Multi-Omics Analysis Reveals Biaxial Regulatory Mechanisms of Cardiac Adaptation by Specialized Racing Training in Yili Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Grouping
2.2. Blood Sample Collection and Preparation
2.3. Echocardiographic Analysis
2.4. Lipids Assessment and Analysis
2.4.1. Data Collection and Processing
2.4.2. Targeted Metabolomics Analysis
2.4.3. Metabolomics Data Analysis
2.5. Transcriptome Sequencing
2.5.1. RNA Extraction and Quality Assessment
2.5.2. mRNA Library Preparation and Sequencing
2.5.3. Quality Control and Data Comparison of mRNA Transcripts
2.5.4. Functional Annotation and Pathway Analysis of Differentially Expressed mRNAs
2.6. miRNA Sequencing
2.6.1. miRNA Isolation, cDNA Library Construction, and Sequencing Identification
2.6.2. Differential Expression Analysis and Target Gene Prediction of miRNAs
2.6.3. Correlation Analysis of Differentially Expressed miRNA-mRNA Targets
2.6.4. Weighted Gene Co-Expression Network Analysis
2.7. RT-qPCR Validation of mRNA and miRNA
2.8. Statistical Analysis
3. Results
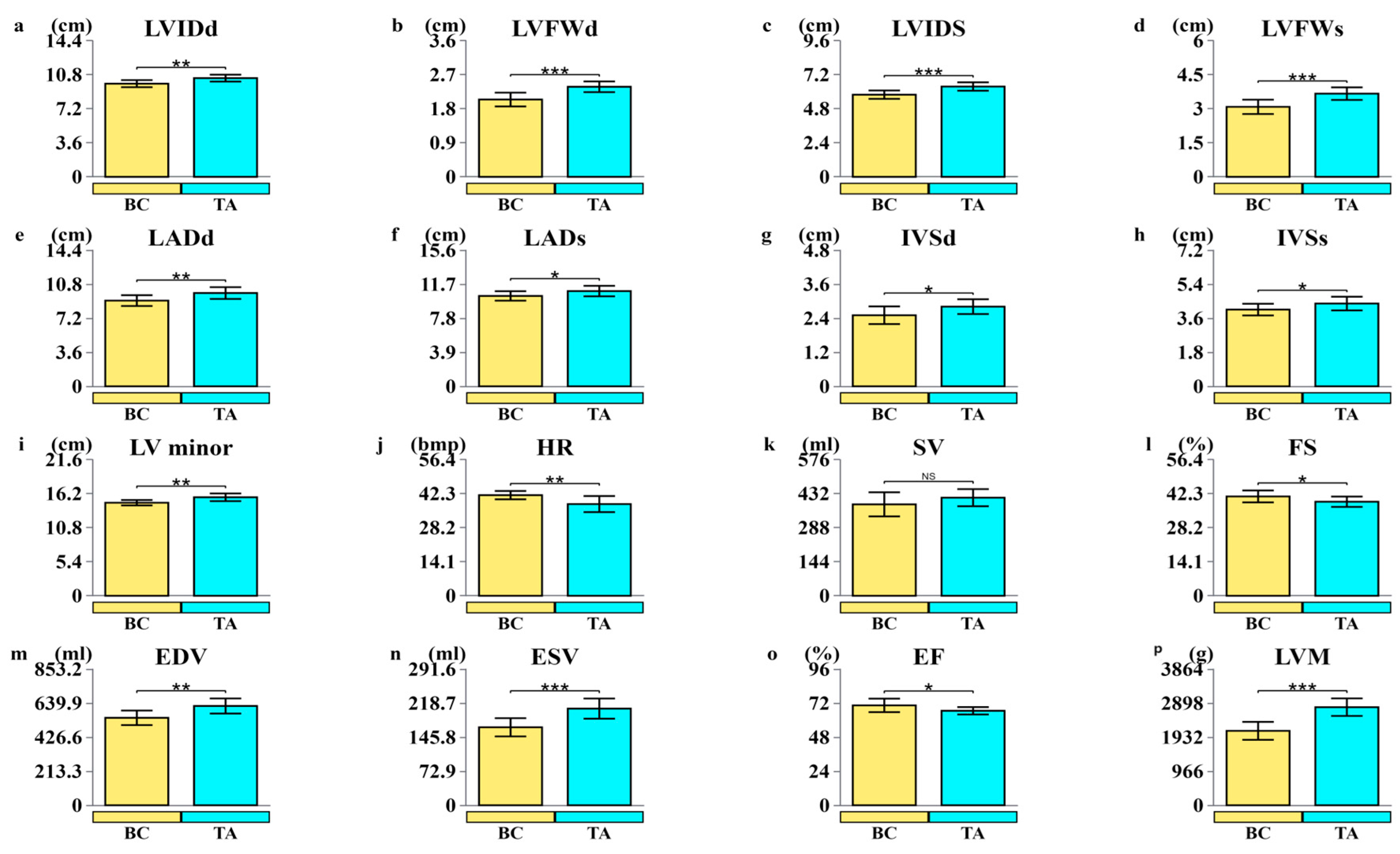
3.1. Structural and Functional Indicators of the Left Ventricle in Yili Horses
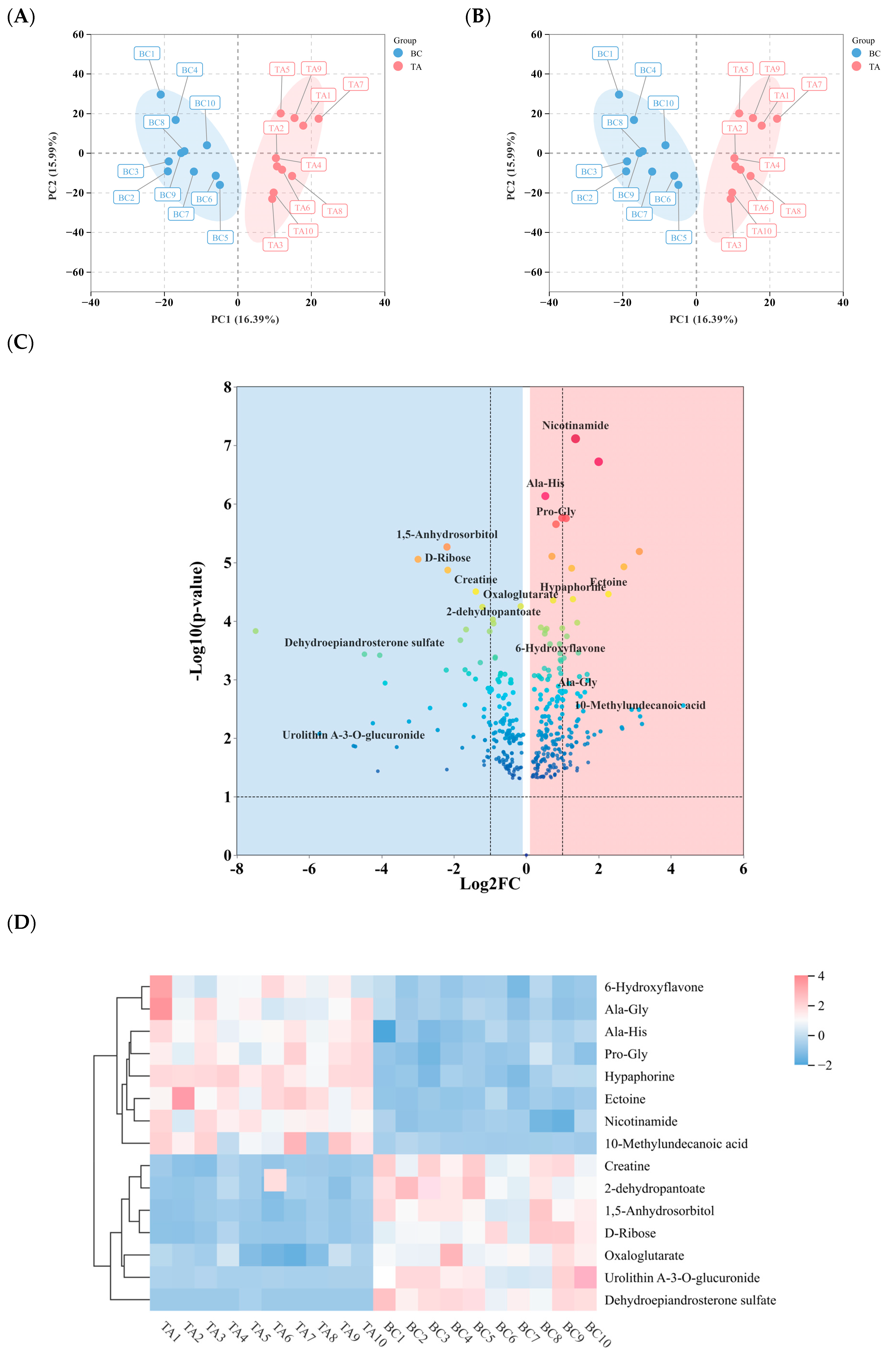
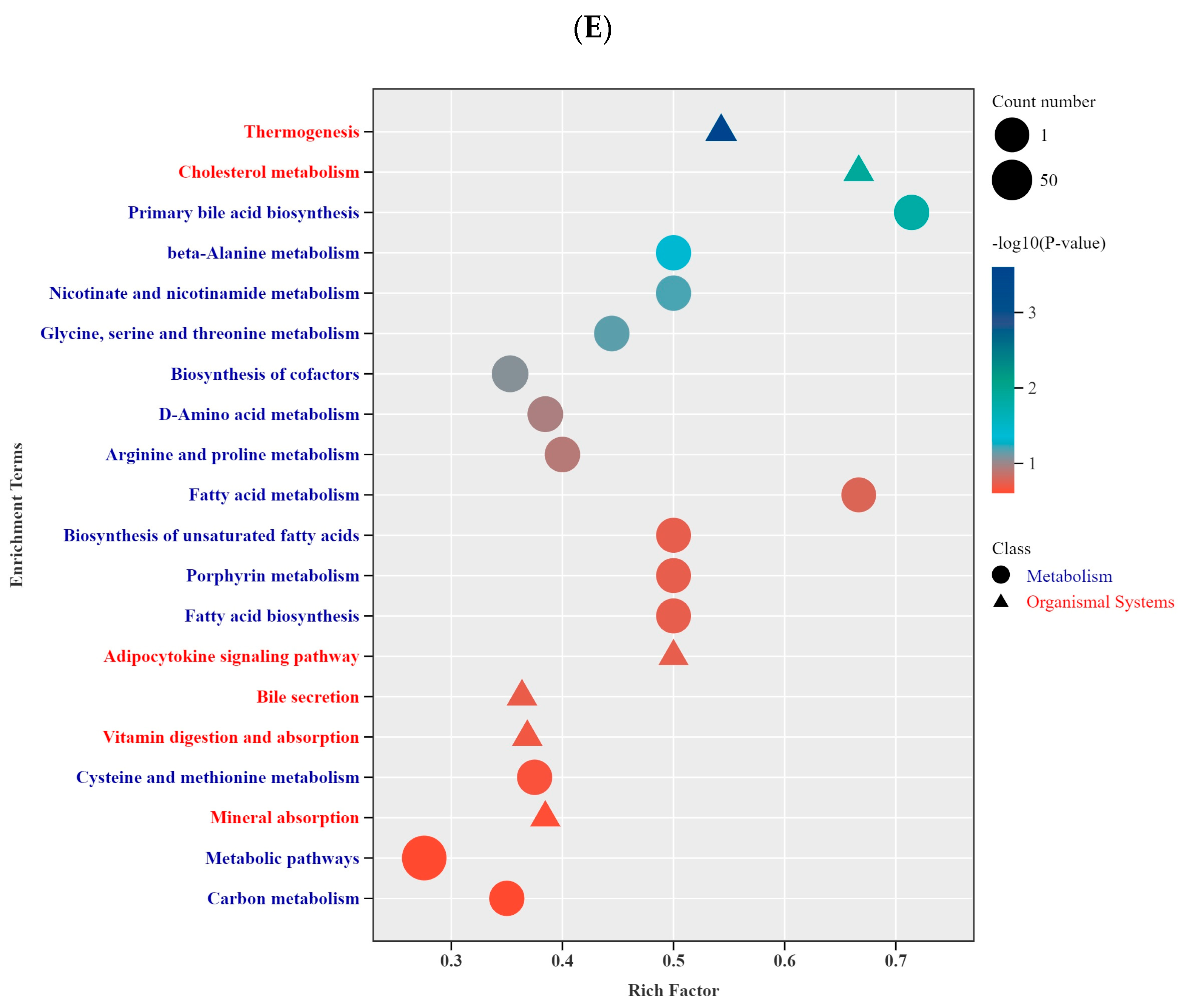
3.2. Metabolomic Differences Before and After Specialized Racing Training
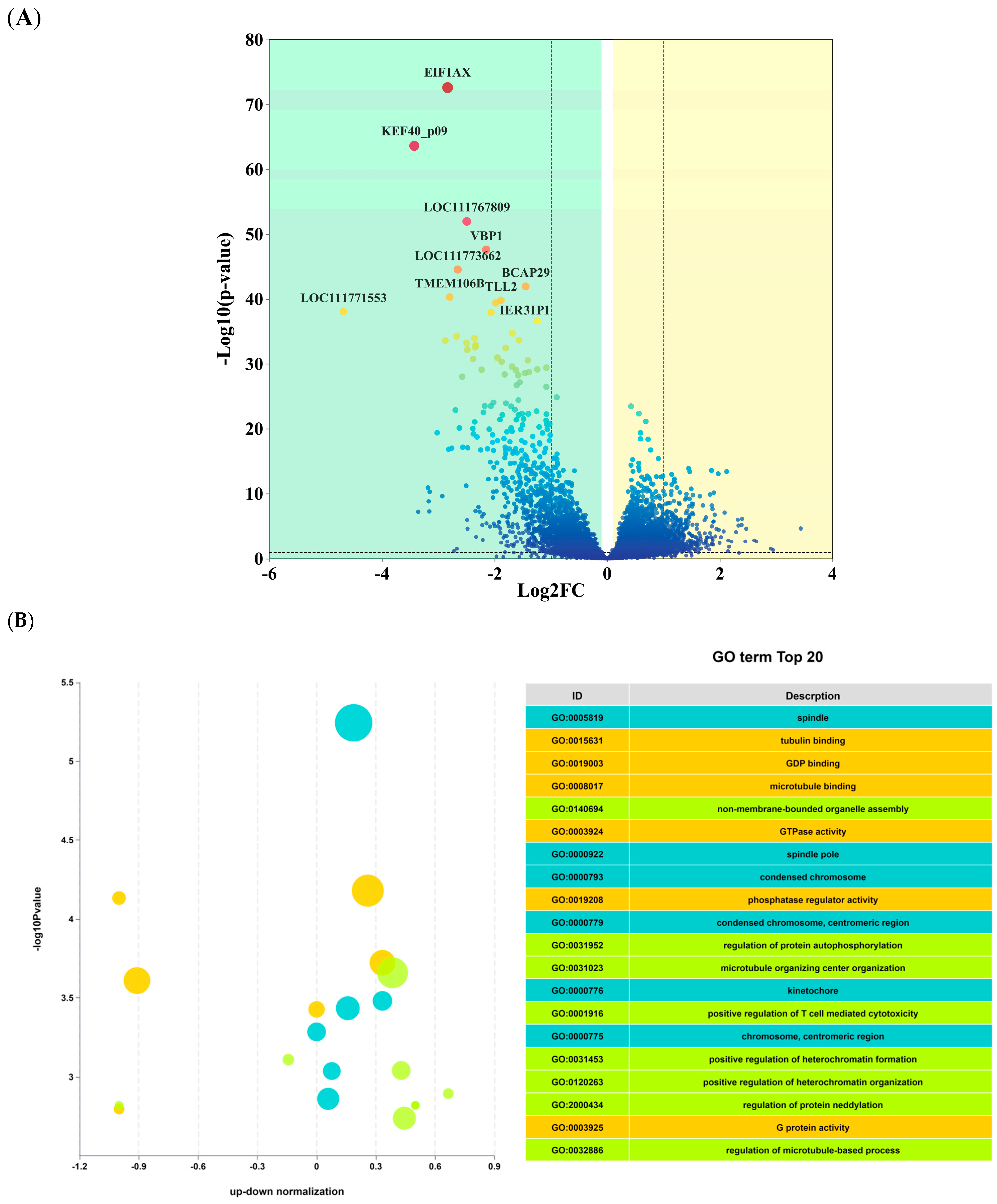
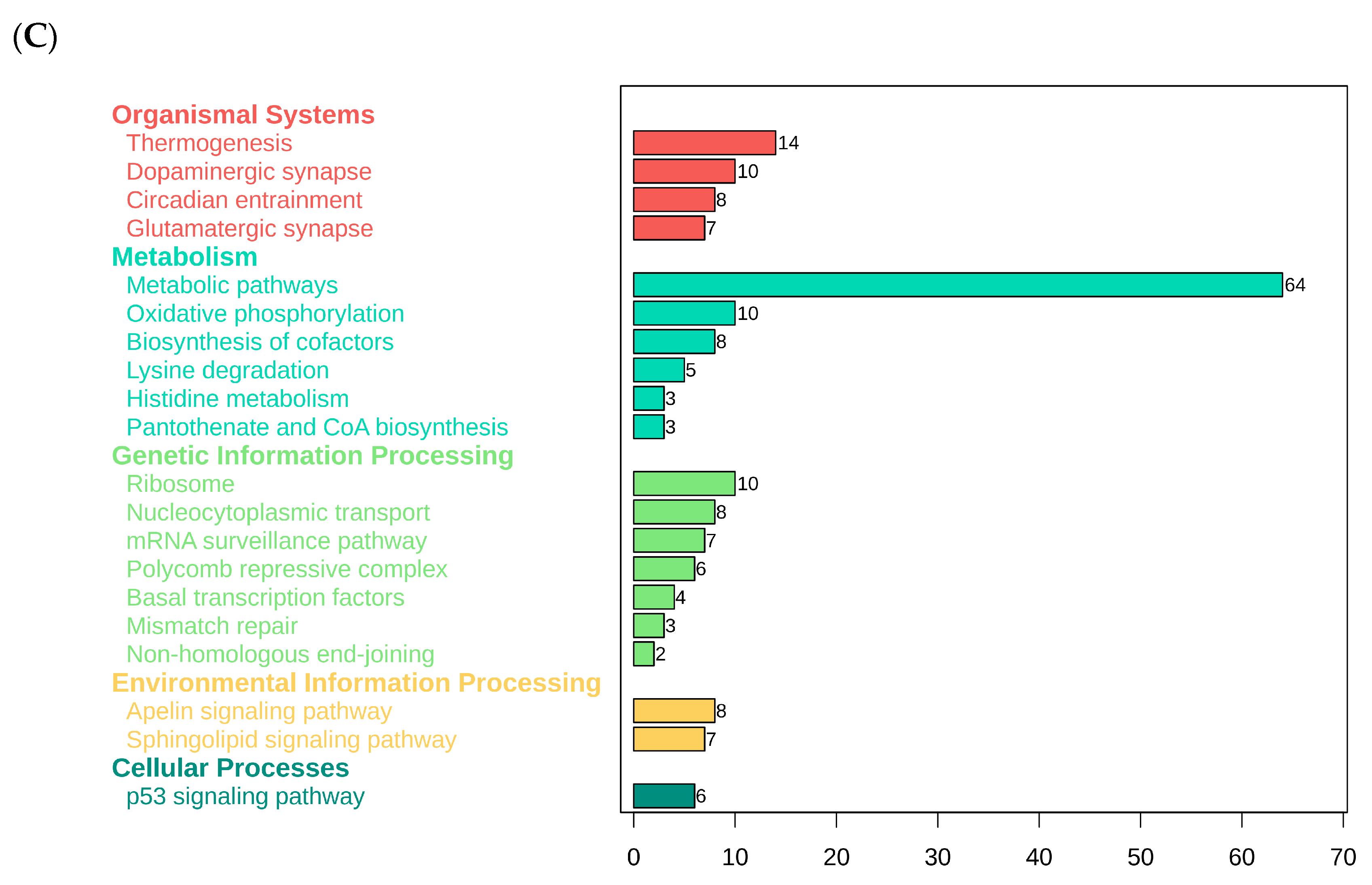
3.3. Functional Classification and Annotation of Differentially Expressed mRNAs
Functional Classification and Annotation of Differentially Expressed Genes
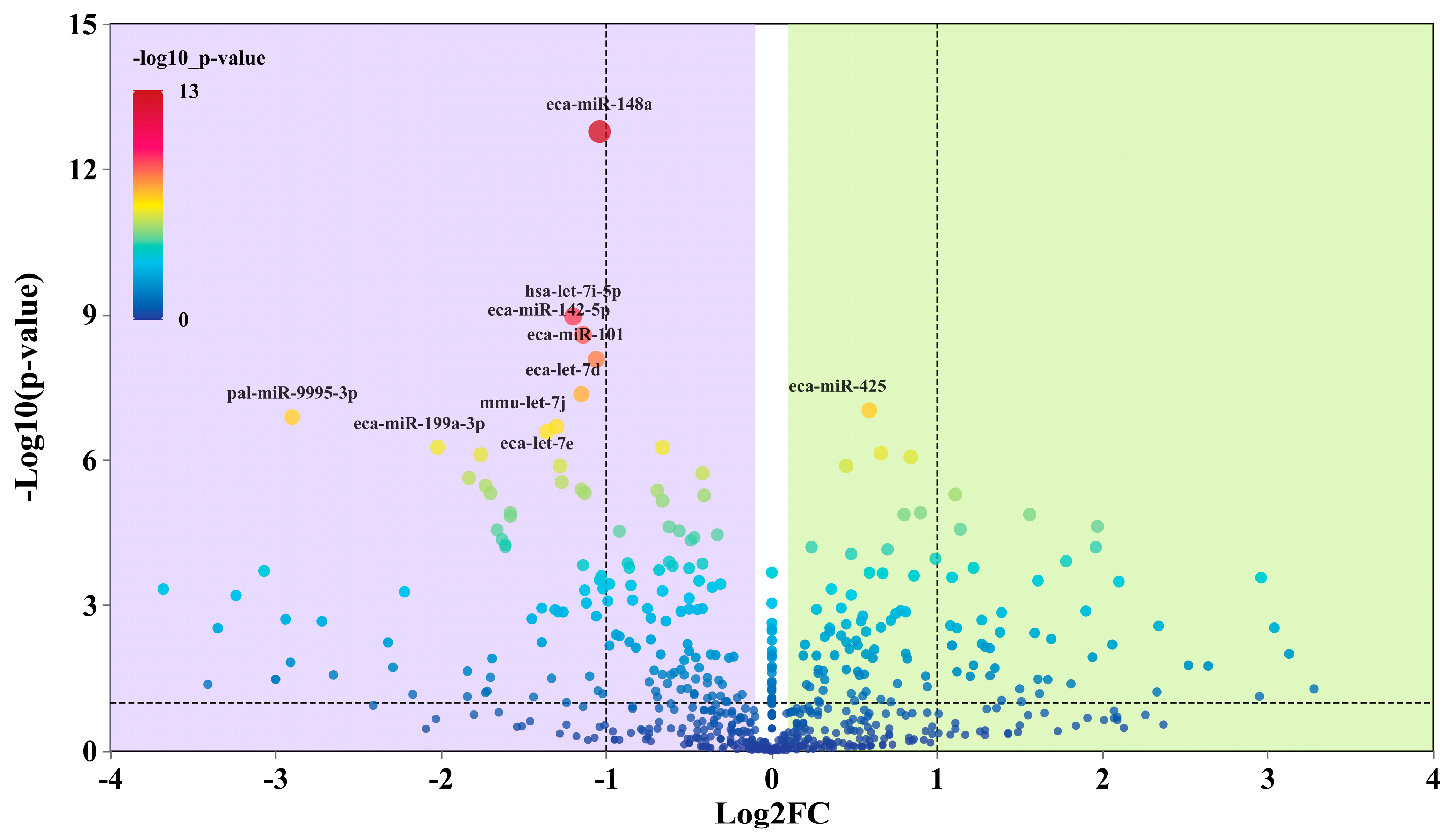
3.4. Differential miRNA Sequencing and Target Gene Functional Analysis
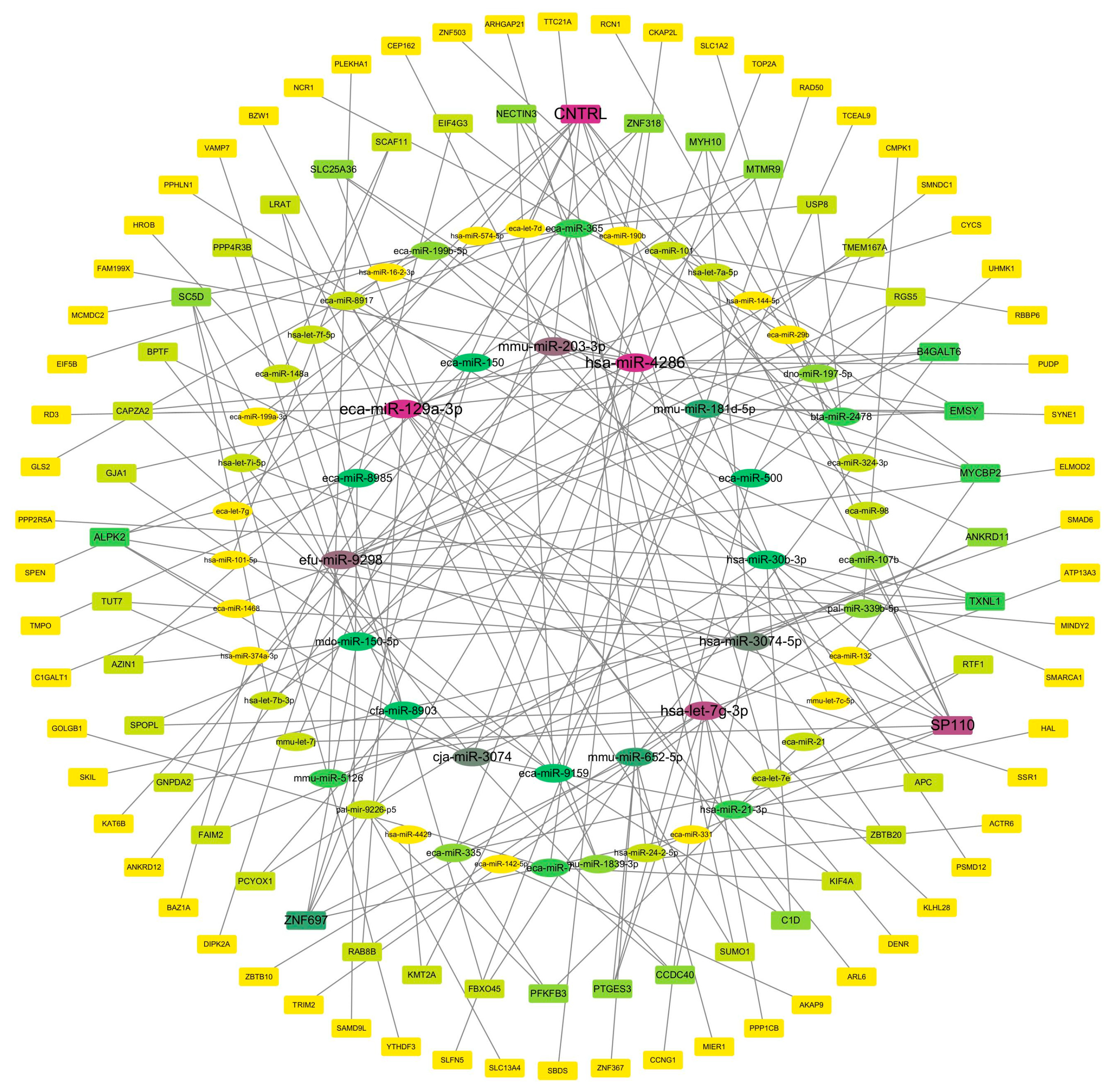
3.5. Integrated miRNA–mRNA Regulatory Network Reveals Upstream Regulatory Factors
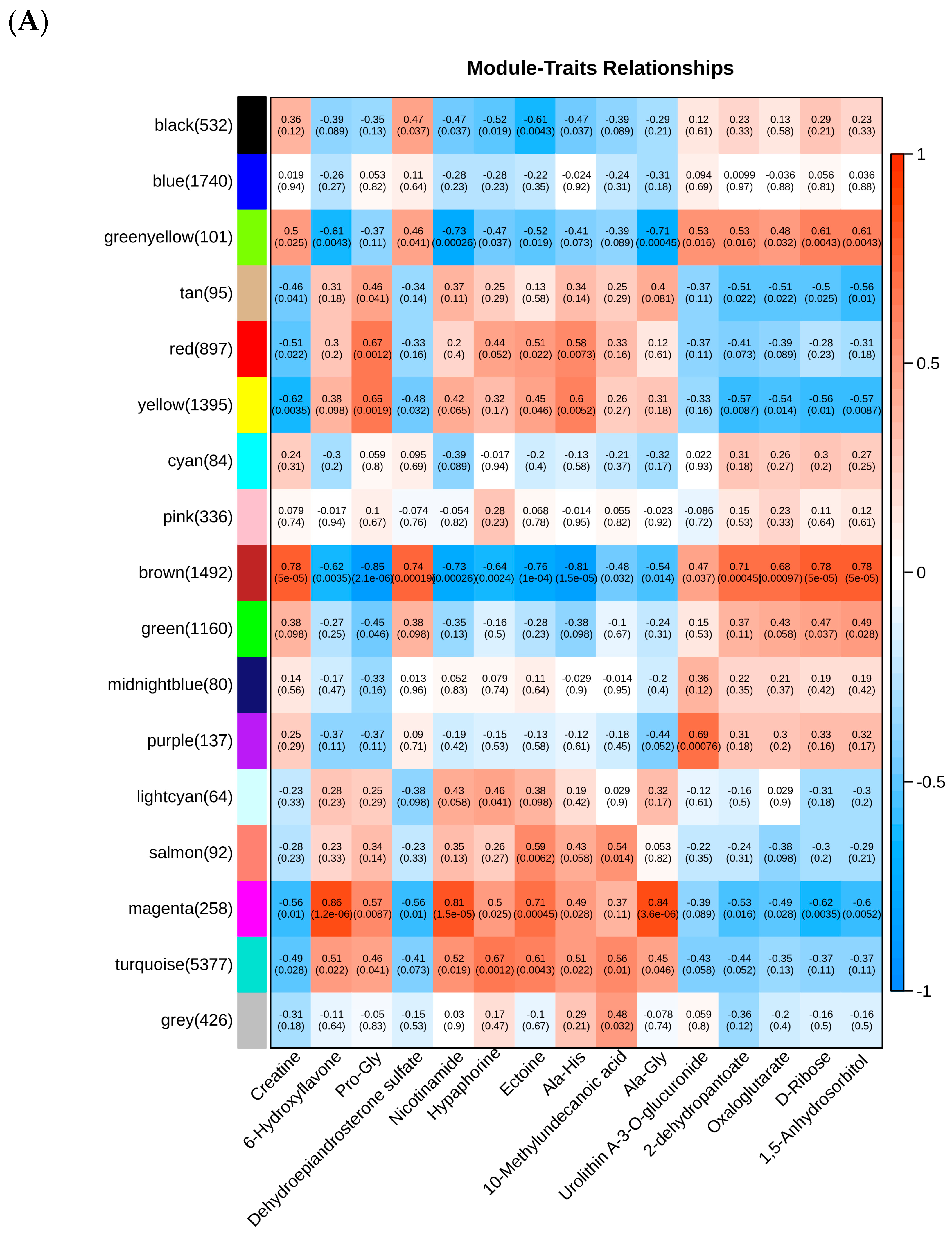
3.6. WGCNA Identification of Hub Genes Associated with Core Lipids
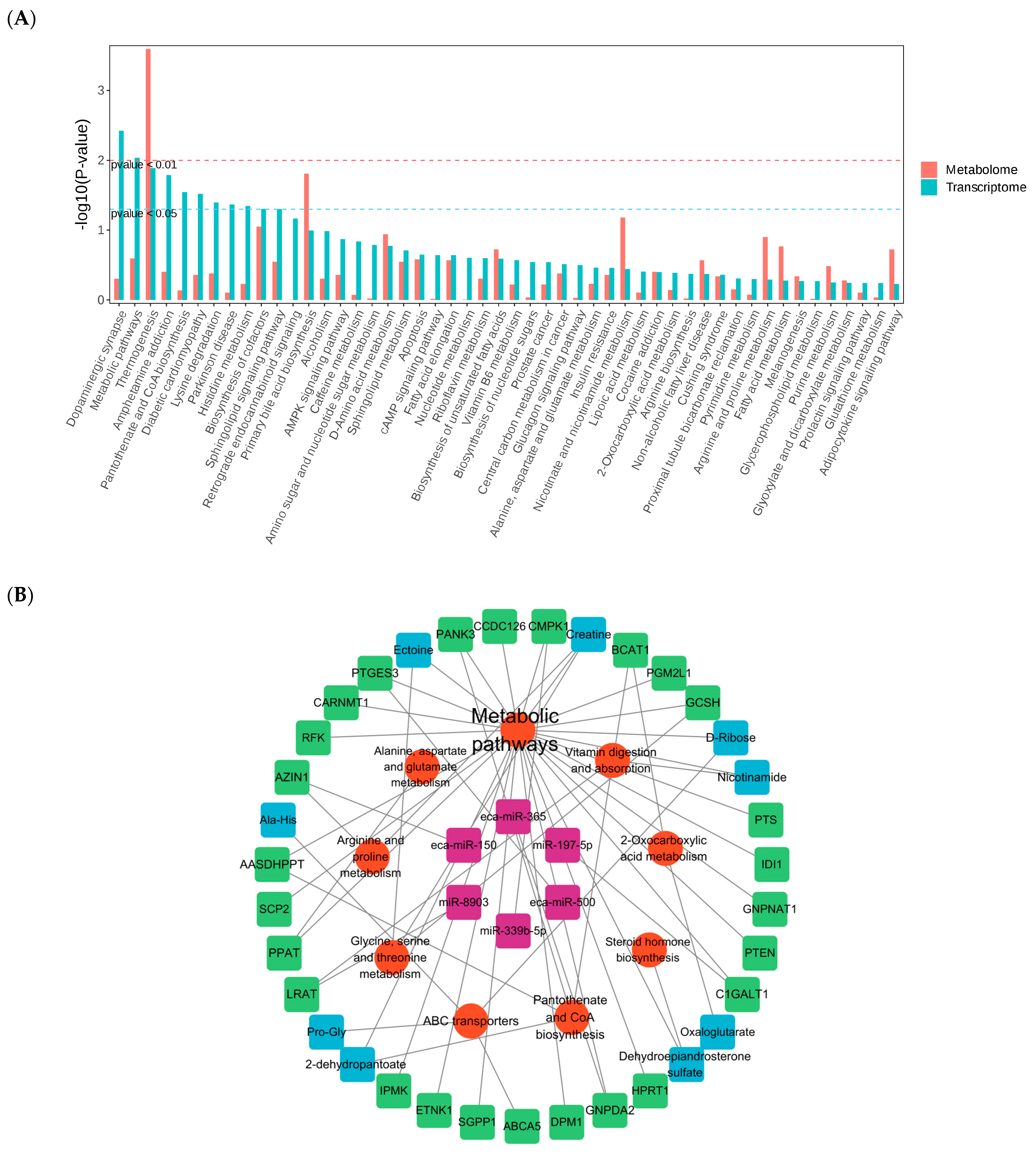
3.7. Multi-Omics Integrated Analysis to Construct a Synergistic Regulation Model for Training Adaptation
4. Discussion
