Deciphering the Contribution of TATA Box and 5′UTR to Defense Signaling in Rice Under Blast Infection
Simple Summary
Abstract
1. Introduction
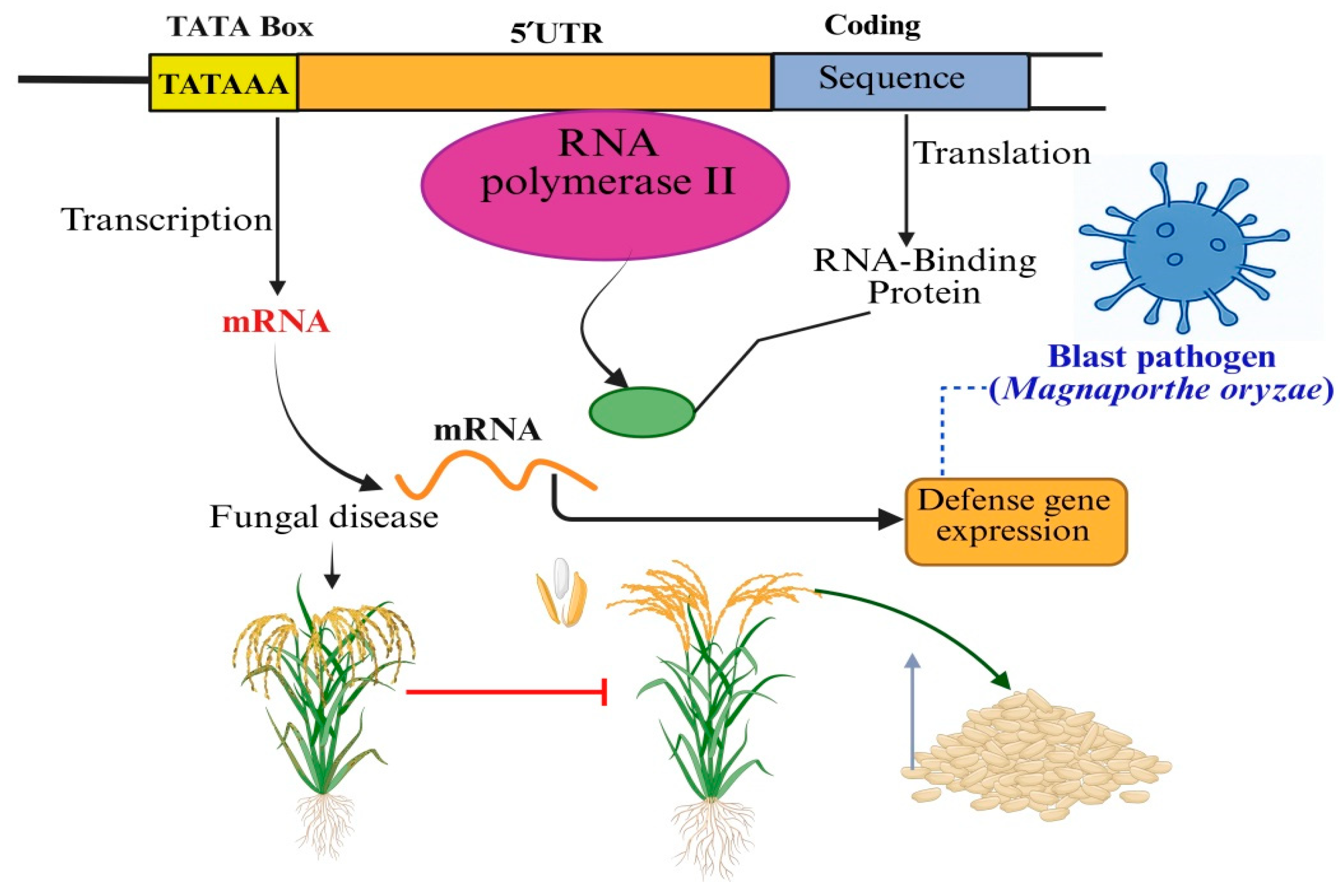
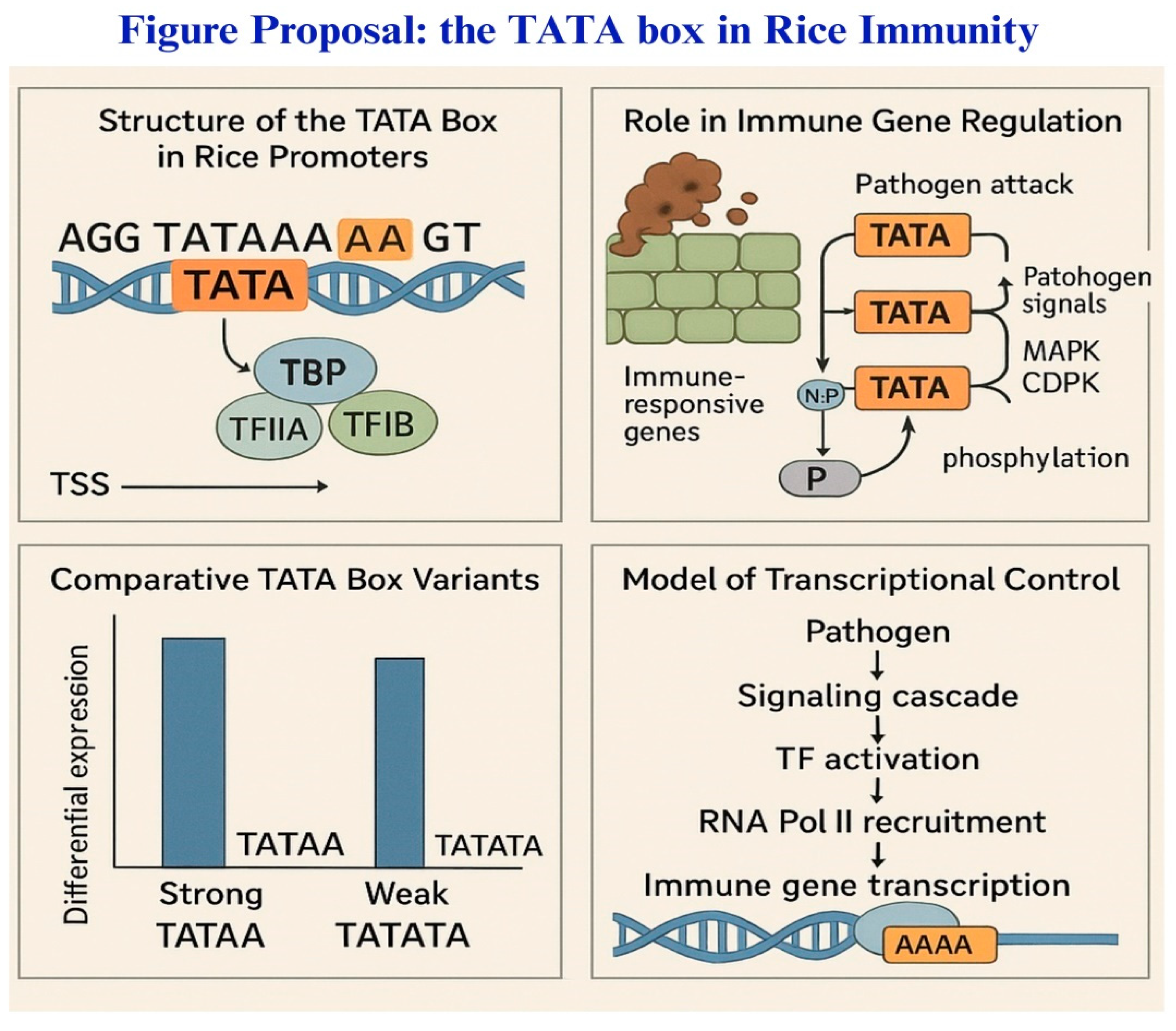
2. The TATA Box: A Transcriptional Control Hub in Rice Immunity
2.1. The 5′UTR: Gate Keeper of Translational Regulation in Defense Responses
2.2. Structural and Functional Features of 5′UTRs
2.3. Functional Importance of the 5′UTR in Stress-Response
2.4. Case Studies of 5′UTR-Mediated Defense Gene Regulation in Rice
2.5. Interaction Between 5′UTRs and RNA-Binding Proteins During Blast Infection
3. Multilayered Regulatory Mechanisms
4. Synergistic Roles of TATA Box and 5′UTR in Rice Blast Defense
4.1. Coordinated Transcriptional and Translational Control of Immune Genes
4.2. Examples of Defense Genes Regulated by Both Elements
4.3. Impact of Natural Polymorphisms on Blast Resistance
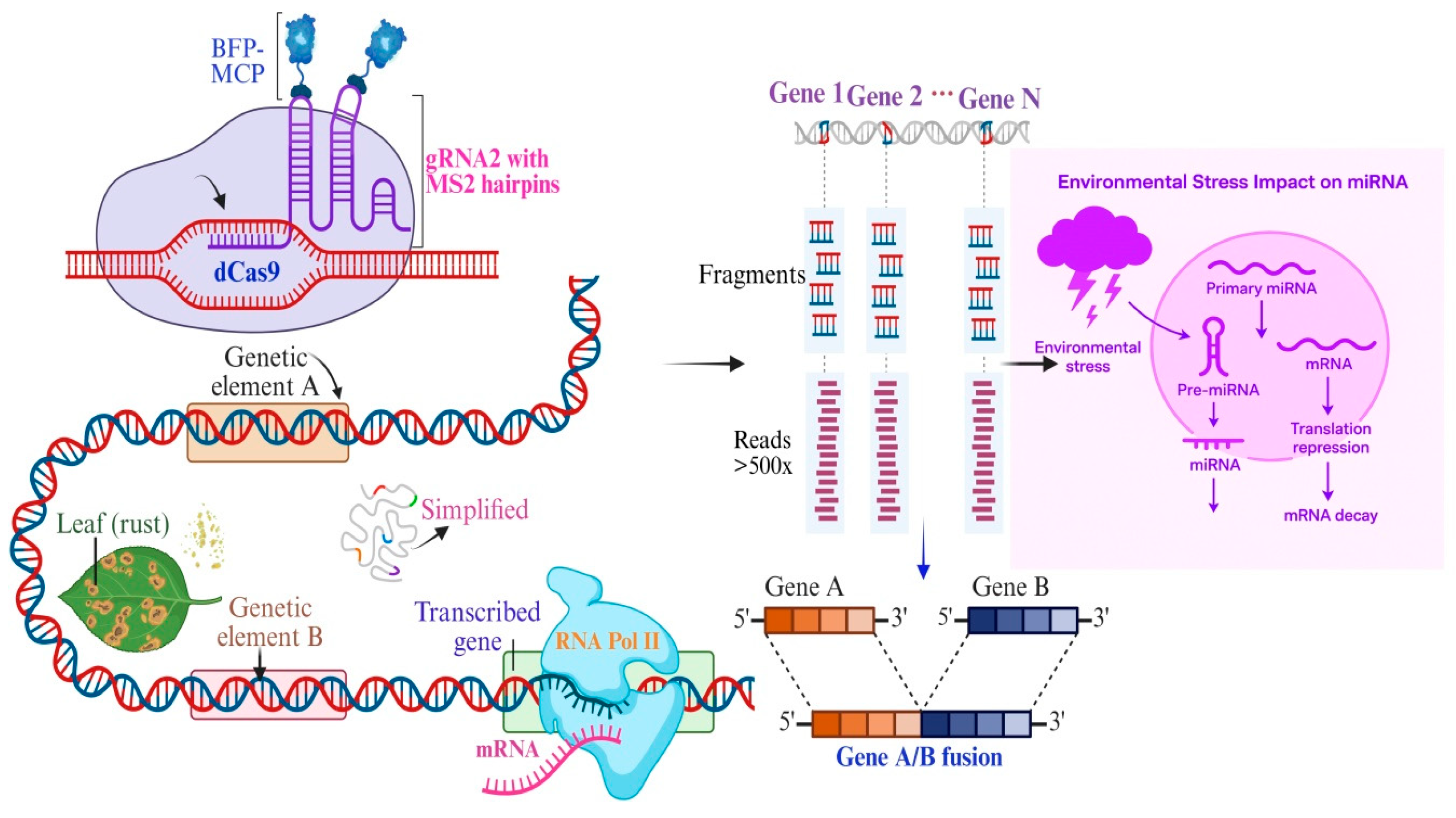
5. Experimental Approaches to Study TATA Box and 5′UTR Functions
5.1. Promoter-Reporter Assays (e.g., GUS, Luciferase)
5.2. CRISPR/Cas9-Mediated Mutagenesis of Regulatory Elements
5.3. Ribosome Profiling to Assess Translational Efficiency
5.4. Comparative Genomics of Resistant and Susceptible Rice Lines
6. Biotechnological Applications for Blast Resistance
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bandumula, N. Rice production in Asia: Key to global food security. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 1323–1328. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.-R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef]
- Mohidem, N.A.; Hashim, N.; Shamsudin, R.; Che Man, H. Rice for food security: Revisiting its production, diversity, rice milling process and nutrient content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sao, R.; Choudhary, D.K.; Thada, A.; Kumar, V.; Mondal, S.; Das, B.K.; Jankuloski, L.; Sharma, D. Advancement in the breeding, biotechnological and genomic tools towards development of durable genetic resistance against the rice blast disease. Plants 2022, 11, 2386. [Google Scholar] [CrossRef]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D. Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef]
- Younas, M.U.; Ahmad, I.; Qasim, M.; Ijaz, Z.; Rajput, N.; Parveen Memon, S.; UL Zaman, W.; Jiang, X.; Zhang, Y.; Zuo, S. Progress in the management of Rice Blast Disease: The role of Avirulence and Resistance genes through gene-for-gene interactions. Agronomy 2024, 14, 163. [Google Scholar] [CrossRef]
- Naz, M.; Chen, Z. 5′UTR Dynamics: Modulating Model Plant Defense Mechanisms against Fungal Threats. J. Agric. Food Chem. 2025, 73, 17303–17314. [Google Scholar] [CrossRef]
- Kumar, V.; Jain, P.; Venkadesan, S.; Karkute, S.G.; Bhati, J.; Abdin, M.Z.; Sevanthi, A.M.; Mishra, D.C.; Chaturvedi, K.K.; Rai, A. Understanding rice-Magnaporthe oryzae interaction in resistant and susceptible cultivars of rice under panicle blast infection using a time-course transcriptome analysis. Genes 2021, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Gondalia, N.; Quiroz, L.F.; Lai, L.; Singh, A.K.; Khan, M.; Brychkova, G.; McKeown, P.C.; Chatterjee, M.; Spillane, C. Harnessing promoter elements to enhance gene editing in plants: Perspectives and advances. Plant Biotechnol. J. 2025, 23, 1375–1395. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Srivastava, A.K.; AbdElgawad, H. Transcriptional and Post-Transcriptional Regulation of Plant Growth, Development, and Stress Responses. J. Plant Growth Regul. 2025, 44, 1317–1322. [Google Scholar] [CrossRef]
- Basso, M.F.; Arraes, F.B.M.; Grossi-de-Sa, M.; Moreira, V.J.V.; Alves-Ferreira, M.; Grossi-de-Sa, M.F. Insights into genetic and molecular elements for transgenic crop development. Front. Plant Sci. 2020, 11, 509. [Google Scholar] [CrossRef]
- Jimmy, J.L.; Karn, R.; Kumari, S.; Sruthilaxmi, C.B.; Pooja, S.; Emerson, I.A.; Babu, S. Rice WRKY13 TF protein binds to motifs in the promoter region to regulate downstream disease resistance-related genes. Funct. Integr. Genom. 2023, 23, 249. [Google Scholar] [CrossRef]
- Wilkie, G.S.; Dickson, K.S.; Gray, N.K. Regulation of mRNA translation by 5′-and 3′-UTR-binding factors. Trends Biochem. Sci. 2003, 28, 182–188. [Google Scholar] [CrossRef]
- Meijer, H.A.; Thomas, A.A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 2002, 367, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Chen, S. Non-target site mechanisms of fungicide resistance in crop pathogens: A review. Microorganisms 2021, 9, 502. [Google Scholar] [CrossRef] [PubMed]
- Tonnessen, B.W.; Bossa-Castro, A.M.; Mauleon, R.; Alexandrov, N.; Leach, J.E. Shared cis-regulatory architecture identified across defense response genes is associated with broad-spectrum quantitative resistance in rice. Sci. Rep. 2019, 9, 1536. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef]
