CPSF1 Is Co-Amplified with MYC but Is Independently Associated with Alternative Polyadenylation in Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis Cohorts
2.2. Somatic Mutation Analysis
2.3. Copy Number Variation Analysis
2.4. Correlation Between Expression and Mutation
2.5. Alternative Polyadenylation (APA) Analysis
2.6. Pathway Enrichment Analysis
2.7. Survival Analysis
3. Results
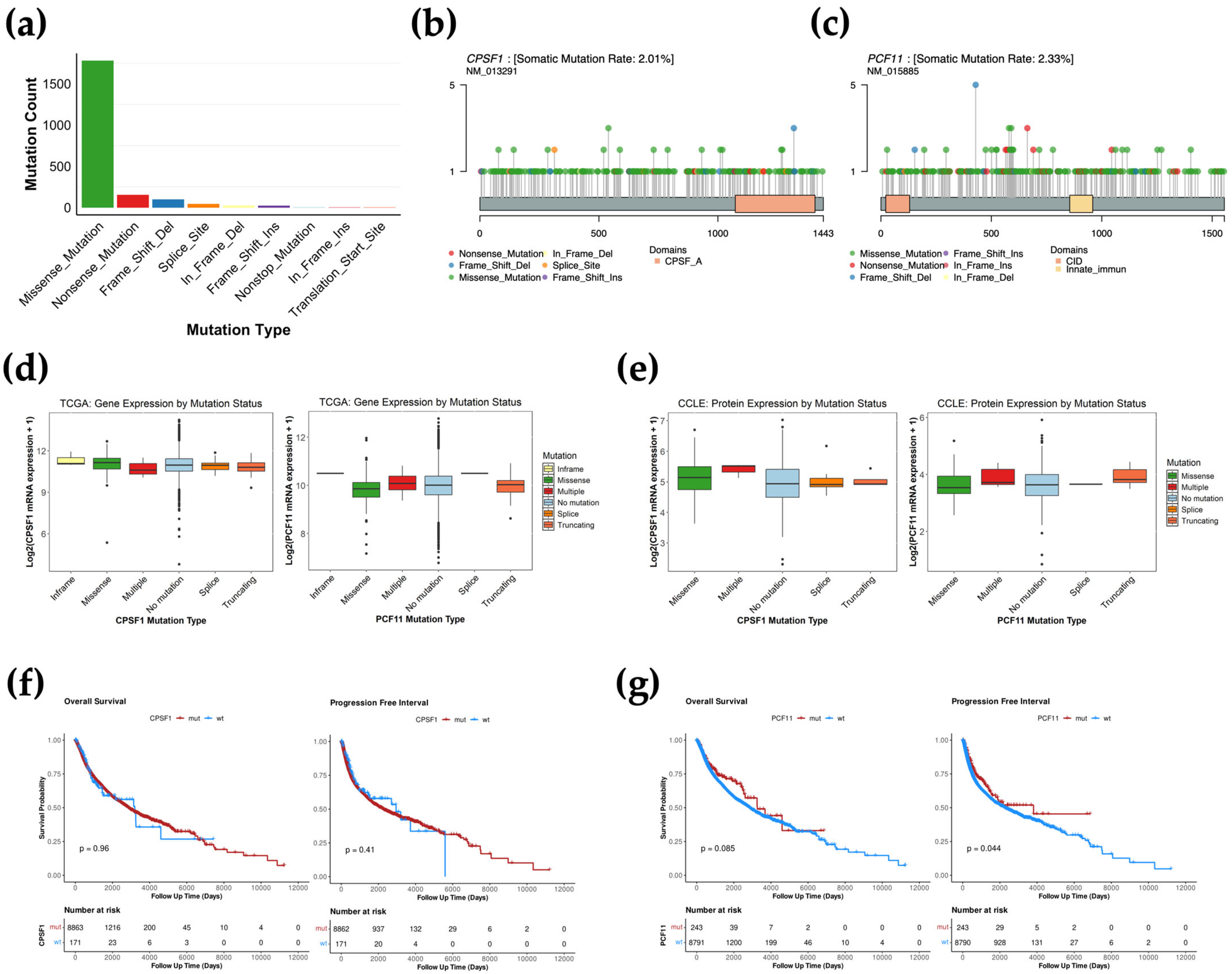
3.1. CPA Mutational Landscape Across TCGA Cancer Types
3.2. CPA Mutations Are Not Associated with Gene Expression Changes
3.3. CPSF1 Is the Most Amplified CPA Gene in Cancer
3.4. High CPSF1 Copy Number Is Associated with Poor Prognosis
3.5. Amplification of CPSF1 Co-Occurs with MYC Amplification but Is an Independent Prognostic Factor
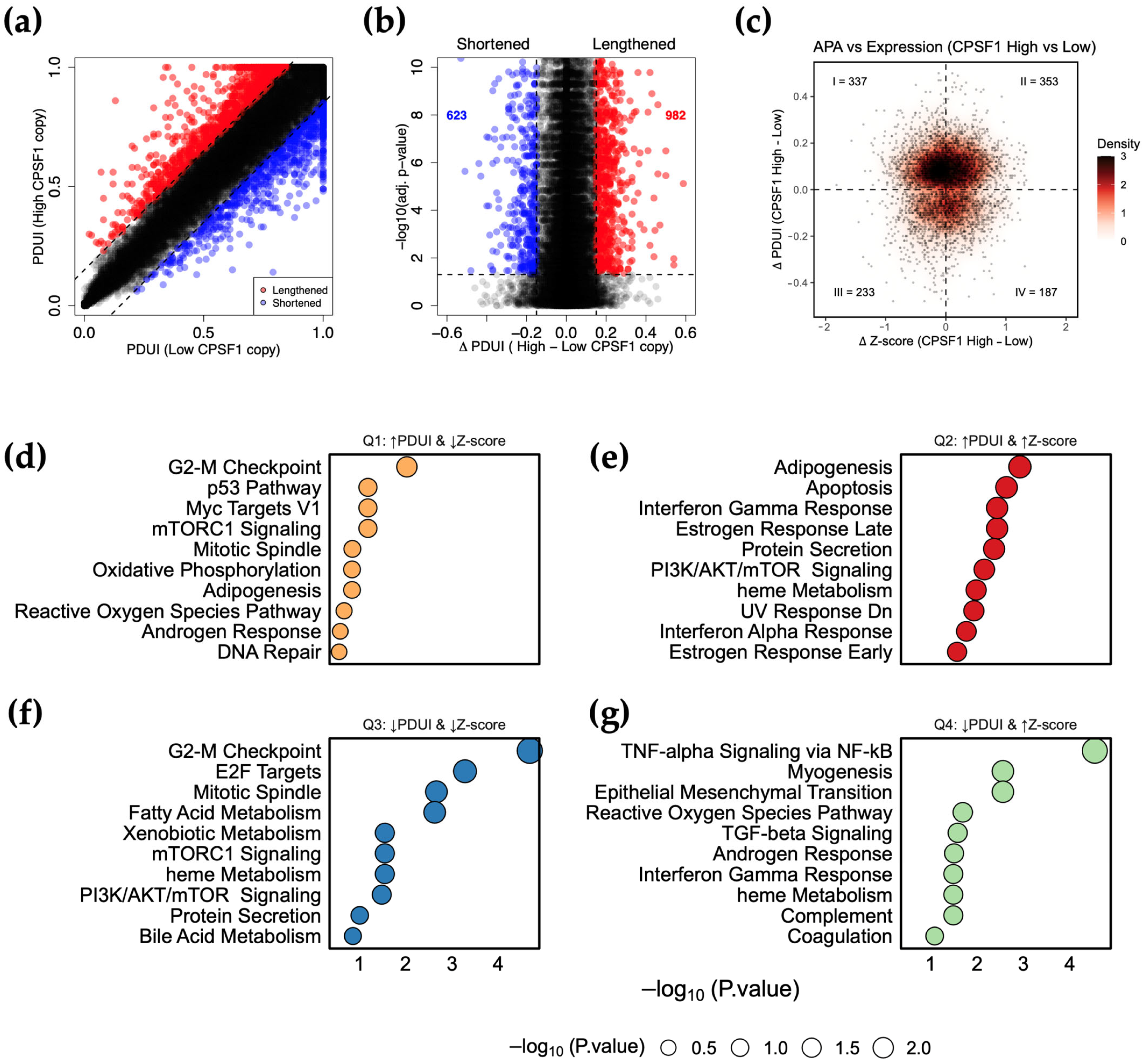
3.6. CPSF1 Amplification Is Associated with Alternative Polyadenylation Events in Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′UTR | 3′Untranslated Region |
| APA | Alternative polyadenylation |
| CCLE | Cancer Cell Line Encyclopedia |
| CFIIm | Mammalian cleavage factor II |
| CFIm | Mammalian cleavage factor I |
| CNV | Copy number variation |
| CPA | Cleavage and polyadenylation |
| CPSF | Cleavage and polyadenylation specificity factor |
| CSTF | Cleavage stimulation factor |
| MAF | Mutation Annotation Format |
| OS | Overall survival |
| PAS | Polyadenylation signal |
| PDUI | Percent Distal Usage Index |
| PFS | Progression-free survival |
| SNP | Single nucleotide polymorphism |