4.1. Structural and Functional Analysis of the Yili Horse Heart
4.2. Metabolomic Differences in BC and TA Under Specialized Racing Training Conditions
4.3. Combined miRNA-mRNA Analysis of BC and TA Under Specific Training Conditions
4.4. Combined Transcriptome-Lipidome Analysis of BC and TA Under Specific Training Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HR | Heart rate |
| EF | Ejection fraction |
| FS | Fractional shortening |
| LVIDd | End-diastolic left ventricular diameter |
| LVIDs | End-systolic left ventricular diameter |
| IVSd | End-diastolic interventricular septal thickness |
| IVSs | End-systolic interventricular septal thickness |
| LVM | Left ventricular myocardial mass |
| LVFWd | End-diastolic left ventricular free wall thickness |
| LVFWs | End-systolic left ventricular free wall thickness |
| LVLD | Left ventricle long axis diameter |
| LADd | End-diastolic left atrial diameter |
| LADs | End-systolic left atrial diameter |
| EDV | End-diastolic left ventricular volume |
| ESV | End-systolic left ventricular volume |
| SV | Stroke volume |
References
- Chang, X.; Zhang, Z.; Yao, X.; Meng, J.; Ren, W.; Zeng, Y. Lipidomics and biochemical profiling of adult Yili horses in a 26 km endurance race: Exploring metabolic adaptations. Front. Vet. Sci. 2025, 12, 159739. [Google Scholar] [CrossRef]
- Le Moyec, L.; Robert, C.; Triba, M.N.; Billat, V.L.; Mata, X.; Schibler, L.; Barrey, E. Protein catabolism and high lipid metabolism associated with long distance exercise are revealed by plasma NMR metabolomics in endurance horses. PLoS ONE 2014, 9, e90730. [Google Scholar] [CrossRef]
- Luck, M.M.; Le Moyec, L.; Barrey, E.; Triba, M.N.; Bouchemal, N.; Savarin, P.; Robert, C. Energetics of endurance exercise in young horses determined by nuclear magnetic resonance metabolomics. Front. Physiol. 2015, 6, 198. [Google Scholar] [CrossRef]
- Tonevitsky, A.G.; Maltseva, D.V.; Abbasi, A.; Samatov, T.R.; Sakharov, D.A.; Shkurnikov, M.U.; Lebedev, A.E.; Galatenko, V.V.; Grigoriev, A.I.; Northoff, H. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 2013, 13, 9. [Google Scholar] [CrossRef]
