef]
- Zeng, D.E.; Hou, P.; Xiao, F.; Liu, Y. Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2015, 24, 56–64. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Pujol, C.J.; Kado, C.I. Genetic and biochemical characterization of the pathway in Pantoea citrea leading to pink disease of pineapple. J. Bacteriol. 2000, 182, 2230–2237. [Google Scholar] [CrossRef]
- Peckham, G.D.; Kaneshiro, W.S.; Luu, V.; Berestecky, J.M.; Alvarez, A.M. Specificity of monoclonal antibodies to strains of Dickeya sp. that cause bacterial heart rot of pineapple. Hybridoma 2010, 29, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, L.; Guo, N.; Zhang, X.; Zhang, C.; Sun, G.; Xie, J. Functional properties of a cysteine proteinase from pineapple fruit with improved resistance to fungal pathogens in Arabidopsis thaliana. Molecules 2014, 19, 2374–2389. [Google Scholar] [CrossRef]
- Santos, C.; Ventura, J.A.; Lima, N. New Insights for Diagnosis of Pineapple Fusariosis by MALDI-TOF MS Technique. Curr. Microbiol. 2016, 73, 206–213. [Google Scholar] [CrossRef]
- He, Y.; Luan, A.; Wu, J.; Lin, W.Z.W. Overcoming key technical challenges in the genetic transformation of pineapple. Trop. Plants 2023, 2, 6. [Google Scholar] [CrossRef]
- Lin, J.; Wu, J.; Zhang, D.; Cai, X.; Du, L.; Lu, L.; Liu, C.; Chen, S.; Yao, Q.; Xie, S.; et al. The GRAS gene family and its roles in pineapple (Ananas comosus L.) developmental regulation and cold tolerance. BMC Plant Biol. 2024, 24, 1204. [Google Scholar] [CrossRef] [PubMed]
- Colmenero-Flores, J.M.; Moreno, L.P.; Smith, C.E.; Covarrubias, A.A. Pvlea-18, a member of a new late-embryogenesis-abundant protein family that accumulates during water stress and in the growing regions of well-irrigated bean seedlings. Plant Physiol. 1999, 120, 93–104. [Google Scholar] [CrossRef]
- Sheoran, I.S.; Sproule, K.A.; Olson, D.J.H.; Ross, A.R.S.; Sawhney, V.K. Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex. Plant Reprod. 2006, 19, 185–196. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Zeng, X.; Xu, Z.; Taranto, A.; Lomas, J.S.; Zhang, Y.; Huang, Y.; Wang, Y.; Yim, W.C.; et al. JCVI: A versatile toolkit for comparative genomics analysis. Imeta 2024, 3, e211. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Sharon, K.; Suvarna, S. Identification of the Calcineurin B-like gene family and gene expression patterns in response to low temperature stress in Prunus mume. Trop. Plants 2017, 3, e010. [Google Scholar]
- Sang, J.; Han, X.; Liu, M.; Qiao, G.; Jiang, J.; Zhuo, R. Selection and validation of reference genes for real-time quantitative PCR in hyperaccumulating ecotype of Sedum alfredii under different heavy metals stresses. PLoS ONE 2013, 8, e82927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Niu, S.; El-Kassaby, Y.A.; Li, W. Genome-wide identification of late embryogenesis abundant protein family and their key regulatory network in Pinus tabuliformis cold acclimation. Tree Physiol. 2023, 43, 1964–1985. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, X.; Wang, Y.; Di, P.; Meng, X.; Peng, W.; Rong, J.; Wang, Y. Genome-wide identification of the LEA gene family in Panax ginseng: Evidence for the role of PgLEA2-50 in plant abiotic stress response. Plant Physiol. Biochem. 2024, 212, 108742. [Google Scholar] [CrossRef]
- Jia, J.S.; Ge, N.; Wang, Q.Y.; Zhao, L.T.; Chen, C.; Chen, J.W. Genome-wide identification and characterization of members of the LEA gene family in Panax notoginseng and their transcriptional responses to dehydration of recalcitrant seeds. BMC Genom. 2023, 24, 126. [Google Scholar] [CrossRef]
