Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design and Horse Grouping
2.3. Echocardiographic Assessment
2.4. Transcriptomic Analysis
2.4.1. RNA Extraction
2.4.2. mRNA and miRNA Library Construction and Sequencing
2.4.3. mRNA Identification
2.4.4. Functional Annotation and Pathway Analysis of DEmRNAs
2.4.5. miRNA Identification
2.4.6. miRNA Target Gene Prediction
2.4.7. Correlation Analysis of DEmRNAs and DEmiRNAs
2.4.8. RT-qPCR Validation of mRNAs and miRNAs
2.5. Metabolomics Analysis
2.5.1. Data Collection and Processing
2.5.2. Targeted Metabolomics Analysis
2.6. Data Analysis
2.7. Statistical Analysis
3. Results
3.1. Cardiac Function Assessment of Yili Horses—Characteristics of Cardiac Remodeling
3.2. miRNA and mRNA Expression Analysis
3.2.1. Overview of Blood Sequencing Data from Yili Horses
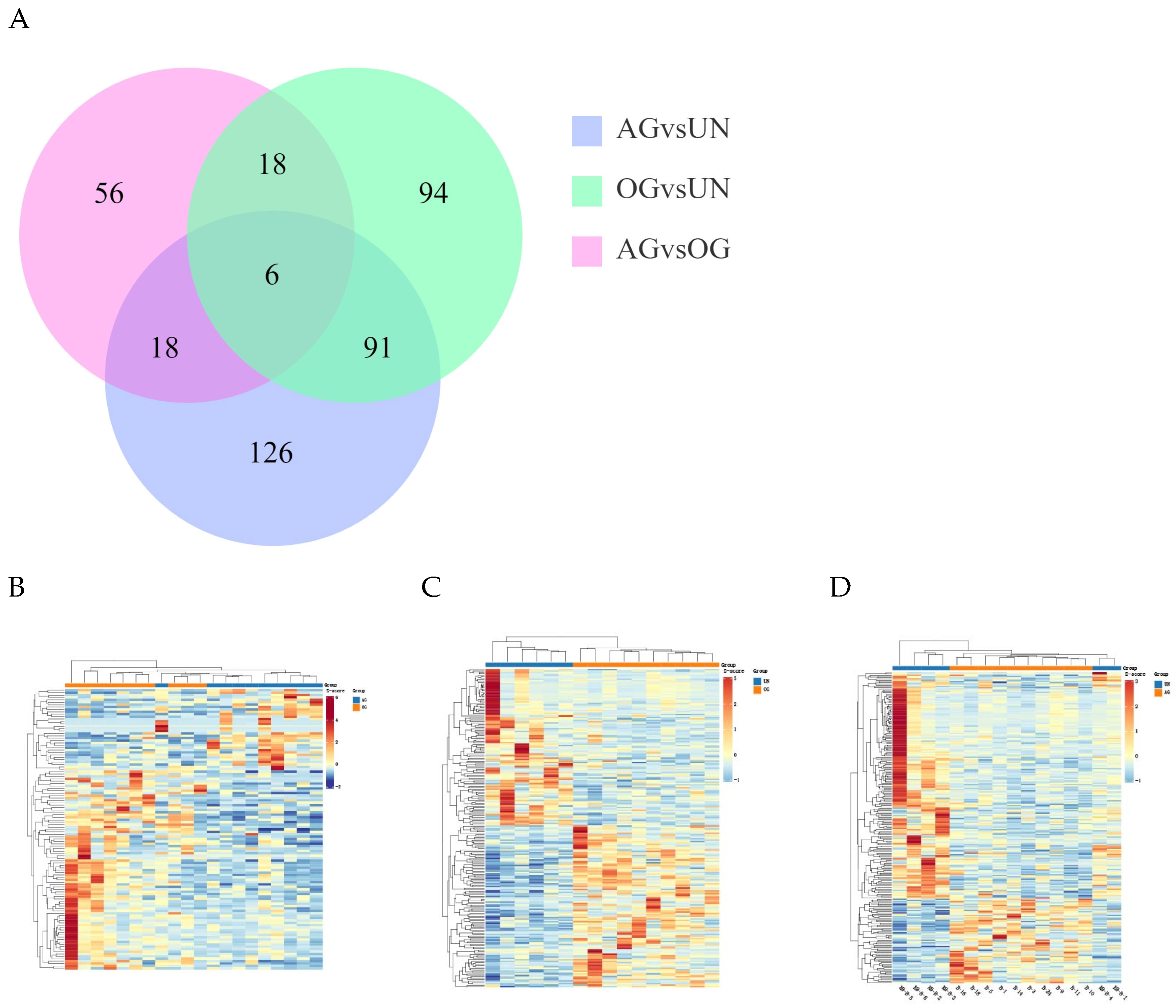
3.2.2. Differential mRNA Analysis
3.2.3. Differential miRNA Analysis
3.3. Differential Expression Analysis
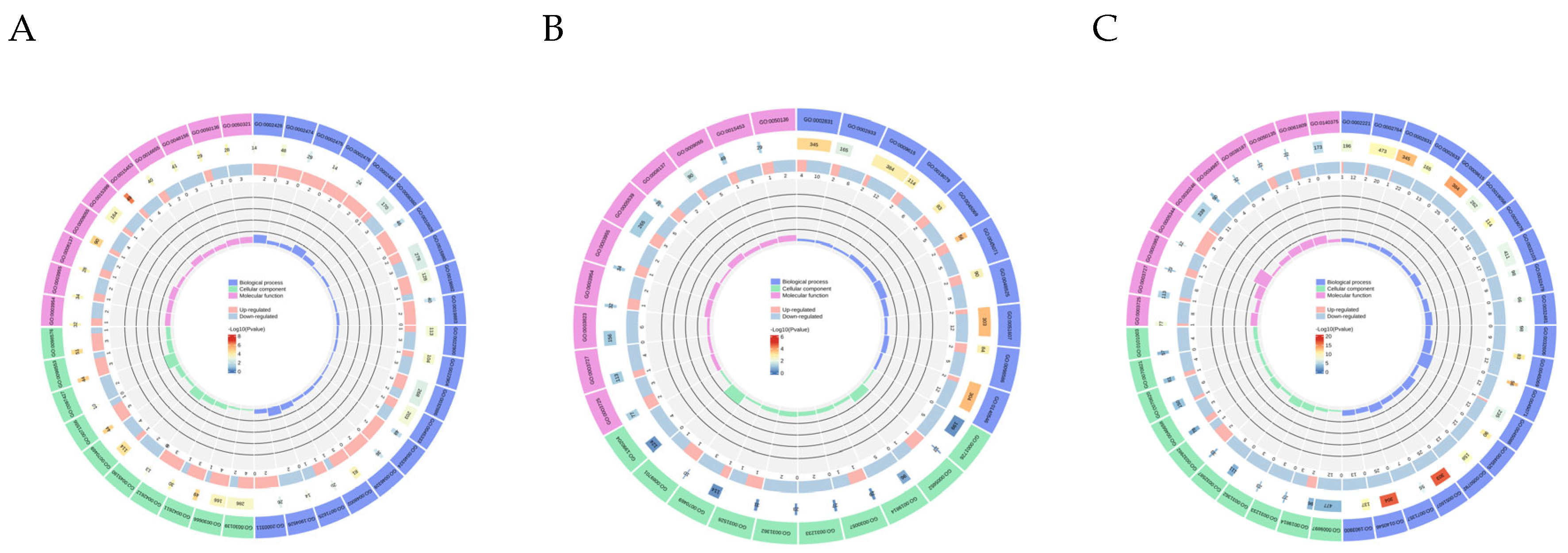
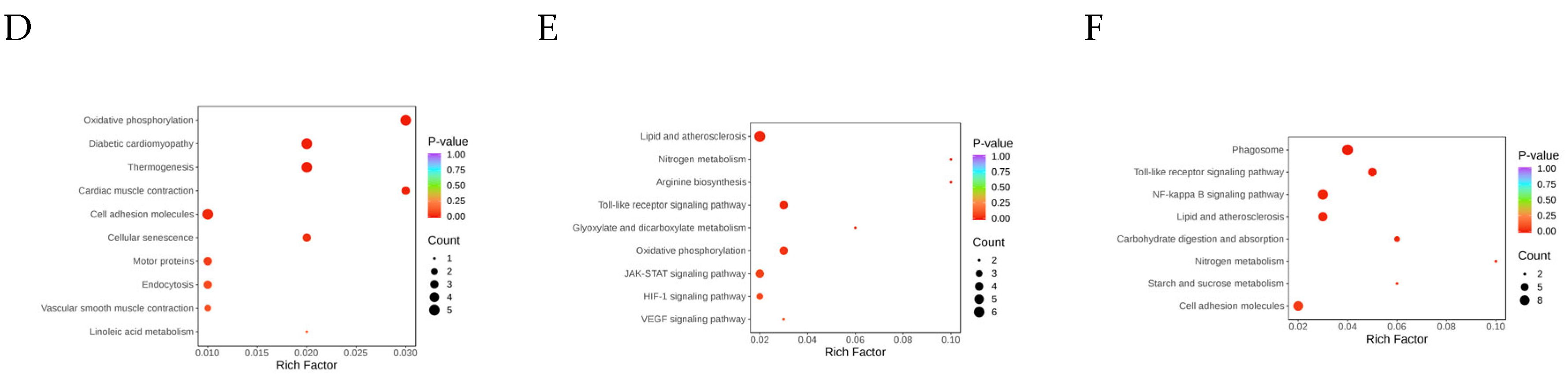
3.3.1. GO Functional Annotation and KEGG Enrichment Analysis of DEGs
3.3.2. GO and KEGG Functional Annotation Analysis
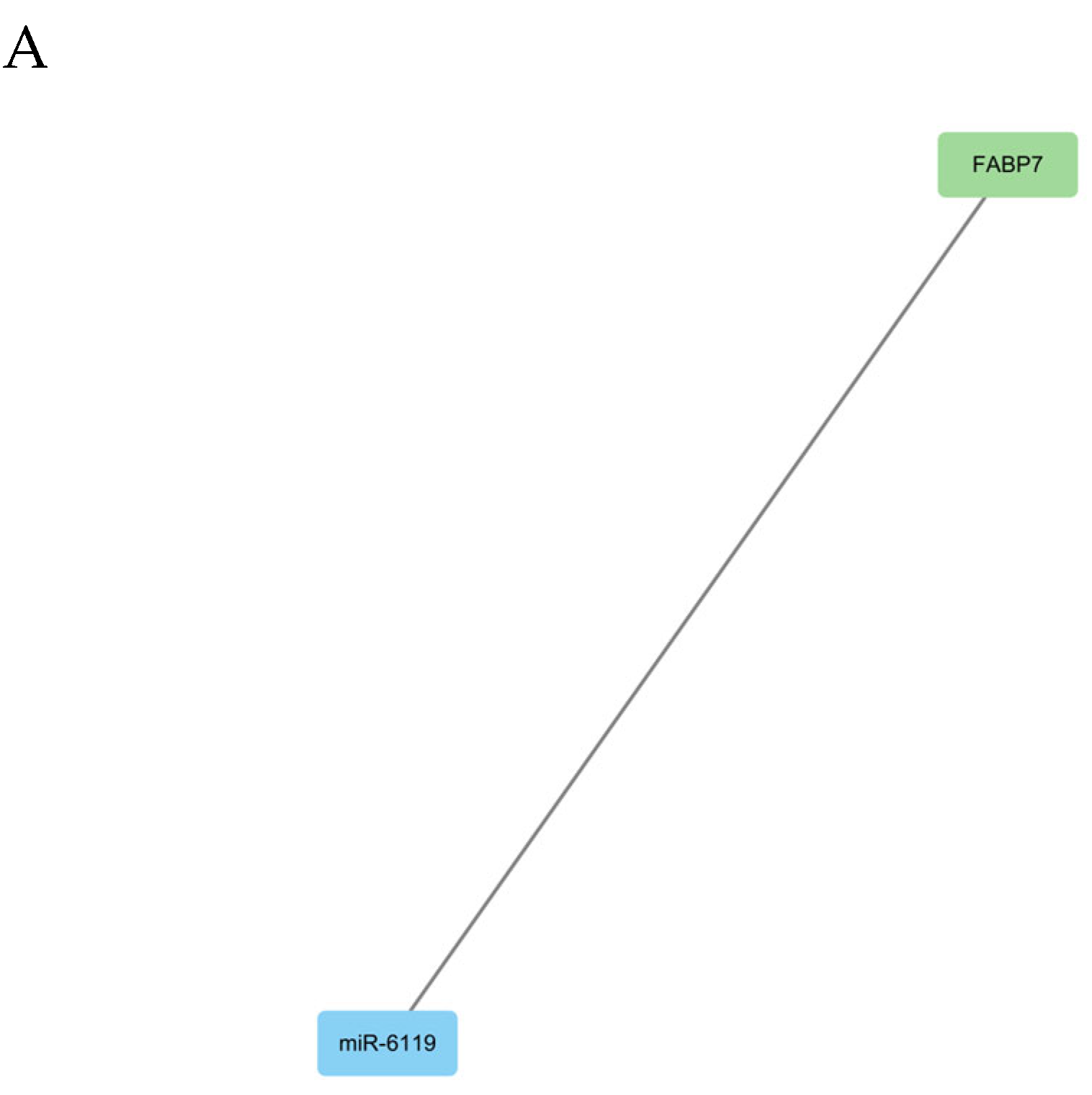
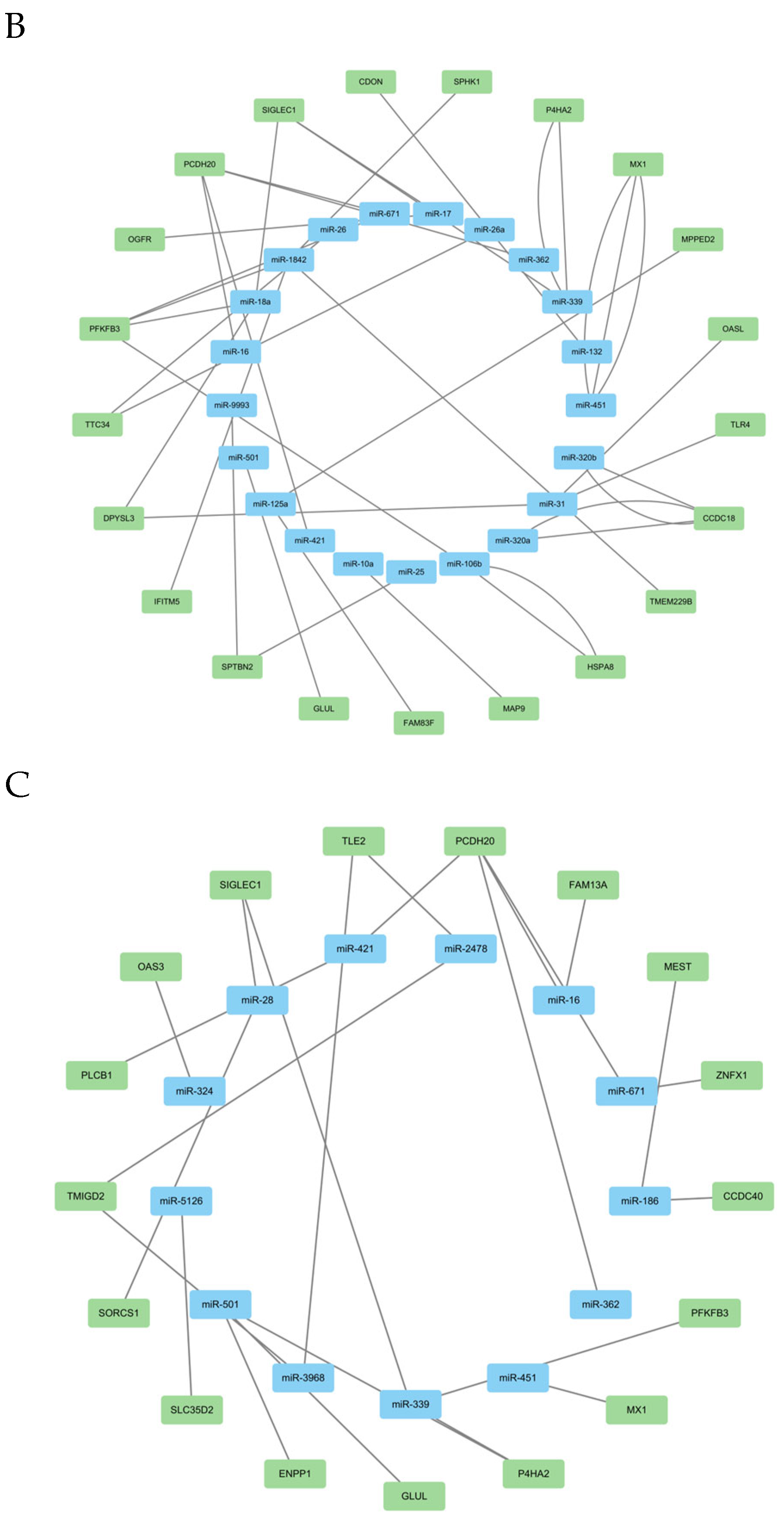
3.3.3. DEmRNA-DEmiRNA Correlation Analysis and Functional Analysis of Target Genes
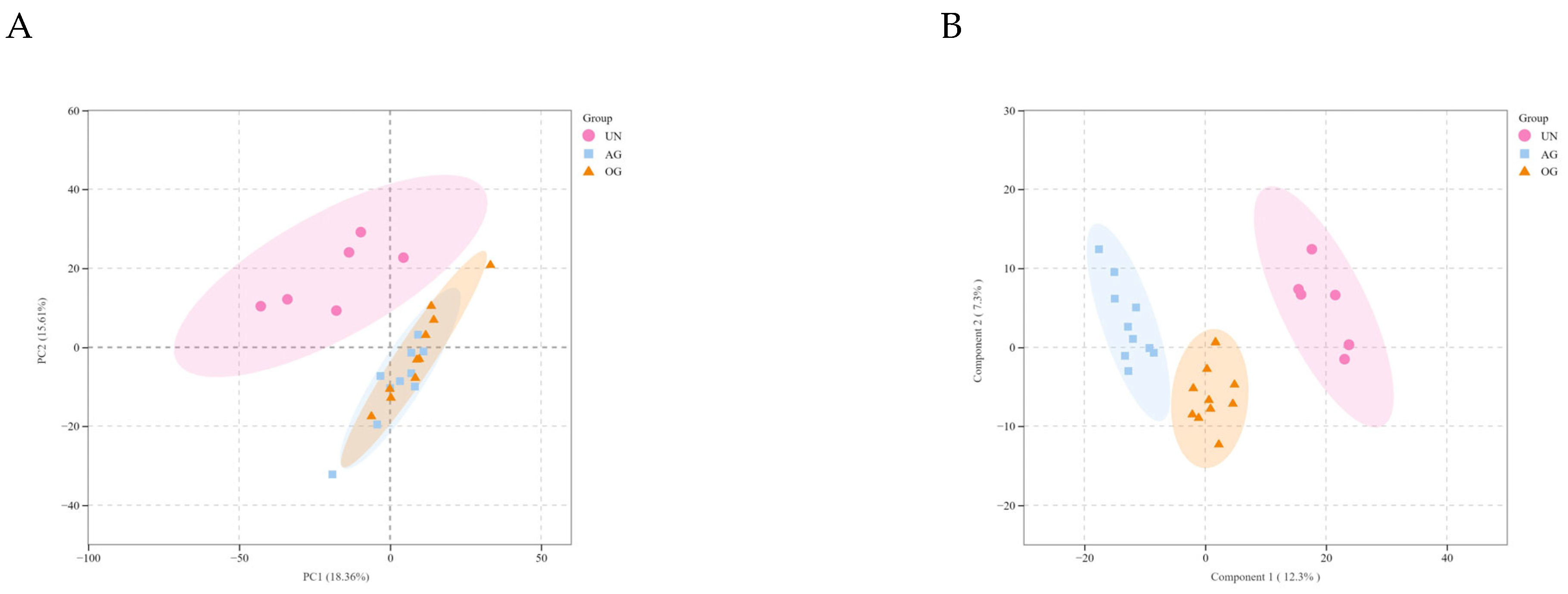
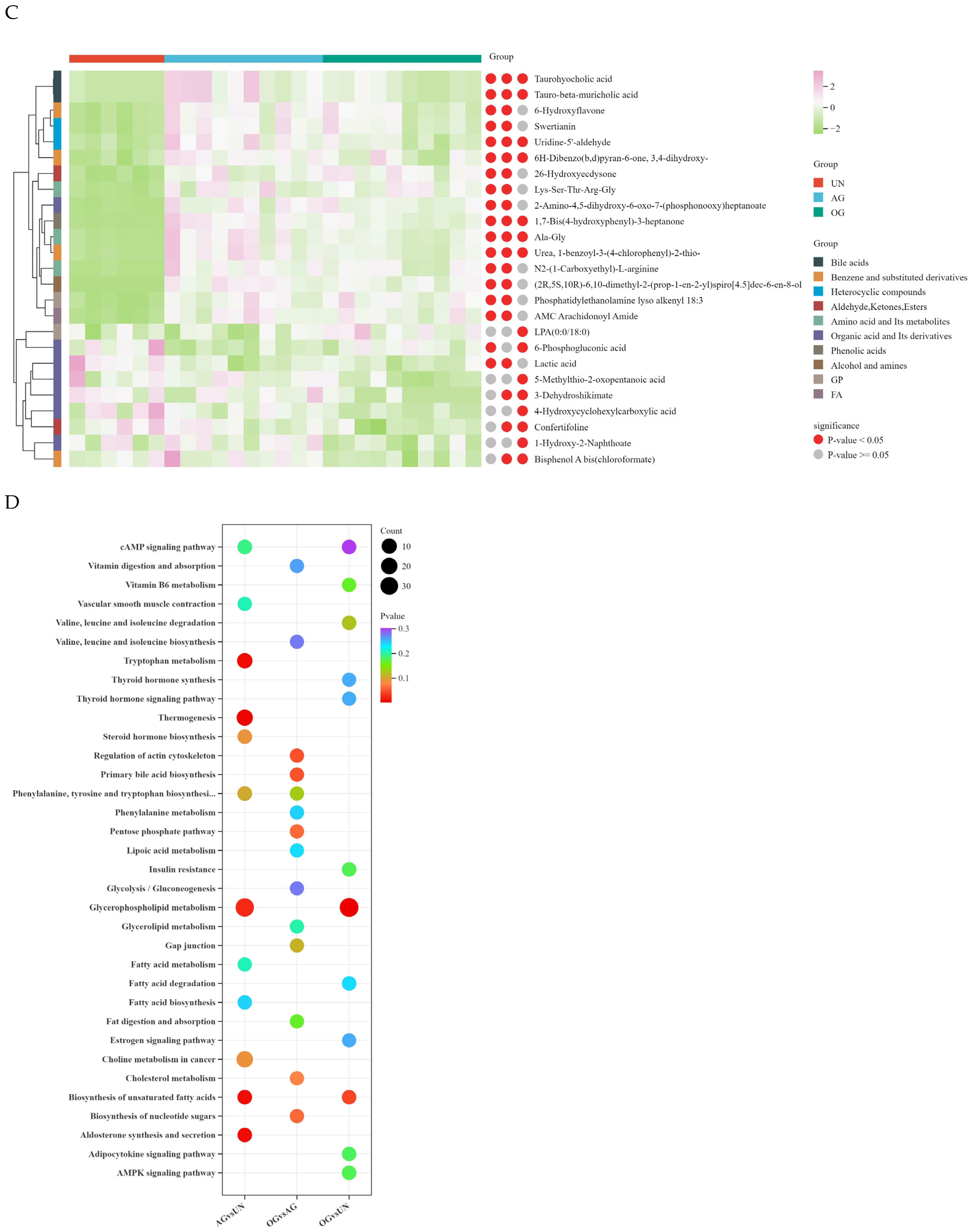
3.4. Broad Targeted Metabolomics