- Yamasaki, S.; Suzuki, A.; Yamano, Y.; Kawabe, H.; Ueno, D.; Demura, T.; Kato, K. Identification of 5′-untranslated regions that function as effective translational enhancers in monocotyledonous plant cells using a novel method of genome-wide analysis. Plant Biotechnol. 2018, 35, 365–373. [Google Scholar] [CrossRef]
- Tang, B.; Liu, C.; Li, Z.; Zhang, X.; Zhou, S.; Wang, G.L.; Chen, X.L.; Liu, W. Multilayer regulatory landscape during pattern-triggered immunity in rice. Plant Biotechnol. J. 2021, 19, 2629–2645. [Google Scholar] [CrossRef]
- Deb, S.; Madhavan, V.N.; Gokulan, C.; Patel, H.K.; Sonti, R.V. Arms and ammunitions: Effectors at the interface of rice and it’s pathogens and pests. Rice 2021, 14, 94. [Google Scholar] [CrossRef]
- Alhoraibi, H.; Bigeard, J.; Rayapuram, N.; Colcombet, J.; Hirt, H. Plant immunity: The MTI-ETI model and beyond. Curr. Issues Mol. Biol. 2019, 30, 39–58. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI continuum in Phytophthora-plant interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef]
- He, Z.; Webster, S.; He, S.Y. Growth–defense trade-offs in plants. Curr. Biol. 2022, 32, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zheng, F.; Wei, S.; Zhang, S.; Li, G.; Cao, P.; Zhao, S. Evolution of disease defense genes and their regulators in plants. Int. J. Mol. Sci. 2019, 20, 335. [Google Scholar] [CrossRef]
- Kumar, S.; Korra, T.; Thakur, R.; Arutselvan, R.; Kashyap, A.S.; Nehela, Y.; Chaplygin, V.; Minkina, T.; Keswani, C. Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress 2023, 8, 100154. [Google Scholar] [CrossRef]
- Jia, Y.; Wei, K.; Qin, J.; Zhai, W.; Li, Q.; Li, Y. The roles of micrornas in the regulation of rice–pathogen interactions. Plants 2025, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Rai, A.; Gupta, S.; Vijayan, J.; Devanna, B.; Ray, S. Rice blast management through host-plant resistance: Retrospect and prospects. Agric. Res. 2012, 1, 37–52. [Google Scholar] [CrossRef]
- Rana, N.; Rahim, M.S.; Kaur, G.; Bansal, R.; Kumawat, S.; Roy, J.; Deshmukh, R.; Sonah, H.; Sharma, T.R. Applications and challenges for efficient exploration of omics interventions for the enhancement of nutritional quality in rice (Oryza sativa L.). Crit. Rev. Food Sci. Nutr. 2020, 60, 3304–3320. [Google Scholar] [CrossRef]
- Mushtaq, M.; Ahmad Dar, A.; Skalicky, M.; Tyagi, A.; Bhagat, N.; Basu, U.; Bhat, B.A.; Zaid, A.; Ali, S.; Dar, T.-U.-H. CRISPR-based genome editing tools: Insights into technological breakthroughs and future challenges. Genes 2021, 12, 797. [Google Scholar] [CrossRef]
- Kababji, A.M.; Butt, H.; Mahfouz, M. Synthetic directed evolution for targeted engineering of plant traits. Front. Plant Sci. 2024, 15, 1449579. [Google Scholar] [CrossRef]
- Nadarajah, K.K.; Abdul Rahman, N.S.N. The role of non-coding RNA in Rice Immunity. Agronomy 2021, 12, 39. [Google Scholar] [CrossRef]
- Wang, H.; Meng, J.; Peng, X.; Tang, X.; Zhou, P.; Xiang, J.; Deng, X. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 2015, 89, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, S.; Zhou, X.; Zhao, C.; Guo, L.; Zhang, J.; Liu, F.; Huo, Q.; Zhao, W.; Guo, Z. Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice. Plant Cell 2023, 35, 2391–2412. [Google Scholar] [CrossRef] [PubMed]
- Ravarani, C.N.; Chalancon, G.; Breker, M.; De Groot, N.S.; Babu, M.M. Affinity and competition for TBP are molecular determinants of gene expression noise. Nat. Commun. 2016, 7, 10417. [Google Scholar] [CrossRef]
- Yang, D.; Li, S.; Xiao, Y.; Lu, L.; Zheng, Z.; Tang, D.; Cui, H. Transcriptome analysis of rice response to blast fungus identified core genes involved in immunity. Plant Cell Environ. 2021, 44, 3103–3121. [Google Scholar] [CrossRef]
- Moro, S.G.; Hermans, C.; Ruiz-Orera, J.; Albà, M.M. Impact of uORFs in mediating regulation of translation in stress conditions. BMC Mol. Cell Biol. 2021, 22, 29. [Google Scholar] [CrossRef]
- Lu, X.; He, Y.; Guo, J.-Q.; Wang, Y.; Yan, Q.; Xiong, Q.; Shi, H.; Hou, Q.; Yin, J.; An, Y.-B. Dynamics of epitranscriptomes uncover translational reprogramming directed by ac4C in rice during pathogen infection. Nat. Plants 2024, 10, 1548–1561. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Plant translational reprogramming for stress resilience. Front. Plant Sci. 2023, 14, 1151587. [Google Scholar] [CrossRef]
- Han, Z.; Li, F.; Qiao, W.; Zheng, X.; Cheng, Y.; Zhang, L.; Huang, J.; Wang, Y.; Lou, D.; Xing, M. Global whole-genome comparison and analysis to classify subpopulations and identify resistance genes in weedy rice relevant for improving crops. Front. Plant Sci. 2023, 13, 1089445. [Google Scholar] [CrossRef]
- Rasool, F.; Uzair, M.; Naeem, M.K.; Rehman, N.; Afroz, A.; Shah, H.; Khan, M.R. Phenylalanine ammonia-lyase (PAL) genes family in wheat (Triticum aestivum L.): Genome-wide characterization and expression profiling. Agronomy 2021, 11, 2511. [Google Scholar] [CrossRef]