- Safdar, A.; Abadi, A.; Akhtar, M.; Hettinga, B.P.; Tarnopolsky, M.A. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE 2017, 4, e5610. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Güller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef]
- Yan, Z.; Okutsu, M.; Akhtar, Y.N.; Lira, V.A. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J. Appl. Physiol. 2011, 110, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liang, X.; Zhou, D.; Lai, L.; Xiao, L.; Liu, L.; Fu, T.; Kong, Y.; Zhou, Q.; Vega, R.B.; et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 2016, 8, 1212–1228. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, K.; Capomaccio, S.; Viglino, A.; Silvestrelli, M.; Beccati, F.; Moscati, L.; Chiaradia, E. Circulating miRNAs as putative biomarkers of exercise adaptation in endurance horses. Front. Physiol. 2018, 9, 429. [Google Scholar] [CrossRef]
- McGivney, B.A.; McGettigan, P.A.; Browne, J.A.; Evans, A.C.; Fonseca, R.G.; Loftus, B.J.; Lohan, A.; MacHugh, D.E.; Murphy, B.A.; Katz, L.M.; et al. Characterization of the equine skeletal muscle transcriptome identifies novel functional responses to exercise training. BMC Genom. 2010, 11, 398. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Z˙ ukowski, K.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol. Genom. 2017, 49, 318–326. [Google Scholar] [CrossRef]
- Wang, T.; Yang, X.; Meng, J. Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses. Biology 2025, 14, 1535. [Google Scholar] [CrossRef]
- Bou, T.; Ding, W.; Liu, H.; Gong, W.; Jia, Z.; Dugarjaviin, M.; Bai, D. A genome-wide landscape of mRNAs, miRNAs, lncRNAs, and circRNAs of skeletal muscles during dietary restriction in Mongolian horses. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 46, 101084. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Meng, J.; Yang, X.; Zeng, Y.; Yao, X.; Ren, W. Differential Metabolomics and Cardiac Function in Trained vs. Untrained Yili Performance Horses. Animals 2025, 15, 2444. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar]