| TC3A | The Cancer 3′UTR Atlas |
| TCGA | The Cancer Genome Atlas |
References
- Boreikaitė, V.; Passmore, L.A. 3′-End Processing of Eukaryotic mRNA: Machinery, Regulation, and Impact on Gene Expression. Annu. Rev. Biochem. 2023, 92, 199–225. [Google Scholar] [CrossRef]
- Mitschka, S.; Mayr, C. Context-Specific Regulation and Function of mRNA Alternative Polyadenylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 779–796. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.J.; Zavolan, M. Alternative Cleavage and Polyadenylation in Health and Disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Hankey, W.; Wagner, E.J.; Li, W.; Wang, Q. Alternative Polyadenylation of mRNA and Its Role in Cancer. Genes Dis. 2021, 8, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Manley, J.L. Alternative Polyadenylation of mRNA Precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Xia, Z.; Donehower, L.A.; Cooper, T.A.; Neilson, J.R.; Wheeler, D.A.; Wagner, E.J.; Li, W. Dynamic Analyses of Alternative Polyadenylation from RNA-Seq Reveal a 3′-UTR Landscape across Seven Tumour Types. Nat. Commun. 2014, 5, 5274. [Google Scholar] [CrossRef]
- Mayr, C.; Bartel, D.P. Widespread Shortening of 3′UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef]
- Chen, S.-L.; Zhu, Z.-X.; Yang, X.; Liu, L.-L.; He, Y.-F.; Yang, M.-M.; Guan, X.-Y.; Wang, X.; Yun, J.-P. Cleavage and Polyadenylation Specific Factor 1 Promotes Tumor Progression Alternative Polyadenylation and Splicing in Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 616835. [Google Scholar] [CrossRef]