- Wei, L.; Wang, D.; Gupta, R.; Kim, S.T.; Wang, Y. A proteomics insight into advancements in the rice–microbe interaction. Plants 2023, 12, 1079. [Google Scholar] [CrossRef]
- Saha, B.; Nayak, J.; Srivastava, R.; Samal, S.; Kumar, D.; Chanwala, J.; Dey, N.; Giri, M.K. Unraveling the involvement of WRKY TFs in regulating plant disease defense signaling. Planta 2024, 259, 7. [Google Scholar] [CrossRef]
- Jia, L.; Mao, Y.; Ji, Q.; Dersh, D.; Yewdell, J.W.; Qian, S.B. Decoding mRNA translatability and stability from the 5′ UTR. Nat. Struct. Mol. Biol. 2020, 27, 814–821. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, T.; Zhu, G.; Wu, B.; Zhang, C.; Zhu, H. LncRNAs exert indispensable roles in orchestrating the interaction among diverse noncoding RNAs and enrich the regulatory network of plant growth and its adaptive environmental stress response. Hortic. Res. 2023, 10, uhad234. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Cao, Q.; Li, Y.; He, M.; Liu, X. Advances in cis-element-and natural variation-mediated transcriptional regulation and applications in gene editing of major crops. J. Exp. Bot. 2023, 74, 5441–5457. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Purohit, J.; Mehta, S.; Parmar, H.; Karippadakam, S.; Rashid, A.; Balamurugan, A.; Bansal, S.; Prakash, G.; Achary, V.M.M. Precision genome editing toolbox: Applications and approaches for improving rice’s genetic resistance to pathogens. Agronomy 2022, 12, 565. [Google Scholar] [CrossRef]
- Kumar, K.; Mandal, S.N.; Neelam, K.; de Los Reyes, B.G. MicroRNA-mediated host defense mechanisms against pathogens and herbivores in rice: Balancing gains from genetic resistance with trade-offs to productivity potential. BMC Plant Biol. 2022, 22, 351. [Google Scholar] [CrossRef] [PubMed]
- Val-Torregrosa, B.; Bundó, M.; San Segundo, B. Crosstalk between nutrient signalling pathways and immune responses in rice. Agriculture 2021, 11, 747. [Google Scholar] [CrossRef]
- Singh, P.K.; Nag, A.; Arya, P.; Kapoor, R.; Singh, A.; Jaswal, R.; Sharma, T.R. Prospects of understanding the molecular biology of disease resistance in rice. Int. J. Mol. Sci. 2018, 19, 1141. [Google Scholar] [CrossRef]
- Bernard, V.; Brunaud, V.; Lecharny, A. TC-motifs at the TATA-box expected position in plant genes: A novel class of motifs involved in the transcription regulation. BMC Genom. 2010, 11, 166. [Google Scholar] [CrossRef]
- Liu, W.X.; Liu, H.L.; Chai, Z.J.; Xu, X.P.; Song, Y.R.; Qu, L.Q. Evaluation of seed storage-protein gene 5′ untranslated regions in enhancing gene expression in transgenic rice seed. Theor. Appl. Genet. 2010, 121, 1267–1274. [Google Scholar] [CrossRef]
- Baldrich, P.; Campo, S.; Wu, M.-T.; Liu, T.-T.; Hsing, Y.-I.C.; Segundo, B.S. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sanuy, F.; Peris-Peris, C.; Tomiyama, S.; Okada, K.; Hsing, Y.-I.; San Segundo, B.; Campo, S. Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus. BMC Plant Biol. 2019, 19, 563. [Google Scholar] [CrossRef]
- Shen, R.; Yao, Q.; Zhong, D.; Zhang, X.; Li, X.; Cao, X.; Dong, C.; Tian, Y.; Zhu, J.-K.; Lu, Y. Targeted insertion of regulatory elements enables translational enhancement in rice. Front. Plant Sci. 2023, 14, 1134209. [Google Scholar] [CrossRef]
- Wang, H.; Chen, M.; Zhang, D.; Meng, X.; Yan, J.; Chu, J.; Li, J.; Yu, H. Shaping rice Green Revolution traits by engineering ATG immediate upstream 5′-UTR sequences of OsSBI and OsHTD1. Plant Biotechnol. J. 2023, 22, 532. [Google Scholar] [CrossRef]
- He, F.; Liu, Q.; Zheng, L.; Cui, Y.; Shen, Z.; Zheng, L. RNA-Seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 2015, 6, 1136. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Luo, S.; Yang, D.; Huang, J.; Jiang, X.; Yu, S.; Fu, J.; Zhou, D.; Chen, X.; He, H. Alternative polyadenylation profiles of susceptible and resistant rice (Oryza sativa L.) in response to bacterial leaf blight using RNA-seq. BMC Plant Biol. 2024, 24, 145. [Google Scholar] [CrossRef]
- Hong, H.; Liu, Y.; Zhang, H.; Xiao, J.; Li, X.; Wang, S. Small RNAs and gene network in a durable disease resistance gene—Mediated defense responses in rice. PLoS ONE 2015, 10, e0137360. [Google Scholar] [CrossRef] [PubMed]
- Baruah, I.; Baldodiya, G.M.; Sahu, J.; Baruah, G. Dissecting the role of promoters of pathogen-sensitive genes in plant defense. Curr. Genom. 2020, 21, 491–503. [Google Scholar] [CrossRef]
- Xing, D.-H.; Lai, Z.-B.; Zheng, Z.-Y.; Vinod, K.; Fan, B.-F.; Chen, Z.-X. Stress-and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 2008, 1, 459–470. [Google Scholar] [CrossRef]
- Mishal, R.; Luna-Arias, J.P. Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation. Gene 2022, 833, 146581. [Google Scholar] [CrossRef]
- Dhatterwal, P.; Sharma, N.; Prasad, M. Decoding the functionality of plant transcription factors. J. Exp. Bot. 2024, 75, 4745–4759. [Google Scholar] [CrossRef]