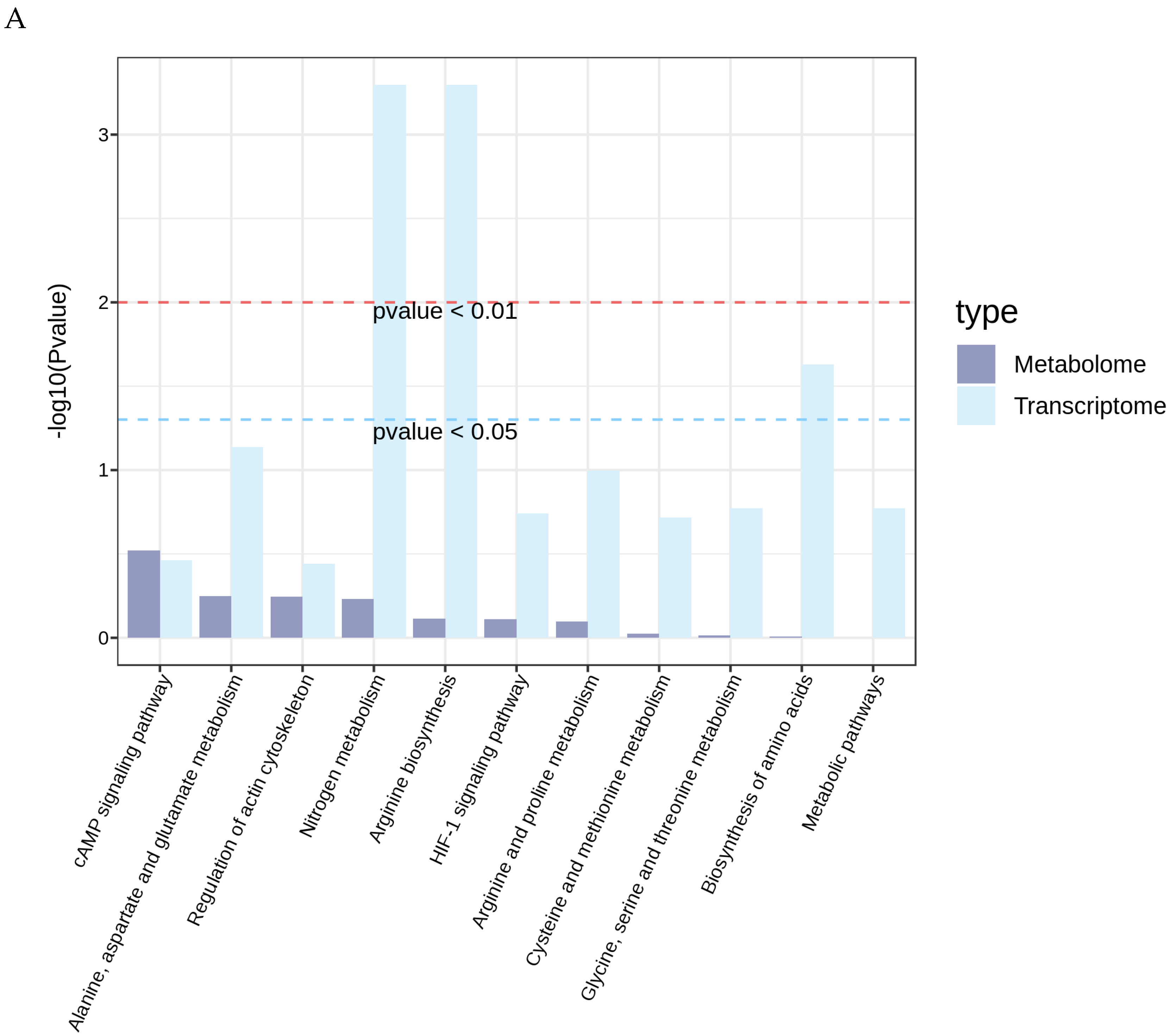
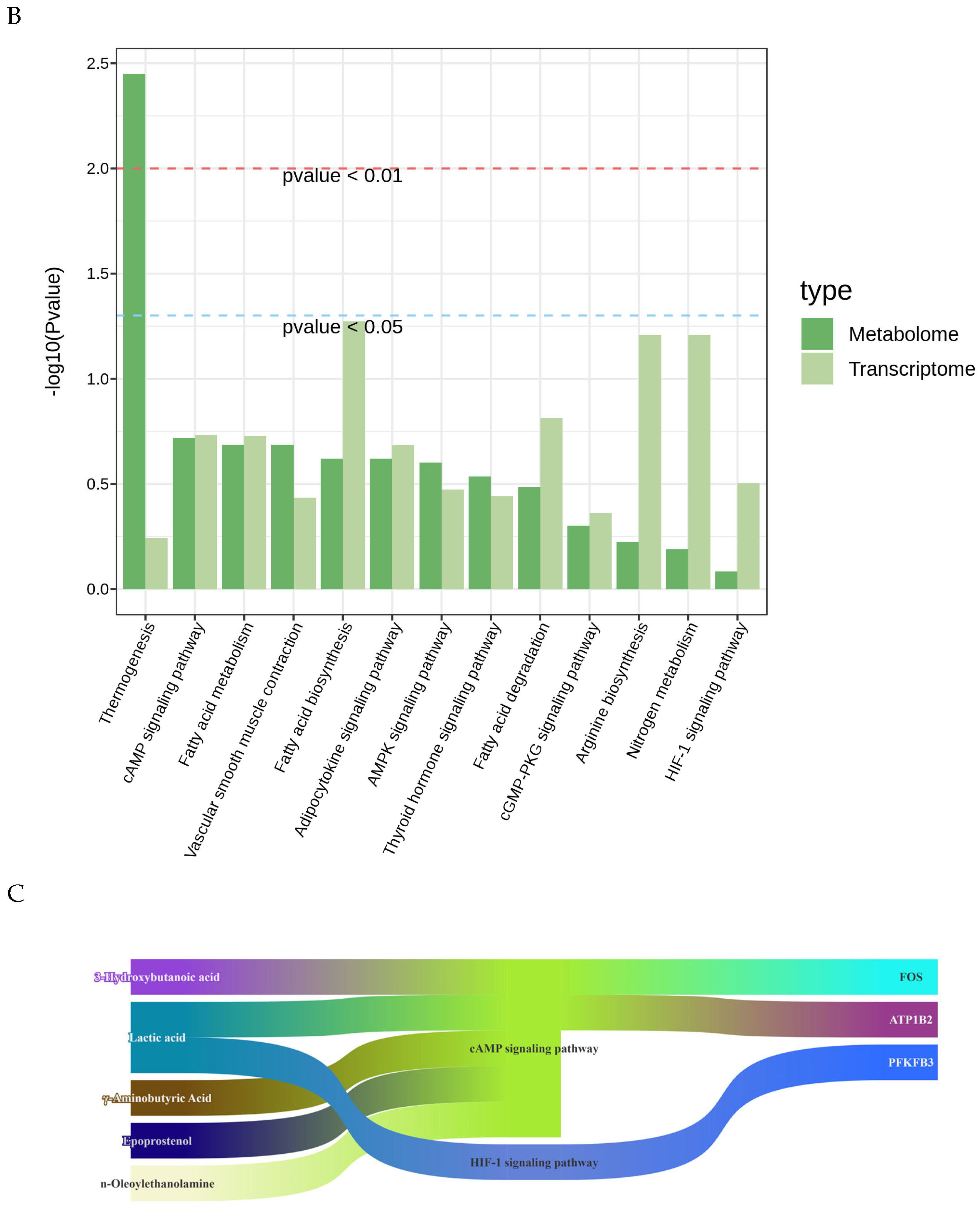
3.5. Integrated Transcriptomic and Metabolomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernandes, T.; Soci, U.; Oliveira, E.M. Eccentric and concentric cardiac hypertrophy induced by exercise training: microRNAs and molecular determinants. Braz. J. Med. Biol. Res. 2011, 44, 836–847. [Google Scholar] [CrossRef]
- Wang, T.; Meng, J.; Yang, X.; Zeng, Y.; Yao, X.; Ren, W. Differential Metabolomics and Cardiac Function in Trained vs. Untrained Yili Performance Horses. Animals 2025, 15, 2444. [Google Scholar] [CrossRef]
- Shave, R.; Howatson, G.; Dickson, D.; Young, L. Exercise-induced cardiac remodeling: Lessons from humans, horses, and dogs. Vet. Sci. 2017, 4, 9. [Google Scholar] [CrossRef]
- Kis, J.; Rózsa, L.; Husvéth, F.; Zsolnai, A.; Anton, I. Role of genes related to performance and reproduction of Thoroughbreds in training and breeding—A review. Acta Vet. Hung. 2021, 69, 315–323. [Google Scholar]
- Alissa, M.; Aldurayhim, M.; Abdulaziz, O.; Alsalmi, O.; Awad, A.; Algopishi, U.B.; Alharbi, S.; Safhi, A.Y.; Khan, K.H.; Uffar, C. From molecules to heart regeneration: Understanding the complex and profound role of non-coding RNAs in stimulating cardiomyocyte proliferation for cardiac repair. Curr. Probl. Cardiol. 2024, 49, 102857. [Google Scholar] [CrossRef]