- Alahmari, A.A.; Chaubey, A.H.; Jonnakuti, V.S.; Tisdale, A.A.; Schwarz, C.D.; Cornwell, A.C.; Maraszek, K.E.; Paterson, E.J.; Kim, M.; Venkat, S.; et al. CPSF3 Inhibition Blocks Pancreatic Cancer Cell Proliferation through Disruption of Core Histone mRNA Processing. RNA 2024, 30, 281–297. [Google Scholar] [CrossRef]
- Masamha, C.P.; Xia, Z.; Yang, J.; Albrecht, T.R.; Li, M.; Shyu, A.-B.; Li, W.; Wagner, E.J. CFIm25 Links Alternative Polyadenylation to Glioblastoma Tumour Suppression. Nature 2014, 510, 412–416. [Google Scholar] [CrossRef]
- Ting, H.; Kim, K.J. Driving Glioblastoma Growth by Alternative Polyadenylation. Cell Res. 2014, 24, 1023–1024. [Google Scholar] [CrossRef][Green Version]
- Chu, Y.; Elrod, N.; Wang, C.; Li, L.; Chen, T.; Routh, A.; Xia, Z.; Li, W.; Wagner, E.J.; Ji, P. Nudt21 Regulates the Alternative Polyadenylation of Pak1 and Is Predictive in the Prognosis of Glioblastoma Patients. Oncogene 2019, 38, 4154–4168. [Google Scholar] [CrossRef]
- Kainov, Y.A.; Aushev, V.N.; Naumenko, S.A.; Tchevkina, E.M.; Bazykin, G.A. Complex Selection on Human Polyadenylation Signals Revealed by Polymorphism and Divergence Data. Genome Biol. Evol. 2016, 8, 1971–1979. [Google Scholar] [CrossRef]
- Kainov, Y.; Hamid, F.; Makeyev, E.V. Recurrent Disruption of Tumour Suppressor Genes in Cancer by Somatic Mutations in Cleavage and Polyadenylation Signals. eLife 2024, 13, 99040. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., III; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Sellers, W.R.