- Shrestha, A.; Khan, A.; Dey, N. Cis–trans engineering: Advances and perspectives on customized transcriptional regulation in plants. Mol. Plant 2018, 11, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, A.; Purohit, J.; Tiwari, K.K.; Deshmukh, R. Targeting transcription factors for plant disease resistance. Curr. Sci. 2019, 117, 1598–1607. [Google Scholar] [CrossRef]
- Li, S.; Peng, Y.; and Panchenko, A.R. DNA methylation: Precise modulation of chromatin structure and dynamics. Curr. Opin. Struct. Biol. 2022, 75, 102430. [Google Scholar] [CrossRef]
- Oliveira-Garcia, E.; Yan, X.; Oses-Ruiz, M.; de Paula, S.; Talbot, N.J. Effector-triggered susceptibility by the rice blast fungus Magnaporthe oryzae. New Phytol. 2024, 241, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Pandey, V.K.; Jha, A.K.; Srivastava, S.; Jakhar, S.; Singh, G.; Rustagi, S.; Malik, S.; Choudhary, P. Intricacies of Plants’ Innate Immune Responses and their Dynamic Relationship with Fungi: A Review. Microbiol. Res. 2024, 285, 127758. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Ortiz-Urquiza, A.; Ying, S.-H.; Zhang, J.-X.; Keyhani, N.O. Interaction between TATA-binding protein (TBP) and multiprotein bridging factor-1 (MBF1) from the filamentous insect pathogenic fungus Beauveria bassiana. PLoS ONE 2015, 10, e0140538. [Google Scholar] [CrossRef]
- Sinha, A.; Narula, K.; Bhola, L.; Sengupta, A.; Choudhary, P.; Nalwa, P.; Kumar, M.; Elagamey, E.; Chakraborty, N.; Chakraborty, S. Proteomic signatures uncover phenotypic plasticity of susceptible and resistant genotypes by wall remodelers in rice blast. Plant Cell Environ. 2024, 47, 3846–3864. [Google Scholar] [CrossRef]
- Kou, Y.; Shi, H.; Qiu, J.; Tao, Z.; Wang, W. Effectors and environment modulating rice blast disease: From understanding to effective control. Trends Microbiol. 2024, 32, 1007–1020. [Google Scholar] [CrossRef]
- Das, A.; Moin, M.; Sahu, A.; Kshattry, M.; Kirti, P.B.; Barah, P. Time-course transcriptome analysis identifies rewiring patterns of transcriptional regulatory networks in rice under Rhizoctonia solani infection. Gene 2022, 828, 146468. [Google Scholar] [CrossRef]
- Kumari, D.; Prasad, B.D.; Dwivedi, P.; Sahni, S.; Kumar, M.; Alamri, S.; Adil, M.F.; Alakeel, K.A. Comprehensive analysis of transcription factor binding sites and expression profiling of rice pathogenesis related genes (OsPR1). Front. Plant Sci. 2024, 15, 1463147. [Google Scholar] [CrossRef]
- Liu, W.; Wang, G.-L. Plant innate immunity in rice: A defense against pathogen infection. Natl. Sci. Rev. 2016, 3, 295–308. [Google Scholar] [CrossRef]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Kummari, D.; Palakolanu, S.R.; Kishor, P.K.; Bhatnagar-Mathur, P.; Singam, P.; Vadez, V.; Sharma, K.K. An update and perspectives on the use of promoters in plant genetic engineering. J. Biosci. 2020, 45, 10-1007. [Google Scholar] [CrossRef]
- Ding, L.; Xu, X.; Kong, W.; Xia, X.; Zhang, S.; Liu, L.-W.; Liu, A.; Zou, L. Genome-wide identification and expression analysis of rice NLR genes responsive to the infections of Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae. Physiol. Mol. Plant Pathol. 2020, 111, 101488. [Google Scholar] [CrossRef]
- Huang, Z.; Xue, Z.; Zhao, X.; Wu, C.; Sun, Y.; Kou, X. Transcription factors, potential regulatory targets in fruit defense responses to pathogens. Postharvest Biol. Technol. 2023, 206, 112589. [Google Scholar] [CrossRef]
- Luo, Y.; Teng, P.; Fabellar, N.; TeBeest, D. The effects of global temperature change on rice leaf blast epidemics: A simulation study in three agroecological zones. Agric. Ecosyst. Environ. 1998, 68, 187–196. [Google Scholar] [CrossRef]
- Ma, S.; Xu, S.; Tao, H.; Huang, Y.; Feng, C.; Huang, G.; Lin, S.; Sun, Y.; Chen, X.; Fabrice Kabore, M.A. OsBRW1, a novel blast-resistant gene, coded a NBS-LRR protein to interact with OsSRFP1 to balance rice growth and resistance. Plant Biotechnol. J. 2025, 23, 250–267. [Google Scholar] [CrossRef]
- Vijayan, J.; Devanna, B.; Singh, N.K.; Sharma, T.R. Cloning and functional validation of early inducible Magnaporthe oryzae responsive CYP76M7 promoter from rice. Front. Plant Sci. 2015, 6, 371. [Google Scholar] [CrossRef]
- Dong, H. Application of genome editing techniques to regulate gene expression in crops. BMC Plant Biol. 2024, 24, 100. [Google Scholar] [CrossRef]
- Meng, X.-B.; Zhao, W.-S.; Lin, R.-M.; Wang, M.; Peng, Y.-L. Identification of a novel rice bZIP-type transcription factor gene, OsbZIP1, involved in response to infection of Magnaporthe grisea. Plant Mol. Biol. Report. 2005, 23, 301–302. [Google Scholar] [CrossRef]
- Lin, R.; Zhao, W.; Meng, X.; Wang, M.; Peng, Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 2007, 172, 120–130. [Google Scholar] [CrossRef]