- Eskandari, A.; Soori, R.; Choobineh, S.; Mazaheri Tirani, Z. Exercise promotes heart regeneration in aged rats by increasing regenerative factors in myocardial tissue. Physiol. Int. 2020, 107, 166–176. [Google Scholar] [CrossRef]
- Fernandes, T.; Baraúna, V.G.; Negrão, C.E.; Phillips, M.I.; Oliveira, E.M. Aerobic exercise training promotes physiological cardiac remodeling involving a set of microRNAs. Am. J. Physiol.-Heart Circ. Physiol. 2015, 309, H543–H552. [Google Scholar] [CrossRef]
- Budhram-Mahadeo, V.S.; Solomons, M.R.; Mahadeo-Heads, E.A. Linking metabolic dysfunction with cardiovascular diseases: Brn-3b/POU4F2 transcription factor in cardiometabolic tissues in health and disease. Cell Death Dis. 2021, 12, 267. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wen, Y.; Li, S.; Lu, X.; Xu, R.; Li, C. Endoplasmic reticulum-mitochondria contacts: A potential therapy target for cardiovascular remodeling-associated diseases. Front. Cell Dev. Biol. 2021, 9, 774989. [Google Scholar] [CrossRef]
- Lew, J.K.-S.; Pearson, J.T.; Saw, E.; Tsuchimochi, H.; Wei, M.; Ghosh, N.; Du, C.-K.; Zhan, D.-Y.; Jin, M.; Umetani, K. Exercise regulates microRNAs to preserve coronary and cardiac function in the diabetic heart. Circ. Res. 2020, 127, 1384–1400. [Google Scholar] [CrossRef]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramírez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef]