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, M.C.; McDonald 3rd, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 Facilitates Sensitive and Confident Localization of the Targets of Focal Somatic Copy-Number Alteration in Human Cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Feng, X.; Li, L.; Wagner, E.J.; Li, W. TC3A: The Cancer 3′UTR Atlas. Nucleic Acids Res. 2018, 46, D1027–D1030. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and Collab-orative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer Analysis Project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The Landscape of Somatic Copy-Number Alteration across Human Cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-Cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018, 6, 282–300.e2. [Google Scholar] [CrossRef]
- Seldon, C.S.; Meiyappan, K.; Hoffman, H.; Guo, J.A.; Goel, N.; Hwang, W.L.; Nguyen, P.L.; Mahal, B.A.; Alshalalfa, M. Genomic Alterations Predictive of Poor Clinical Outcomes in Pan-Cancer. Oncotarget 2022, 13, 1069–1077. [Google Scholar] [CrossRef]
- Schaafsma, E.; Zhao, Y.; Zhang, L.; Li, Y.; Cheng, C. MYC Activity Inference Captures Diverse Mechanisms of Aberrant MYC Pathway Activation in Human Cancers. Mol. Cancer Res. 2021, 19, 414–428. [Google Scholar] [CrossRef]
- Luzzatto, L. Somatic Mutations in Cancer Development. Environ. Health 2011, 10 (Suppl. S1), S12. [Google Scholar] [CrossRef] [PubMed]