- Ryczek, N.; Łyś, A.; Makałowska, I. The functional meaning of 5′ UTR in protein-coding genes. Int. J. Mol. Sci. 2023, 24, 2976. [Google Scholar] [CrossRef]
- Rasekhian, M.; Roohvand, F.; Habtemariam, S.; Marzbany, M.; Kazemimanesh, M. The role of 3′UTR of RNA viruses on mRNA stability and translation enhancement. Mini Rev. Med. Chem. 2021, 21, 2389–2398. [Google Scholar] [CrossRef]
- Ahmed, F.; Benedito, V.A.; Zhao, P.X. Mining functional elements in messenger RNAs: Overview, challenges, and perspectives. Front. Plant Sci. 2011, 2, 84. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Ivanov, I.P.; Hinnebusch, A.G. Translational regulation by uORFs and start codon selection stringency. Genes Dev. 2023, 37, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Raman, V.; Simon, S.A.; Romag, A.; Demirci, F.; Mathioni, S.M.; Zhai, J.; Meyers, B.C.; Donofrio, N.M. Physiological stressors and invasive plant infections alter the small RNA transcriptome of the rice blast fungus, Magnaporthe oryzae. BMC Genom. 2013, 14, 326. [Google Scholar] [CrossRef]
- Traubenik, S.; Ferrari, M.; Blanco, F.A.; Zanetti, M.E. Translational regulation in pathogenic and beneficial plant–microbe interactions. Biochem. J. 2021, 478, 2775–2788. [Google Scholar] [CrossRef] [PubMed]
- Tora, L.; Timmers, H.T.M. The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem. Sci. 2010, 35, 309–314. [Google Scholar] [CrossRef]
- Hitti, E.; Al-Yahya, S.; Al-Saif, M.; Mohideen, P.; Mahmoud, L.; Polyak, S.J.; Khabar, K.S. A versatile ribosomal protein promoter-based reporter system for selective assessment of RNA stability and post-transcriptional control. RNA 2010, 16, 1245–1255. [Google Scholar] [CrossRef]
- Jabre, I.; Reddy, A.S.; Kalyna, M.; Chaudhary, S.; Khokhar, W.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 2019, 47, 2716–2726. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, S.; Mori, M.; Sugano, S.; Takatsuji, H. Transcription factor WRKY62 plays a role in pathogen defense and hypoxia-responsive gene expression in rice. Plant Cell Physiol. 2016, 57, 2541–2551. [Google Scholar] [CrossRef]
- Liu, D.; He, J.; Li, Q.; Zhang, X.; Wang, Y.; Sun, Q.; Wang, W.; Zhang, M.; Wang, Y.; Xu, H. A WRKY transcription factor confers broad-spectrum resistance to biotic stresses and yield stability in rice. Proc. Natl. Acad. Sci. USA 2025, 122, e2411164122. [Google Scholar] [CrossRef]
- Jimmy, J.L.; Babu, S. Role of Os WRKY transcription factors in rice disease resistance. Trop. Plant Pathol. 2015, 40, 355–361. [Google Scholar] [CrossRef]
- Tao, Z.; Yan, F.; Hahn, M.; Ma, Z. Regulatory roles of epigenetic modifications in plant-phytopathogen interactions. Crop Health 2023, 1, 6. [Google Scholar] [CrossRef]
- Tan, Y.-y.; Du, H.; Wu, X.; Liu, Y.-h.; Jiang, M.; Song, S.-y.; Wu, L.; Shu, Q.-y. Gene editing: An instrument for practical application of gene biology to plant breeding. J. Zhejiang Univ. Sci. B 2020, 21, 460. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, S.; Ebana, K.; Yamamoto, T.; Yano, M. Integration of Genomics into Rice Breeding. Rice 2010, 3, 131–137. [Google Scholar] [CrossRef]
- Samanta, S.; Roychoudhury, A. Molecular crosstalk of jasmonate with major phytohormones and plant growth regulators during diverse stress responses. J. Plant Growth Regul. 2025, 44, 62–88. [Google Scholar] [CrossRef]
- Kamzeeva, P.N.; Alferova, V.A.; Korshun, V.A.; Varizhuk, A.M.; Aralov, A.V. 5ation regulation in eukaryotes: Current understanding and methodological challenges. Int. J. Mol. Sci. 2025, 26, 1187. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Y.; Gu, P.; Naz, M. Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation. Plants 2025, 14, 2002. [Google Scholar] [CrossRef]
- Hardy, E.C.; Balcerowicz, M. Untranslated yet indispensable—UTRs act as key regulators in the environmental control of gene expression. J. Exp. Bot. 2024, 75, 4314–4331. [Google Scholar] [CrossRef]
- Liu, S.; Li, B.; Liang, Q.; Liu, A.; Qu, L.; Yang, J. Classification and function of RNA–protein interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1601. [Google Scholar] [CrossRef] [PubMed]
- Mutsuro-Aoki, H.; Teramura, H.; Tamukai, R.; Fukui, M.; Kusano, H.; Schepetilnikov, M.; Ryabova, L.A.; Shimada, H. Dissection of a rice OsMac1 mRNA 5′UTR to uncover regulatory elements that are responsible for its efficient translation. PLoS ONE 2021, 16, e0253488. [Google Scholar] [CrossRef] [PubMed]
- Khanale, V.; Bhattacharya, A.; Prashar, M.; Char, B. Genome editing interventions to combat rice blast disease. Plant Biotechnol. Rep. 2023, 17, 1–13. [Google Scholar] [CrossRef]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. CMLS 2012, 69, 3613–3634. [Google Scholar] [CrossRef]
- Nasir, F.; Tian, L.; Chang, C.; Li, X.; Gao, Y.; Tran, L.-S.P.; Tian, C. Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection. Semin. Cell Dev. Biol. 2018, 83, 95–105. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.; Mieulet, D.; Obata, T.; Fernie, A.R.; Guiderdoni, E.; Mueller-Roeber, B. MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J. 2013, 76, 258–273. [Google Scholar] [CrossRef]