- Hou, A.; Xu, X.; Zhang, Y.; He, H.; Feng, Y.; Fan, W.; Tan, R.; Gong, L.; Chen, J. Excessive fatty acids activate PRMT5/MDM2/Drosha pathway to regulate miRNA biogenesis and lipid metabolism. Liver Int. 2024, 44, 1634–1650. [Google Scholar] [CrossRef]
- Wang, T. A Multi-Omics-Based Study on the Mechanism of Exercise-Induced Cardiac Remodeling and Anti-Fibrosis in Yili Horses; Xinjiang Agricultural University: Xinjiang, China, 2025. [Google Scholar]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Nam, J.-W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA. org resource: Targets and expression. Nucleic Acids Res. 2008, 36 (Suppl. S1), D149–D153. [Google Scholar] [CrossRef]
- Enright, A.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D. MicroRNA targets in Drosophila. Genome Biol. 2003, 4, P8. [Google Scholar] [CrossRef]
- Vega, R.B.; Konhilas, J.P.; Kelly, D.P.; Leinwand, L.A. Molecular mechanisms underlying cardiac adaptation to exercise. Cell Metab. 2017, 25, 1012–1026. [Google Scholar] [CrossRef]
- Durando, M.M.; Corley, K.T.; Boston, R.C.; Birks, E.K. Cardiac output determination by use of lithium dilution during exercise in horses. Am. J. Vet. Res. 2008, 69, 1054–1060. [Google Scholar] [CrossRef]
- Hinchcliff, K.W.; Kaneps, A.J.; Geor, R.J. Equine Exercise Physiology: The Science of Exercise in the Athletic Horse; Elsevier Health Sciences: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Edenfield, K.M.; Reifsteck, F.; Carek, S.; Harmon, K.G.; Asken, B.M.; Dillon, M.C.; Street, J.; Clugston, J.R. Echocardiographic