- Gutman, T.; Goren, G.; Efroni, O.; Tuller, T. Estimating the Predictive Power of Silent Mutations on Cancer Classification and Prognosis. npj Genom. Med. 2021, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Miladi, M.; Dukare, S.; Boulay, K.; Caudron-Herger, M.; Groß, M.; Backofen, R.; Diederichs, S. A Pan-Cancer Analysis of Synonymous Mutations. Nat. Commun. 2019, 10, 2569. [Google Scholar] [CrossRef] [PubMed]
- LaForce, G.R.; Farr, J.S.; Liu, J.; Akesson, C.; Gumus, E.; Pinkard, O.; Miranda, H.C.; Johnson, K.; Sweet, T.J.; Ji, P.; et al. Sup-pression of Premature Transcription Termination Leads to Reduced mRNA Isoform Diversity and Neurodegeneration. Neuron 2022, 110, 1340–1357.e7. [Google Scholar] [CrossRef]
- Ouyang, J.; Sun, W.; Xiao, X.; Li, S.; Jia, X.; Zhou, L.; Wang, P.; Zhang, Q. CPSF1 Mutations Are Associated with Early-Onset High Myopia and Involved in Retinal Ganglion Cell Axon Projection. Hum. Mol. Genet. 2019, 28, 1959–1970. [Google Scholar] [CrossRef]
- Arnadottir, G.A.; Oddsson, A.; Jensson, B.O.; Gisladottir, S.; Simon, M.T.; Arnthorsson, A.O.; Katrinardottir, H.; Fridriksdottir, R.; Ivarsdottir, E.V.; Jonasdottir, A.; et al. Population-Level Deficit of Homozygosity Unveils CPSF3 as an Intellectual Disability Syndrome Gene. Nat. Commun. 2022, 13, 705. [Google Scholar] [CrossRef]
- Shlien, A.; Malkin, D. Copy Number Variations and Cancer. Genome Med. 2009, 1, 62. [Google Scholar] [CrossRef]