- Guo, X.; Ning, J.; Chen, Y.; Liu, G.; Zhao, L.; Fan, Y.; Sun, S. Recent advances in differential expression analysis for single-cell RNA-seq and spatially resolved transcriptomic studies. Brief. Funct. Genom. 2024, 23, 95–109. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Irani, F.-B.A.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The miRNA. org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Szyf, M. The genome-and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can. J. Psychiatry 2013, 58, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Meng, J.; Peng, X.; Huang, J.; Huang, Y.; Yan, X.; Li, X.; Yang, X.; Chang, X.; Zeng, Y.; et al. Metabolomics analysis and mRNA/miRNA profiling reveal potential cardiac regulatory mechanisms in Yili racehorses under different training regimens. PLoS ONE 2025, 20, e0322468. [Google Scholar] [CrossRef]
- Contrepois, K.; Wu, S.; Moneghetti, K.J.; Hornburg, D.; Ahadi, S.; Tsai, M.S.; Metwally, A.A.; Wei, E.; Lee-McMullen, B.; Quijada, J.V.; et al. Molecular Choreography of Acute Exercise. Cell 2020, 181, 1112–1130. [Google Scholar] [CrossRef]
- Nie, Y.; Sato, Y.; Garner, R.T.; Kargl, C.; Kaung, S.; Gilpin, C.; Gavin, T.P. Skeletal muscle-derived exosomes regulate endothelial cell functions via reactive oxygen species-activated nuclear factor-κB signaling. Exp. Physiol. 2019, 104, 1262–1273. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Tanihata, J.; Ishiyama, A.; OYA, Y.; Rüegg, U.; Takeda, S.; Hashido, K. Characterization and Functional Analysis of Extracellular Vesicles and Muscle-Abundant miRNAs (miR-1, miR-133a, and miR-206) in C2C12 Myocytes and mdx Mice. PLoS ONE 2017, 12, e0167811. [Google Scholar] [CrossRef]
- Sohel, H.M. Extracellular/Circulating MicroRNAs: Rele.ase Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Kis, J.; Rózsa, L.; Husvéth, F.; Zsolnai, A.; Anton, I. Role of genes related to performance and reproduction of Thoroughbreds in training and breeding—A review. Acta Vet. Hung. 2021, 69, 315–323. [Google Scholar]