measurements of left ventricular end-diastolic diameter and interventricular septal diameter in collegiate football athletes at preparticipation evaluation referenced to body surface area. BMJ Open Sport Exerc. Med. 2019, 5, e000488. [Google Scholar] [CrossRef]
- Hosseini, M.; Piri, M.; Agha-Alinejad, H.; Haj-Sadeghi, S. The effect of endurance, resistance and concurrent training on the heart structure of female students. Biol. Sport 2012, 29, 17–21. [Google Scholar] [CrossRef]
- Wang, J.J.; Chang, S.C.; Tran, A.; Sanchez, J.; Yang, M.; Omi, K. MYH14 Protects Against of Isoproterenol-induced Left Ventricular Hypertrophy and Fibrosis. Circ. Res. 2019, 125 (Suppl. S1), A263. [Google Scholar] [CrossRef]
- Bazzazi, H.; Popel, A.S. Computational investigation of sphingosine kinase 1 (SphK1) and calcium dependent ERK1/2 activation downstream of VEGFR2 in endothelial cells. PLoS Comput. Biol. 2017, 13, e1005332. [Google Scholar] [CrossRef]
- Min, J.; Zeng, T.; Roux, M.; Lazar, D.; Chen, L.; Tudzarova, S. The role of HIF1α-PFKFB3 pathway in diabetic retinopathy. J. Clin. Endocrinol. Metab. 2021, 106, 2505–2519. [Google Scholar] [CrossRef]
- Emini Veseli, B.; Perrotta, P.; Van Wielendaele, P.; Lambeir, A.M.; Abdali, A.; Bellosta, S.; Monaco, G.; Bultynck, G.; Martinet, W.; De Meyer, G.R. Small molecule 3PO inhibits glycolysis but does not bind to 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3 (PFKFB3). FEBS Lett. 2020, 594, 3067–3075. [Google Scholar] [CrossRef]