- Steele, C.D.; Abbasi, A.; Islam, S.M.A.; Bowes, A.L.; Khandekar, A.; Haase, K.; Hames-Fathi, S.; Ajayi, D.; Verfaillie, A.; Dhami, P.; et al. Signatures of Copy Number Alterations in Human Cancer. Nature 2022, 606, 984–991. [Google Scholar] [CrossRef]
- Sakai, A.; Ando, M.; Fukusumi, T.; Ren, S.; Liu, C.; Qualliotine, J.; Haft, S.; Sadat, S.; Saito, Y.; Guo, T.W.; et al. Aberrant Expression of CPSF1 Promotes Head and Neck Squamous Cell Carcinoma via Regulating Alternative Splicing. PLoS ONE 2020, 15, e0233380. [Google Scholar] [CrossRef]
- Tietz, K.T.; McCluskey, B.M.; Miller, C.R.; Li, Y.; Munro, S.A.; Dehm, S.M. CPSF1 Inhibition Promotes Widespread Use of In-tergenic Polyadenylation Sites and Impairs Glycolysis in Prostate Cancer Cells. Cell Rep. 2025, 44, 115211. [Google Scholar] [CrossRef]
- Wang, L.; Lang, G.-T.; Xue, M.-Z.; Yang, L.; Chen, L.; Yao, L.; Li, X.-G.; Wang, P.; Hu, X.; Shao, Z.-M. Dissecting the Heterogeneity of the Alternative Polyadenylation Profiles in Triple-Negative Breast Cancers. Theranostics 2020, 10, 10531–10547. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Liu, W.; Guan, X.; Xie, X.; Zhang, Y. CPSF3 Is a Promising Prognostic Biomarker and Predicts Recurrence of Non-Small Cell Lung Cancer. Oncol. Lett. 2019, 18, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Y.; Liu, D.; Yang, L. Targeting Cleavage and Polyadenylation Specific Factor 1 via shRNA Inhibits Cell Proliferation in Human Ovarian Cancer. J. Biosci. 2017, 42, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, G.; Zhao, Z.; Li, D.; Song, Y.; Zhan, Q. Deregulated Expression and Subcellular Localization of CPSF6, a circR-NA-Binding Protein, Promote Malignant Development of Esophageal Squamous Cell Carcinoma. Chin. J. Cancer Res. Chung-Kuo Yen Cheng Yen Chiu 2022, 34, 11–27. [Google Scholar]