- Malik, S.; Roeder, R.G. Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators. Nat. Rev. Genet. 2023, 24, 767–782. [Google Scholar] [CrossRef]
- Sharma, S.; Kapoor, S.; Ansari, A.; Tyagi, A.K. The general transcription factors (GTFs) of RNA polymerase II and their roles in plant development and stress responses. Crit. Rev. Biochem. Mol. Biol. 2024, 59, 267–309. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; and Barna, M. Functional 5nal 5a, M. Functional 5onal 5. The general transcription factors (GTFs) of RNA polymeras. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Lopes, N.d.S.; Santos, A.S.; De Novais, D.P.S.; Pirovani, C.P.; Micheli, F. Pathogenesis-related protein 10 in resistance to biotic stress: Progress in elucidating functions, regulation and modes of action. Front. Plant Sci. 2023, 14, 1193873. [Google Scholar] [CrossRef]
- Wang, H.L.V.; Chekanova, J.A. Small RNAs: Essential regulators of gene expression and defenses against environmental stresses in plants. Wiley Interdiscip. Rev. RNA 2016, 7, 356–381. [Google Scholar] [CrossRef]
- Hannan Parker, A.; Wilkinson, S.W.; Ton, J. Epigenetics: A catalyst of plant immunity against pathogens. New Phytol. 2022, 233, 66–83. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Chang, H.; Zhan, M.; Qin, Q.-M.; Zhang, B.; Li, Z.; Liu, Y. Mining Magnaporthe oryzae sRNAs with potential Transboundary regulation of Rice genes associated with growth and defense through expression profile analysis of the pathogen-infected Rice. Front. Genet. 2019, 10, 296. [Google Scholar] [CrossRef]
- Meng, Q.; Gupta, R.; Min, C.W.; Kwon, S.W.; Wang, Y.; Je, B.I.; Kim, Y.-J.; Jeon, J.-S.; Agrawal, G.K.; Rakwal, R. Proteomics of Rice—Magnaporthe oryzae interaction: What have we learned so far? Front. Plant Sci. 2019, 10, 1383. [Google Scholar] [CrossRef]
- Saidi, A.; Hajibarat, Z.; Hajibarat, Z. Identification of responsive genes and analysis of genes with bacterial-inducible cis-regulatory elements in the promoter regions in Oryza sativa L. Acta Agric. Slov. 2020, 116, 115–123. [Google Scholar] [CrossRef]
- Sultana, R.; Imam, Z.; Kumar, R.R.; Banu, V.S.; Nahakpam, S.; Bharti, R.; Bharadwaj, C.; Singh, A.K.; Pasala, R.K.; Singh, D.R. Signaling and defence mechanism of jasmonic and salicylic acid response in pulse crops: Role of WRKY transcription factors in stress response. J. Plant Growth Regul. 2025, 44, 5–21. [Google Scholar] [CrossRef]
- Singh, J.; Gupta, S.K.; Devanna, B.N.; Singh, S.; Upadhyay, A.; Sharma, T.R. Blast resistance gene Pi54 over-expressed in rice to understand its cellular and sub-cellular localization and response to different pathogens. Sci. Rep. 2020, 10, 5243. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, H.; Bi, Y.; Song, F. Rice E3 ubiquitin ligases: From key modulators of host immunity to potential breeding applications. Plant Commun. 2024, 5, 101128. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Aftab, T. Phytohormones, plant growth regulators and signaling molecules: Cross-talk and stress responses. Plant Cell Rep. 2021, 40, 1301–1303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Dabi, T.; Lamb, C. TATA box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 1995, 7, 1681–1689. [Google Scholar]
- Ballini, E.; Morel, J.-B.; Droc, G.; Price, A.; Courtois, B.; Notteghem, J.-L.; Tharreau, D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant-Microbe Interact. 2008, 21, 859–868. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Variar, M.; Prashanthi, S.; Singh, A.; Singh, U.; Chand, D.; Singh, N. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef]
- Huq, M.A.; Akter, S.; Nou, I.S.; Kim, H.T.; Jung, Y.J.; Kang, K.K. Identification of functional SNPs in genes and their effects on plant phenotypes. J. Plant Biotechnol. 2016, 43, 1–11. [Google Scholar] [CrossRef]
- Younas, M.U.; Qasim, M.; Ahmad, I.; Feng, Z.; Iqbal, R.; Jiang, X.; Zuo, S. Exploring the molecular mechanisms of rice blast resistance and advances in breeding for disease tolerance. Mol. Biol. Rep. 2024, 51, 1093. [Google Scholar] [CrossRef]
- Sharma, S.K.; Sharma, D.; Meena, R.P.; Yadav, M.K.; Hosahatti, R.; Dubey, A.K.; Sharma, P.; Kumar, S.; Pramesh, D.; Nabi, S.U. Recent insights in rice blast disease resistance. In Blast Disease of Cereal Crops: Evolution and Adaptation in Context of Climate Change; Springer: Cham, Switzerland, 2021; pp. 89–123. [Google Scholar]
- Chen, Y.; Sun, A.; Wang, M.; Zhu, Z.; Ouwerkerk, P.B. Functions of the CCCH type zinc finger protein OsGZF1 in regulation of the seed storage protein GluB-1 from rice. Plant Mol. Biol. 2014, 84, 621–634. [Google Scholar] [CrossRef]