- Huang, A.; Wang, Z.; Tang, H.; Jia, Z.; Ji, X.; Yang, X.; Jiang, W. Bardoxolone Methyl Ameliorates Myocardial Ischemia/Reperfusion Injury by Activating the Nrf2/HO-1 Signaling Pathway. Cardiovasc. Ther. 2023, 2023, 5693732. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Bajpai, S.; Chaturvedi, P.; Wirtz, D.; Semenza, G.L. Hypoxia-Inducible Factor 1 (HIF-1) Promotes Extracellular Matrix Remodeling Under Hypoxic Conditions by Inducing P4HA1, P4HA2, and PLOD2 Expression in Fibroblasts; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Zhang, L.; Wang, Y.; Zhao, M.; Xi, L. Effect of Exercise on the Expression of Cardiac FOS and Heat Shock Protein 70 in Rats. J. Beijing Sport Univ. 2010, 33, 64–66+70. [Google Scholar]
- Han, J.; Ye, S.; Zou, C.; Chen, T.; Wang, J.; Li, J.; Jiang, L.; Xu, J.; Huang, W.; Wang, Y. Angiotensin II causes biphasic STAT3 activation through TLR4 to initiate cardiac remodeling. Hypertension 2018, 72, 1301–1311. [Google Scholar] [CrossRef]
- Jiang, D.-S.; Liu, Y.; Zhou, H.; Zhang, Y.; Zhang, X.-D.; Zhang, X.-F.; Chen, K.; Gao, L.; Peng, J.; Gong, H. Interferon regulatory factor 7 functions as a novel negative regulator of pathological cardiac hypertrophy. Hypertension 2014, 63, 713–722. [Google Scholar] [CrossRef]