- Ge, Y.; Huang, J.; Chen, R.; Fu, Y.; Ling, T.; Ou, X.; Rong, X.; Cheng, Y.; Lin, Y.; Zhou, F.; et al. Downregulation of CPSF6 Leads to Global mRNA 3′UTR Shortening and Enhanced Antiviral Immune Responses. PLoS Pathog. 2024, 20, e1012061. [Google Scholar] [CrossRef]
- Binothman, N.; Hachim, I.Y.; Lebrun, J.-J.; Ali, S. CPSF6 Is a Clinically Relevant Breast Cancer Vulnerability Target: Role of CPSF6 in Breast Cancer. eBioMedicine 2017, 21, 65–78. [Google Scholar] [CrossRef]
- Tan, S.; Zhang, M.; Shi, X.; Ding, K.; Zhao, Q.; Guo, Q.; Wang, H.; Wu, Z.; Kang, Y.; Zhu, T.; et al. CPSF6 Links Alternative Polyadenylation to Metabolism Adaption in Hepatocellular Carcinoma Progression. J. Exp. Clin. Cancer Res. 2021, 40, 85. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-Cancer Patterns of Somatic Copy Number Alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- Choschzick, M.; Lassen, P.; Lebeau, A.; Marx, A.H.; Terracciano, L.; Heilenkötter, U.; Jaenicke, F.; Bokemeyer, C.; Izbicki, J.; Sauter, G.; et al. Amplification of 8q21 in Breast Cancer Is Independent of MYC and Associated with Poor Patient Outcome. Mod. Pathol. 2010, 23, 603–610. [Google Scholar] [CrossRef]
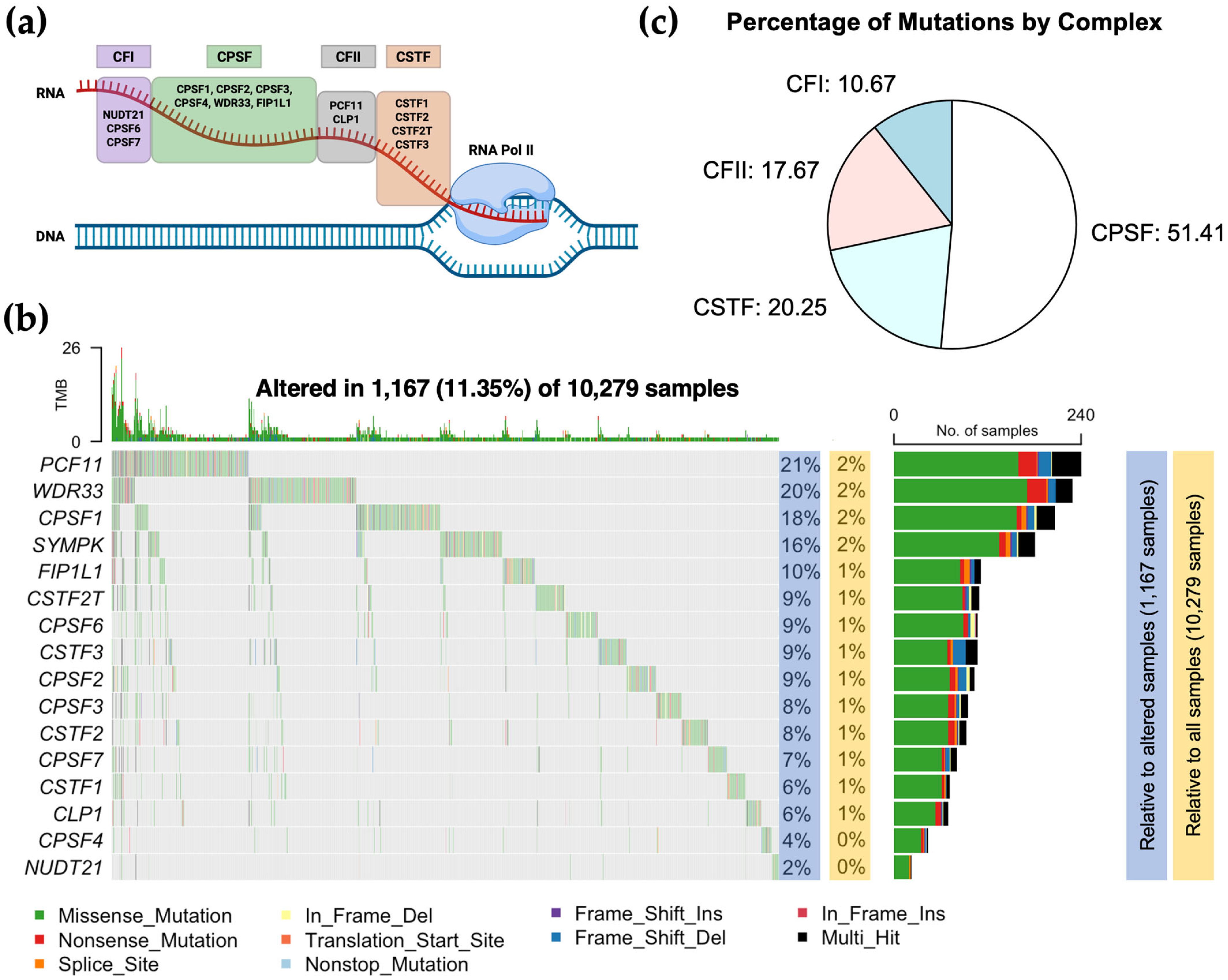
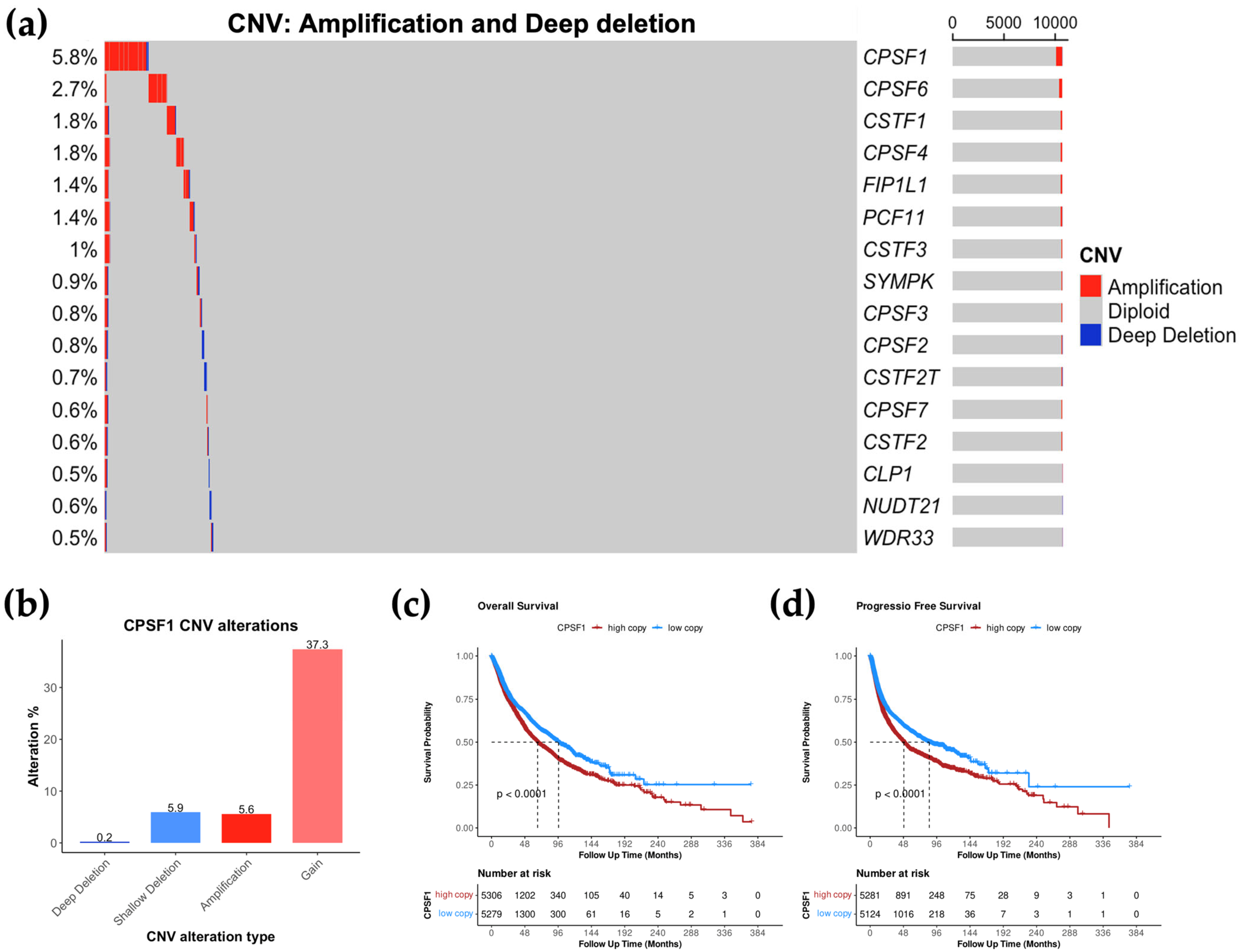
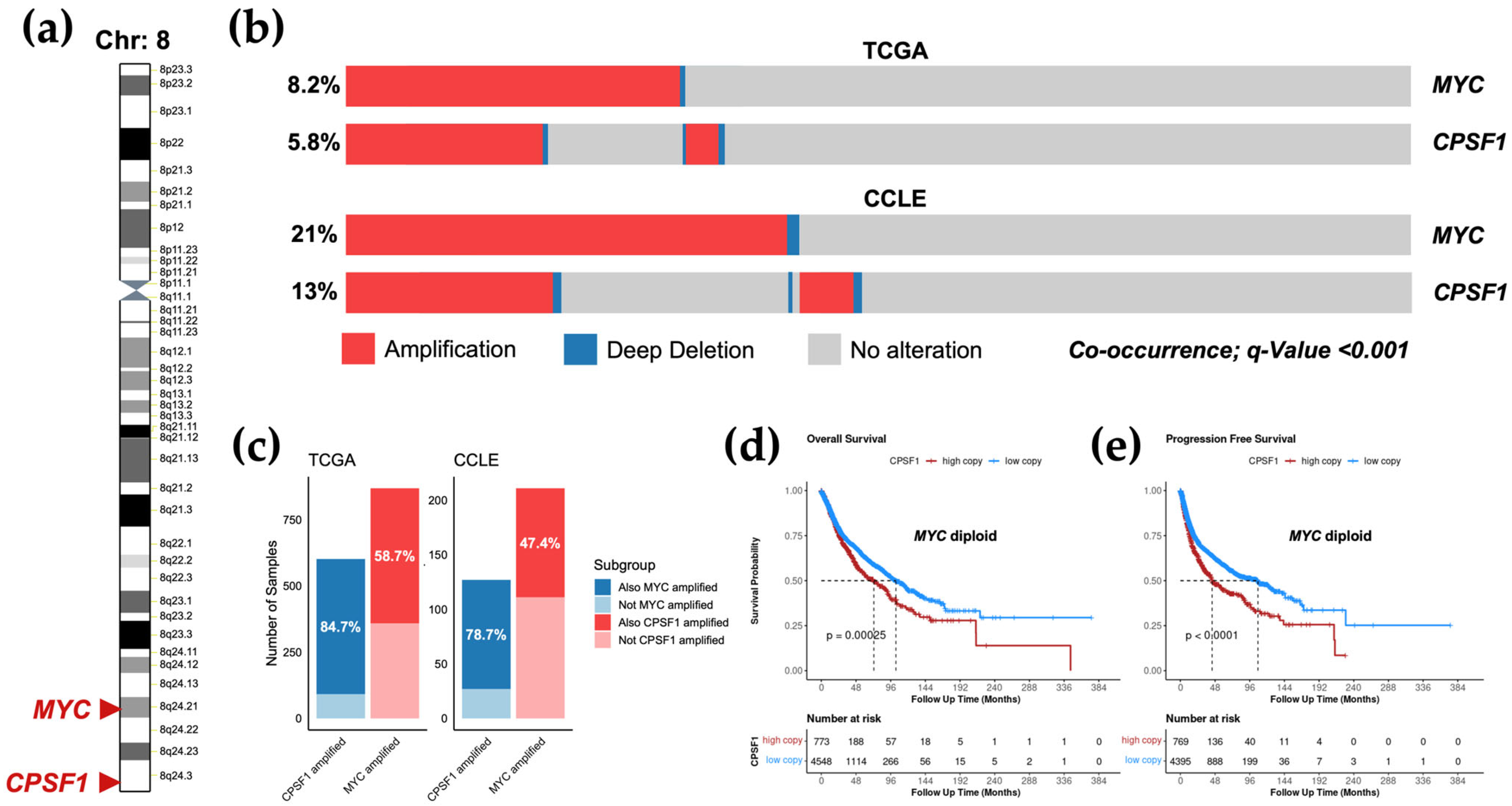