- Nissen, S.D.; Weis, R.; Krag-Andersen, E.K.; Hesselkilde, E.M.; Isaksen, J.L.; Carstensen, H.; Linz, D.; Sanders, P.; Hopster-Iversen, C.; Jespersen, T. Electrocardiographic characteristics of trained and untrained standardbred racehorses. J. Vet. Intern. Med. 2022, 36, 1119–1130. [Google Scholar] [CrossRef]
- Mosteoru, S.; Gaiţă, L.; Gaiţă, D. Sport as Medicine for Dyslipidemia (and Other Risk Factors). Curr. Atheroscler. Rep. 2023, 25, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.C.; Candow, D.G.; Smith - Ryan, A.E.; Hirsch, K.R.; Roberts, M.D.; VanDusseldorp, T.A.; Stratton, M.T.; Kiani, M.; Little, J.P. Supplements and Nutritional Interventions to Augment High-Intensity Interval Training Physiological and Performance Adaptations—A Narrative Review. Nutrients 2020, 12, 390. [Google Scholar] [CrossRef]
- Hao, X.; Zha, C.; Shao, F.; Wang, L.; Tan, B. Amino acids regulate energy utilization through the mammalian target of rapamycin complex 1 and adenosine monophosphate-activated protein -kinase pathway in porcine enterocytes. Anim. Nutr. 2020, 6, 98–106. [Google Scholar]
- Moukette, B.; Kawaguchi, S.; Sepulveda, M.; Hayasaka, T.; Aonuma, T.; Liangpunsakul, S.; Yang, L.; Dharmakumar, R.; Conway, S.; Kim, I. MiR-150 blunts cardiac dysfunction in mice with cardiomyocyte loss of β1-adrenergic receptor/β-arrestin signaling and controls a unique transcriptome. Cell Death Discov. 2022, 8, 504. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, Y.; Huang, S.; Chen, Y.; Chen, X.; Li, M.; Si, X.; He, X.; Zheng, H.; Zhong, L.; et al. Inhibition of AZIN2-sv induces neovascularization and improves prognosis after myocardial infarction by blocking ubiquitin-dependent talin1 degradation and activating the Akt pathway. EBioMedicine 2019, 39, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.S.; Yang, B.G.; Zhang, X.Y.; Wang, Z.Y.; Li, S.; Kang, W.G. Role and Mechanism of miR-199b-5p in Myocardial Fibrosis and Endoplasmic Reticulum Stress in Rats with Myocardial Infarction. Lab. Med. Clin. 2025, 22, 914–919. [Google Scholar]