- Villao-Uzho, L.; Chávez-Navarrete, T.; Pacheco-Coello, R.; Sánchez-Timm, E.; Santos-Ordóñez, E. Plant promoters: Their identification, characterization, and role in gene regulation. Genes 2023, 14, 1226. [Google Scholar] [CrossRef]
- Römer, P.; Recht, S.; Strauß, T.; Elsaesser, J.; Schornack, S.; Boch, J.; Wang, S.; Lahaye, T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010, 187, 1048–1057. [Google Scholar] [CrossRef]
- Brooks, E.G.; Elorriaga, E.; Liu, Y.; Duduit, J.R.; Yuan, G.; Tsai, C.-J.; Tuskan, G.A.; Ranney, T.G.; Yang, X.; Liu, W. Plant promoters and terminators for high-precision bioengineering. BioDesign Res. 2023, 5, 0013. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhu, J.; Zhao, D.; Liu, Q.; Yang, Q.; Zhang, T. Targeting cis-regulatory elements for rice grain quality improvement. Front. Plant Sci. 2021, 12, 705834. [Google Scholar] [CrossRef] [PubMed]
- Ofori, A.D.; Zheng, T.; Titriku, J.K.; Appiah, C.; Xiang, X.; Kandhro, A.G.; Ahmed, M.I.; Zheng, A. The Role of Genetic Resistance in Rice Disease Management. Int. J. Mol. Sci. 2025, 26, 956. [Google Scholar] [CrossRef]
- Wang, L.; Fu, J.; Shen, Q.; Wang, Q. OsWRKY10 extensively activates multiple rice diterpenoid phytoalexin biosynthesis genes to enhance rice blast resistance. Plant J. 2023, 115, 758–771. [Google Scholar] [CrossRef]
- Le, V.T.; Kim, M.-S.; Jung, Y.-J.; Kang, K.-K.; Cho, Y.-G. Research trends and challenges of using CRISPR/Cas9 for improving rice productivity. Agronomy 2022, 12, 164. [Google Scholar] [CrossRef]
- Zhu, X.-T.; Zhou, R.; Che, J.; Zheng, Y.-Y.; ul Qamar, M.T.; Feng, J.-W.; Zhang, J.; Gao, J.; Chen, L.-L. Ribosome profiling reveals the translational landscape and allele-specific translational efficiency in rice. Plant Commun. 2023, 4, 100457. [Google Scholar] [CrossRef]
- Ma, Y.; Guo, W.; Mou, Q.; Shao, X.; Lyu, M.; Garcia, V.; Kong, L.; Lewis, W.; Ward, C.; Yang, Z. Spatial imaging of glycoRNA in single cells with ARPLA. Nat. Biotechnol. 2024, 42, 608–616. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, G.; Liu, M.; Wang, L.; Lou, Y.; Baldwin, I.; Li, R. Multiomic analyses reveal key sectors of jasmonate-mediated defense responses in rice. Plant Cell 2024, 36, 3362–3377. [Google Scholar] [CrossRef]
- Samadder, P.; Sivamani, E.; Lu, J.; Li, X.; Qu, R. Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol. Genet. Genom. 2008, 279, 429–439. [Google Scholar] [CrossRef]
- Sharma, R.; De Vleesschauwer, D.; Sharma, M.K.; Ronald, P.C. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 2013, 6, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S. Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [CrossRef]
- Antony, A.; Veerappapillai, S.; Karuppasamy, R. Deciphering early responsive signature genes in rice blast disease: An integrated temporal transcriptomic study. J. Appl. Genet. 2024, 65, 665–681. [Google Scholar] [CrossRef]
- Kumar, G.R.; Sakthivel, K.; Sundaram, R.M.; Neeraja, C.N.; Balachandran, S.M.; Rani, N.S.; Viraktamath, B.C.; Madhav, M.S. Allele mining in crops: Prospects and potentials. Biotechnol. Adv. 2010, 28, 451–461. [Google Scholar] [CrossRef]
- Pan, S.; Wang, H.; Zhang, H.; Tang, Z.; Xu, L.; Yan, Z.; Hu, Y. UTR-Insight: Integrating deep learning for efficient 5′ UTR discovery and design. BMC Genom. 2025, 26, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sapkota, M.; van der Knaap, E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: Unlocking the neglected potential for crop improvement. Hortic. Res. 2020, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Rajput, N.; Younas, M.U.; Qasim, M.; Memon, S.P.; Memon, S.; El-Rahman, M.A.; Aghayeva, S.; Ercisli, S.; Iqbal, R.; Zuo, S. Understanding rice blast: Investigating biotechnological methods to speed up the development of robust rice cultivars. Genet. Resour. Crop Evol. 2025, 72, 1333–1352. [Google Scholar] [CrossRef]

| Cis-Element | Core Motif | Transcriptional Regulator | Biological Function in Rice Defense Against Blast Pathogen | Reference |
|---|---|---|---|---|
| TATA box | TATA(A/T)A(A/T) | TATA-binding protein (TBP), TFIID complex | Initiates transcription by recruiting RNA polymerase II; essential for activating defense gene promoters (e.g., PR, WRKY, NBS-LRR genes) during pathogen attack. | [35] |
| 5′UTR | Variable, includes upstream open reading frames (uORFs), secondary structures | RNA-binding proteins, translational regulators | Modulates mRNA translation efficiency and stability; influences timing and strength of defense gene expression in response to pathogen-induced signals (e.g., salicylic acid, jasmonic acid). | [51] |
| 5′UTR (uORFs) | Upstream open reading frames | Translational machinery components | uORFs in 5′UTRs can regulate downstream gene expression post-transcriptionally, affecting defense responses. | [52] |
| 5′UTR (miRNA targets) | miRNA binding sites | miRNAs (e.g., miR7695) | miRNAs can bind to 5′UTRs, modulating translation of defense-related genes. | [53] |