| Gene | Amplification | Gain | Diploid | Shallow Deletion | Deep Deletion |
|---|---|---|---|---|---|
| CPSF1 | 5.6 | 37.3 | 51 | 5.9 | 0.2 |
| CSTF1 | 1.8 | 36.8 | 58 | 3.4 | 0 |
| CPSF4 | 1.8 | 34.7 | 57.2 | 6.3 | 0 |
| CPSF6 | 2.7 | 18.5 | 69.6 | 9.2 | 0 |
| SYMPK | 0.7 | 17.2 | 64.2 | 17.7 | 0.2 |
| CPSF3 | 0.7 | 17.8 | 72.2 | 9.1 | 0.1 |
| NUDT21 | 0.2 | 12.7 | 60.8 | 25.9 | 0.4 |
| CPSF2 | 0.4 | 12.3 | 62.8 | 24.1 | 0.4 |
| CPSF7 | 0.5 | 12.7 | 71.7 | 15 | 0.1 |
| CSTF3 | 0.9 | 10.7 | 69.7 | 18.6 | 0.1 |
| CSTF2T | 0.3 | 9.3 | 65.5 | 24.5 | 0.4 |
| FIP1L1 | 1.3 | 9.5 | 69 | 20.2 | 0.1 |
| CLP1 | 0.3 | 12.3 | 72.2 | 15 | 0.1 |
| WDR33 | 0.3 | 13.6 | 75.7 | 10.2 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alahmari, A.A. CPSF1 Is Co-Amplified with MYC but Is Independently Associated with Alternative Polyadenylation in Cancer. Biology 2025, 14, 1637. https://doi.org/10.3390/biology14121637
Alahmari AA. CPSF1 Is Co-Amplified with MYC but Is Independently Associated with Alternative Polyadenylation in Cancer. Biology. 2025; 14(12):1637. https://doi.org/10.3390/biology14121637
Chicago/Turabian StyleAlahmari, Abdulrahman A. 2025. "CPSF1 Is Co-Amplified with MYC but Is Independently Associated with Alternative Polyadenylation in Cancer" Biology 14, no. 12: 1637. https://doi.org/10.3390/biology14121637
APA StyleAlahmari, A. A. (2025). CPSF1 Is Co-Amplified with MYC but Is Independently Associated with Alternative Polyadenylation in Cancer. Biology, 14(12), 1637. https://doi.org/10.3390/biology14121637