- Wang, J.; Yang, X. Functions and Mechanisms of Non-Coding RNAs in Maintaining Cardiac Homeostasis. Life Sci. 2018, 30, 1193–1201. [Google Scholar]
- Sawako, S.; Divya, V.; Tomoaki, T.; Prives, C. GLS2 shapes ferroptosis in hepatocellular carcinoma. Oncotarget 2023, 14, 14900–14903. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, H.; Yang, T.; Li, Y.; Gao, C.; Jiao, T.; Cai, Y.; Zhao, S. The expression regulatory network in the lung tissue of Tibetan pigs provides insight into hypoxia-sensitive pathways in high-altitude hypoxia. Front. Genet. 2021, 12, 691592. [Google Scholar] [CrossRef]
- Liang, X.; Chen, M.; Wang, D.; Wen, J.; Chen, J. Vitamin A deficiency indicating as low expression of LRAT may be a novel biomarker of primary hypertension. Clin. Exp. Hypertens. 2021, 43, 151–163. [Google Scholar] [CrossRef]
- Sabater-Arcis, M.; Bargiela, A.; Furling, D.; Artero, R. miR-7 Restores Phenotypes in Myotonic Dystrophy Muscle Cells by Repressing Hyperactivated Autophagy. Mol. Ther. Nucleic Acids 2020, 19, 278–292. [Google Scholar] [CrossRef]
- Tomé, M.; López-Romero, P.; Albo, C.; Artero, R. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011, 18, 985–995. [Google Scholar] [CrossRef]
- Fochi, S.; Giuriato, G.; De Simone, T.; Gomez-Lira, M.; Tamburin, S.; Del Piccolo, L.; Schena, F.; Venturelli, M.; Romanelli, M.G. Regulation of microRNAs in Satellite Cell Renewal, Muscle Function, Sarcopenia and the Role of Exercise. Int. J. Mol. Sci. 2020, 21, 6732. [Google Scholar] [CrossRef]
- Zhang, K.; Zheng, X.; Sun, Y.; Feng, X.; Wu, X.; Liu, W.; Gao, C.; Yan, Y.; Tian, W.; Wang, Y. TOP2A modulates signaling via the AKT/mTOR pathway to promote ovarian cancer cell proliferation. Cancer Biol. Ther. 2024, 25, 2325126. [Google Scholar] [CrossRef]
- Wu, M.V.; Bikopoulos, G.; Hung, S.; Ceddia, R.B. Thermogenic capacity is antagonistically regulated in classical brown and white subcutaneous fat depots by high-fat diet and endurance training in rats: Impact on whole-body energy expenditure. J. Biol. Chem. 2014, 289, 34129–34140. [Google Scholar] [CrossRef]