- Ma, W.; Tian, Y.; Shi, L.; Liang, J.; Ouyang, Q.; Li, J.; Chen, H.; Sun, H.; Ji, H.; Liu, X. N-Acetyltransferase 10 represses Uqcr11 and Uqcrb independently of ac4C modification to promote heart regeneration. Nat. Commun. 2024, 15, 2137. [Google Scholar] [CrossRef] [PubMed]
- Jinzheng, M.; Chao, S.; Jie, Y.; Minxin, W. Research advances on changes of myocardial metabolic patterns under physiological and pathological conditions. Chin. J. Cardiovasc. Med. 2022, 27, 383–388. [Google Scholar]


| TERM | Untrained Group (UN) | Ordinary Group (OG) | Agility Group (AG) |
|---|---|---|---|
| RVDd | 2.38 ± 0.13 a | 2.6 ± 0.22 a | 2.6 ± 0.25 a |
| IVSd | 2.14 ± 0.22 b | 2.41 ± 0.22 a | 2.51 ± 0.31 a |
| LVIDd | 8.77 ± 0.22 c | 9.56 ± 0.27 b | 10.04 ± 0.19 a |
| LVFWd | 1.79 ± 0.26 b | 1.97 ± 0.22 a | 2.03 ± 0.17 a |
| RVDs | 1.66 ± 0.13 a | 1.72 ± 0.24 a | 1.76 ± 0.26 a |
| IVSs | 3.52 ± 0.25 b | 3.92 ± 0.29 a | 4.06 ± 0.31 a |
| LVIDS | 5.35 ± 0.11 b | 5.71 ± 0.17 a | 5.86 ± 0.13 a |
| LVFWs | 2.64 ± 0.26 b | 3 ± 0.34 a | 3.09 ± 0.31 a |
| LADd | 8.17 ± 0.49 b | 8.63 ± 0.46 a | 9.07 ± 0.57 a |
| LADs | 9.32 ± 0.52 b | 10.23 ± 0.49 a | 10.36 ± 0.54 a |
| AODd | 4.62 ± 0.24 c | 5.11 ± 0.14 b | 5.35 ± 0.18 a |
| PADd | 3.89 ± 0.23 b | 4.2 ± 0.24 a | 4.22 ± 0.12 a |
| PADs | 4.29 ± 0.22 b | 4.75 ± 0.36 a | 4.83 ± 0.23 a |
| EF | 0.61 ± 0.06 b | 0.65 ± 0.07 ab | 0.68 ± 0.06 a |
| FS | 0.39 ± 0.01 c | 0.4 ± 0.01 b | 0.42 ± 0.02 a |
| LVminor | 14 ± 0.57 b | 14.92 ± 0.41 a | 14.77 ± 0.43 a |