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses. Biomolecules 2021, 11, 1662. [Google Scholar] [CrossRef]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and Collinearity in Plant Genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Y.; Xu, Z.; Luan, A.; Mao, Q.; Feng, J.; Xie, T.; Gong, X.; Wang, X.; Chen, H.; et al. Transcriptome Profiling of the Pineapple under Low Temperature to Facilitate Its Breeding for Cold Tolerance. PLoS ONE 2016, 11, e0163315. [Google Scholar] [CrossRef]
- Zhang, J.; Duan, Z.; Zhang, D.; Zhang, J.; Di, H.; Wu, F.; Wang, Y. Co-transforming bar and CsLEA enhanced tolerance to drought and salt stress in transgenic alfalfa (Medicago sativa L.). Biochem. Biophys. Res. Commun. 2016, 472, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Ling, H.; Yang, J.; Li, Y.; Guo, S. LEA proteins from Gastrodia elata enhance tolerance to low temperature stress in Escherichia coli. Gene 2018, 646, 136–142. [Google Scholar] [CrossRef]
- Rodriguez-Salazar, J.; Moreno, S.; Espin, G. LEA proteins are involved in cyst desiccation resistance and other abiotic stresses in Azotobacter vinelandii. Cell Stress Chaperones 2017, 22, 397–408. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Charfeddine, S.; Saidi, M.N.; Charfeddine, M.; Gargouri-Bouzid, R. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol. Biol. Rep. 2015, 42, 1163–1174. [Google Scholar] [CrossRef]
- Du, D.; Zhang, Q.; Cheng, T.; Pan, H.; Yang, W.; Sun, L. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol. Biol. Rep. 2013, 40, 1937–1946. [Google Scholar] [CrossRef]
- Yu, X.; Qu, M.; Shi, Y.; Hao, C.; Guo, S.; Fei, Z.; Gao, L. Chromosome-scale genome assemblies of wild tomato relatives Solanum habrochaites and Solanum galapagense reveal structural variants associated with stress tolerance and terpene biosynthesis. Hortic. Res. 2022, 9, uhac139. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Tong, Q.; Wang, Y.; Wang, Z.; Xu, G.; Elias, G.K.; Li, S.; Liang, Z. Transcriptomic Analysis of the Grapevine LEA Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for VamDHN3. Plant Cell Physiol. 2020, 61, 775–786. [Google Scholar] [CrossRef]
- Zan, T.; Li, L.; Li, J.; Zhang, L.; Li, X. Genome-wide identification and characterization of late embryogenesis abundant protein-encoding gene family in wheat: Evolution and expression profiles during development and stress. Gene 2020, 736, 144422. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Yao, W.; Zhao, K.; Liu, L.; Fan, G.; Zhou, B.; Jiang, T. Genome-wide search and structural and functional analyses for late embryogenesis-abundant (LEA) gene family in poplar. BMC Plant Biol. 2021, 21, 110. [Google Scholar] [CrossRef]
- Zhao, Y.; Hao, Y.; Dong, Z.; Tang, W.; Wang, X.; Li, J.; Wang, L.; Hu, Y.; Fang, L.; Guan, X.; et al. Identification and expression analysis of LEA gene family members in pepper (Capsicum annuum L.). FEBS Open Bio 2023, 13, 2246–2262. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Zhu, L.; Wang, L.; Zhang, H.; Zhang, X.; Xu, S.; Xue, J. Identification of the Maize LEA Gene Family and Its Relationship with Kernel Dehydration. Plants 2023, 12, 3674. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Kang, Y.; Liu, W.; Li, S.; Wang, Z.; Xia, X.; Chen, X.; Qian, L.; Xiong, X.; et al. Genome-wide characterization of LEA gene family reveals a positive role of BnaA.LEA6.a in freezing tolerance in rapeseed (Brassica napus L.). BMC Plant Biol. 2024, 24, 433. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Guo, B.; Wang, B.; Li, X.; Yang, T.; Li, N.; Wang, J.; Yu, Q. The LEA gene family in tomato and its wild relatives: Genome-wide identification, structural characterization, expression profiling, and role of SlLEA6 in drought stress. BMC Plant Biol. 2022, 22, 596. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Zhu, H.B.; Jin, G.L.; Liu, H.L.; Wu, W.R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Huang, R.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.; He, L. Genome-wide identification, evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). BMC Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chi, H.; Liu, C.; Zhang, T.; Han, L.; Li, L.; Pei, X.; Long, Y. Genome-wide identification and functional characterization of LEA genes during seed development process in linseed flax (Linum usitatissimum L.). BMC Plant Biol. 2021, 21, 193. [Google Scholar] [CrossRef]
- Candat, A.; Paszkiewicz, G.; Neveu, M.; Gautier, R.; Logan, D.C.; Avelange-Macherel, M.H.; Macherel, D. The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 2014, 26, 3148–3166. [Google Scholar] [CrossRef] [PubMed]
- Grelet, J.; Benamar, A.; Teyssier, E.; Avelange-Macherel, M.H.; Grunwald, D.; Macherel, D. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005, 137, 157–167. [Google Scholar] [CrossRef]
- Evans, K.V.; Ransom, E.; Nayakoti, S.; Wilding, B.; Mohd Salleh, F.; Grzina, I.; Erber, L.; Tse, C.; Hill, C.; Polanski, K.; et al. Expression of the Arabidopsis redox-related LEA protein, SAG21 is regulated by ERF, NAC and WRKY transcription factors. Sci. Rep. 2024, 14, 7756. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B.C. Plant Small RNAs: Their Biogenesis, Regulatory Roles, and Functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Hull, G.; Guilleminot, J.; Devic, M.; Delseny, M. Differential expression of the Arabidopsis genes coding for Em-like proteins. J. Exp. Bot. 2000, 51, 1211–1220. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Z.; Cai, X.; Wu, D.; Gong, H.; Wang, J.; Zhang, Y.; Yao, Q.; Wang, L.; Liang, Y.; Zhang, Y.; et al. Genome-Wide Analysis of the LEA Gene Family in Pineapple (Ananas comosus L.) Reveals Its Potential Roles in Cold Stress Response and Reproductive Development. Biology 2025, 14, 1655. https://doi.org/10.3390/biology14121655
Hou Z, Cai X, Wu D, Gong H, Wang J, Zhang Y, Yao Q, Wang L, Liang Y, Zhang Y, et al. Genome-Wide Analysis of the LEA Gene Family in Pineapple (Ananas comosus L.) Reveals Its Potential Roles in Cold Stress Response and Reproductive Development. Biology. 2025; 14(12):1655. https://doi.org/10.3390/biology14121655
Chicago/Turabian StyleHou, Zhimin, Xinkai Cai, Denghang Wu, Haichao Gong, Jing Wang, Yinan Zhang, Qinglong Yao, Lulu Wang, Yuqin Liang, Yangmei Zhang, and et al. 2025. "Genome-Wide Analysis of the LEA Gene Family in Pineapple (Ananas comosus L.) Reveals Its Potential Roles in Cold Stress Response and Reproductive Development" Biology 14, no. 12: 1655. https://doi.org/10.3390/biology14121655
APA StyleHou, Z., Cai, X., Wu, D., Gong, H., Wang, J., Zhang, Y., Yao, Q., Wang, L., Liang, Y., Zhang, Y., Qin, Y., Wang, X., & Zheng, P. (2025). Genome-Wide Analysis of the LEA Gene Family in Pineapple (Ananas comosus L.) Reveals Its Potential Roles in Cold Stress Response and Reproductive Development. Biology, 14(12), 1655. https://doi.org/10.3390/biology14121655