- Mercer, K.E.; Maurer, A.; Pack, L.M.; Ono-Moore, K.; Spray, B.J.; Campbell, C.; Chandler, C.J.; Burnett, D.; Souza, E.; Casazza, G.; et al. Exercise training and diet-induced weight loss increase markers of hepatic bile acid (BA) synthesis and reduce serum total BA concentrations in obese women. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E864–E873. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Chiu, W.-C.; Chiu, Y.-S. Effect of Inonotus obliquus Extract Supplementation on Endurance Exercise and Energy-Consuming Processes through Lipid Transport in Mice. Nutrients 2022, 14, 5007. [Google Scholar] [CrossRef] [PubMed]
- Mashnafi, S.; Plat, J.; Mensink, R.P.; Joris, P.J.; Kleinloog, J.P.D.; Baumgartner, S. Effects of an 8-week aerobic exercise program on plasma markers for cholesterol absorption and synthesis in older overweight and obese men. Lipids Health Dis. 2021, 20, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, L.; Tao, Y.; Lv, Q.; Wang, M.; Yu, P.; Li, D.; Zhu, Q.; Wang, Y.; Shang, M.; et al. Proline metabolic reprogramming modulates cardiac remodeling induced by pressure overload in the heart. Sci. Adv. 2024, 10, eadl3549. [Google Scholar] [CrossRef]
- Dimina, L.; Tremblay-Franco, M.; Deveaux, A.; Tardivel, C.; Fouillet, H.; Polakof, S.; Martin, J.-C.; Mariotti, F. Plasma metabolome analysis suggests that L-arginine supplementation affects microbial activity resulting in a decrease in trimethylamine N-oxide—A randomized controlled trial in healthy overweight adults with cardiometabolic risk factors. Curr. Dev. Nutr. 2023, 7, 102038. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Konhilas, J.P.; Kelly, D.P.; Leinwand, L.A. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab. 2017, 25, 1012–1026. [Google Scholar] [CrossRef]