| 5′UTR (synthetic enhancers) | Synthetic sequences (e.g., AMVE) | Engineered regulatory elements | Insertion of synthetic enhancers into 5′UTRs can boost translation of defense genes. | [54] |
| 5′UTR (AUS modifications) | ATG upstream sequences | Genome editing tools (e.g., CRISPR) | Modifying immediate upstream sequences of start codons can influence translation efficiency of defense genes. | [55] |
| 5′UTR (lncRNA interactions) | Long non-coding RNA binding sites | lncRNAs | lncRNAs interacting with 5′UTRs can affect stability and translation of defense-related mRNAs. | [56] |
| 5′UTR (alternative polyadenylation) | Polyadenylation signals | Polyadenylation machinery | Alternative polyadenylation in 5′UTRs can lead to transcript variants affecting defense gene expression. | [57] |
| 5′UTR (miRNA-mediated regulation) | miRNA binding sites | miRNAs (e.g., miR393b) | miRNAs can regulate secretion pathways by targeting 5′UTRs, influencing defense protein exocytosis. | [31] |
| 5′UTR (sRNA networks) | Small RNA binding sites | sRNAs | sRNAs can modulate gene networks by interacting with 5′UTRs, affecting defense responses. | [58] |
| Target Gene(s)/TF | Regulatory Element or Mechanism | Key Findings | Approach Used | Reference |
|---|---|---|---|---|
| OsPR1 family | Promoter and TFBS (TATA box, W-box, GCC-box) | Identified TF binding motifs in OsPR1 promoters linked to blast response | In silico promoter and TFBS analysis + expression profiling | [72] |
| OsBRW1, OsSRFP1 | NBS-LRR gene regulation | OsBRW1 interacts with OsSRFP1 to regulate immunity and growth balance | Functional genomics + protein interaction assays | [79] |
| Multiple resistance genes | Promoter, TFs, genome editing targets | Reviews precision genome editing for blast resistance | CRISPR-Cas toolbox review | [46] |
| CYP76M7 | Promoter (early inducible) | Identified promoter activated by M. oryzae infection | Cloning + promoter::GUS assay | [80] |
| Multiple crop genes | Gene expression regulation via editing | Overview of genome editing regulating gene expression in crops incl. rice | Review (CRISPR, base editing, epigenome editing) | [81] |
| OsbZIP1 | bZIP TF gene | Novel bZIP TF induced by M. grisea, likely regulating defense genes | Expression analysis + TF identification | [82] |
| OsNAC19 | NAC TF gene | OsNAC19 expression induced upon blast infection | Expression profiling + gene characterization | [83] |
| Regulatory Layer | Component | Function | Role in Fungal Defense | Hormonal Integration | References |
|---|---|---|---|---|---|
| Transcription Initiation | TATA box | Core promoter element; recruits RNA polymerase II and general TFs | Enables rapid and accurate transcription of pathogenesis-related (PR) genes | Coordinates with SA/JA-induced TFs (e.g., WRKYs) | [72,82] |
| TF Binding and Specificity | W-box, TCA-motif, ABRE | Binding sites for WRKY, TGA, and bZIP TFs | Drives defense gene activation in response to Magnaporthe oryzae | SA and ABA responsive; WRKY-TGA cross-talk | [46,83] |
| Translational Regulation | 5′UTR uORFs | Modulate translation efficiency and ribosome scanning | Fine-tunes protein production during infection stress | ABA/SA modulated translational buffering | [79,81] |
| mRNA Stability and Localization | GC-rich motifs, hairpin loops in 5′UTR | Affect mRNA stability and translatability | Ensures sustained expression of defense proteins under biotic stress | JA/ABA signals influence mRNA decay/stabilization | [80] |
| Energy Efficiency | Combined promoter and 5′UTR elements | Orchestrate transcriptional and translational economy | Prevents overexpression, allowing timely, cost-efficient defense | Feedback regulation via SA-JA antagonism | [81] |
| Signal Amplification | Promoter-5′UTR synergy | Multiplexed regulation via cis-element clusters | Rapid upregulation of secondary metabolism and PR proteins | Hormonal signal amplification through TF cascades | [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Naz, M.; Zhang, Y.; Afzal, M.R. Deciphering the Contribution of TATA Box and 5′UTR to Defense Signaling in Rice Under Blast Infection. Biology 2025, 14, 1522. https://doi.org/10.3390/biology14111522
Fan X, Naz M, Zhang Y, Afzal MR. Deciphering the Contribution of TATA Box and 5′UTR to Defense Signaling in Rice Under Blast Infection. Biology. 2025; 14(11):1522. https://doi.org/10.3390/biology14111522
Chicago/Turabian StyleFan, Xiaoru, Misbah Naz, Yong Zhang, and Muhammad Rahil Afzal. 2025. "Deciphering the Contribution of TATA Box and 5′UTR to Defense Signaling in Rice Under Blast Infection" Biology 14, no. 11: 1522. https://doi.org/10.3390/biology14111522
APA StyleFan, X., Naz, M., Zhang, Y., & Afzal, M. R. (2025). Deciphering the Contribution of TATA Box and 5′UTR to Defense Signaling in Rice Under Blast Infection. Biology, 14(11), 1522. https://doi.org/10.3390/biology14111522