| SV | 285.07 ± 17.98 c | 350.93 ± 22.67 b | 398.9 ± 24.78 a |
| EDV | 423.35 ± 23.81 c | 511.47 ± 31.39 b | 569.21 ± 23.9 a |
| ESV | 138.28 ± 6.42 b | 160.54 ± 10.9 a | 170.31 ± 8.8 a |
| CO | 12,830.48 ± 1030.97 b | 18,248.57 ± 2172.95 a | 18,309.83 ± 1867.84 a |
| LVM | 1422.91 ± 246.18 c | 1895.89 ± 211.19 b | 2160.79 ± 225.55 a |
| HR | 55.23 ± 2.73 a | 46.97 ± 2.66 b | 45.93 ± 4.28 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Yang, X.; Ren, W.; Meng, J.; Yao, X.; Chu, H.; Yao, R.; Zhai, M.; Zeng, Y. Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses. Biology 2025, 14, 1535. https://doi.org/10.3390/biology14111535
Wang T, Yang X, Ren W, Meng J, Yao X, Chu H, Yao R, Zhai M, Zeng Y. Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses. Biology. 2025; 14(11):1535. https://doi.org/10.3390/biology14111535
Chicago/Turabian StyleWang, Tongliang, Xixi Yang, Wanlu Ren, Jun Meng, Xinkui Yao, Hongzhong Chu, Runchen Yao, Manjun Zhai, and Yaqi Zeng. 2025. "Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses" Biology 14, no. 11: 1535. https://doi.org/10.3390/biology14111535
APA StyleWang, T., Yang, X., Ren, W., Meng, J., Yao, X., Chu, H., Yao, R., Zhai, M., & Zeng, Y. (2025). Integrating miRNA, mRNA, and Targeted Metabolomics Analyses to Explore the Regulatory Mechanism of Cardiac Remodeling in Yili Horses. Biology, 14(11), 1535. https://doi.org/10.3390/biology14111535