- Roffe-Vazquez, D.N.; Huerta-Delgado, A.S.; Castillo, E.C.; Villarreal-Calderón, J.R.; Gonzalez-Gil, A.M.; Enriquez, C.; Garcia-Rivas, G.; Elizondo-Montemayor, L. Correlation of vitamin D with inflammatory cytokines, atherosclerotic parameters, and lifestyle factors in the setting of heart failure: A 12-month follow-up study. Int. J. Mol. Sci. 2019, 20, 5811. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, M.; Cao, D.; Zheng, A.; Zhang, Y.; Su, Y.; Wang, J.; Xu, Y.; Zhou, M.; Tang, Y.; et al. Inhibition of fap promotes cardiac repair by stabilizing BNP. Circ. Res. 2023, 132, 586–600. [Google Scholar] [CrossRef] [PubMed]
- Arnold, Z.; Dostal, C.; Szabó, P.L.; Aykac, I.; Goncalves, A.I.A.; Sousa, S.L.; Baydar, S.; Budde, H.; Váradi, B.; Nadasy, G.L.; et al. Tenascin-C drives cardiovascular dysfunction in a mouse model of diabetic cardiomyopathy. Cardiovasc. Diabetol. 2025, 24, 235. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Q.; Zhou, A.; Zhang, L.; Li, X. Identification of plasma lipid metabolism and potential biomarkers in patients with different coronary occlusive acute myocardial infarction. Front. Cell Dev. Biol. 2025, 13, 1575431. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Li, M.; Ren, W.; Meng, J.; Yao, X.; Chu, H.; Yao, R.; Zhai, M.; Zeng, Y. Multi-Omics Analysis Reveals Biaxial Regulatory Mechanisms of Cardiac Adaptation by Specialized Racing Training in Yili Horses. Biology 2025, 14, 1609. https://doi.org/10.3390/biology14111609
Wang T, Li M, Ren W, Meng J, Yao X, Chu H, Yao R, Zhai M, Zeng Y. Multi-Omics Analysis Reveals Biaxial Regulatory Mechanisms of Cardiac Adaptation by Specialized Racing Training in Yili Horses. Biology. 2025; 14(11):1609. https://doi.org/10.3390/biology14111609
Chicago/Turabian StyleWang, Tongliang, Mengying Li, Wanlu Ren, Jun Meng, Xinkui Yao, Hongzhong Chu, Runchen Yao, Manjun Zhai, and Yaqi Zeng. 2025. "Multi-Omics Analysis Reveals Biaxial Regulatory Mechanisms of Cardiac Adaptation by Specialized Racing Training in Yili Horses" Biology 14, no. 11: 1609. https://doi.org/10.3390/biology14111609
APA StyleWang, T., Li, M., Ren, W., Meng, J., Yao, X., Chu, H., Yao, R., Zhai, M., & Zeng, Y. (2025). Multi-Omics Analysis Reveals Biaxial Regulatory Mechanisms of Cardiac Adaptation by Specialized Racing Training in Yili Horses. Biology, 14(11), 1609. https://doi.org/10.3390/biology14111609





