Identification of the YABBY Gene Family in Cerasus humilis and Analysis of Expression Patterns During Different Growth Stages
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Description of Plant Material
2.3. Identification and Physicochemical Properties of the YABBY Gene in C. humilis
2.4. Phylogenetic Analysis
2.5. Gene Structure and Conserved Motif Analysis
2.6. Promoter Analysis
2.7. Chromosome Localization and Collinearity Analysis
2.8. Gene Expression Analysis
2.9. Real-Time Quantitative Analysis
2.10. Analysis of Potential Protein Interaction
2.11. Determination of Total Sugar and Total Flavone Content
2.12. Determination of Antioxidant Capacity
2.13. Statistical Analysis
3. Results
3.1. Identification of the ChYABBY Gene and Its Physical and Chemical Properties
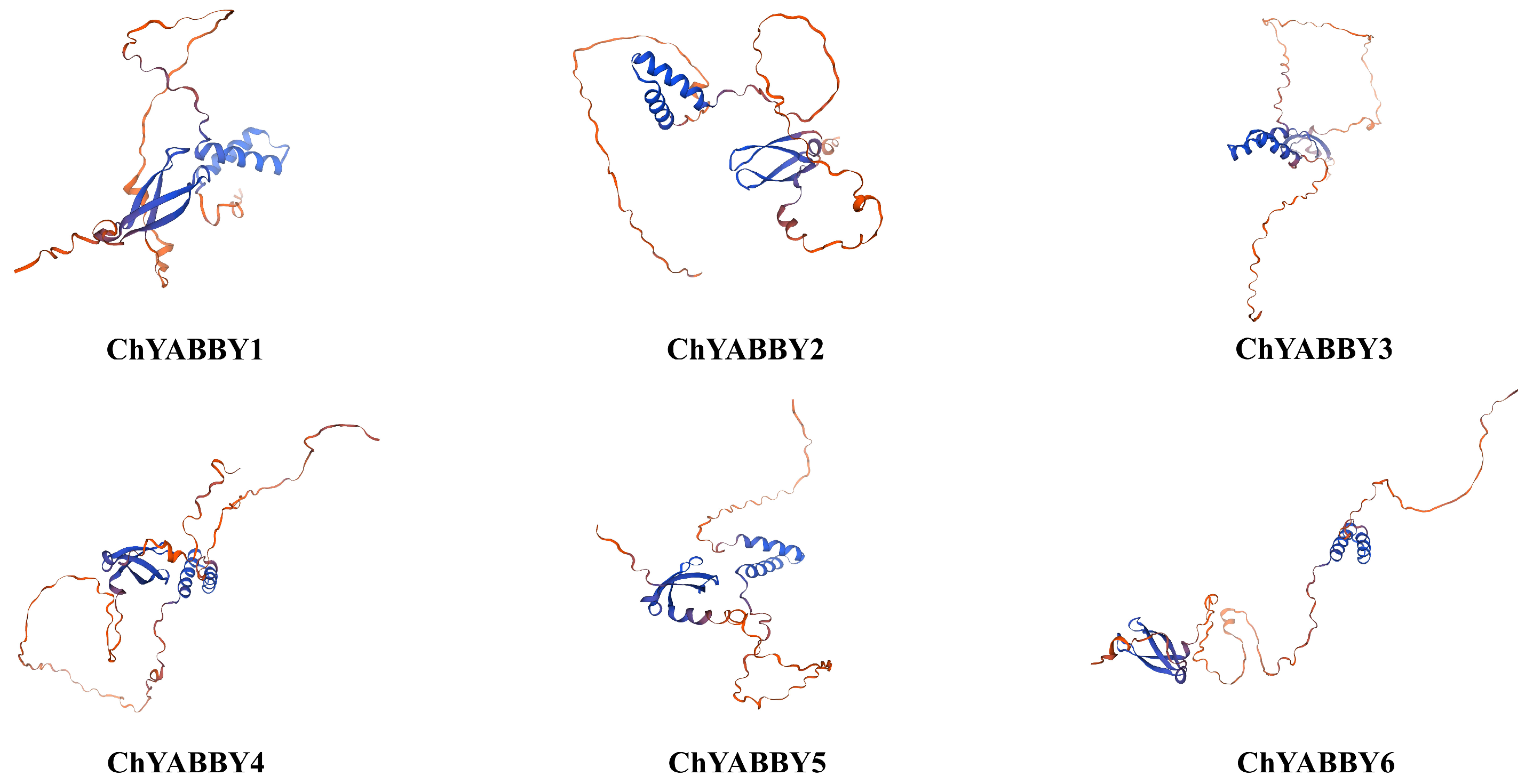
3.2. Structural Analysis of ChYABBY Protein and ChYABBY Gene
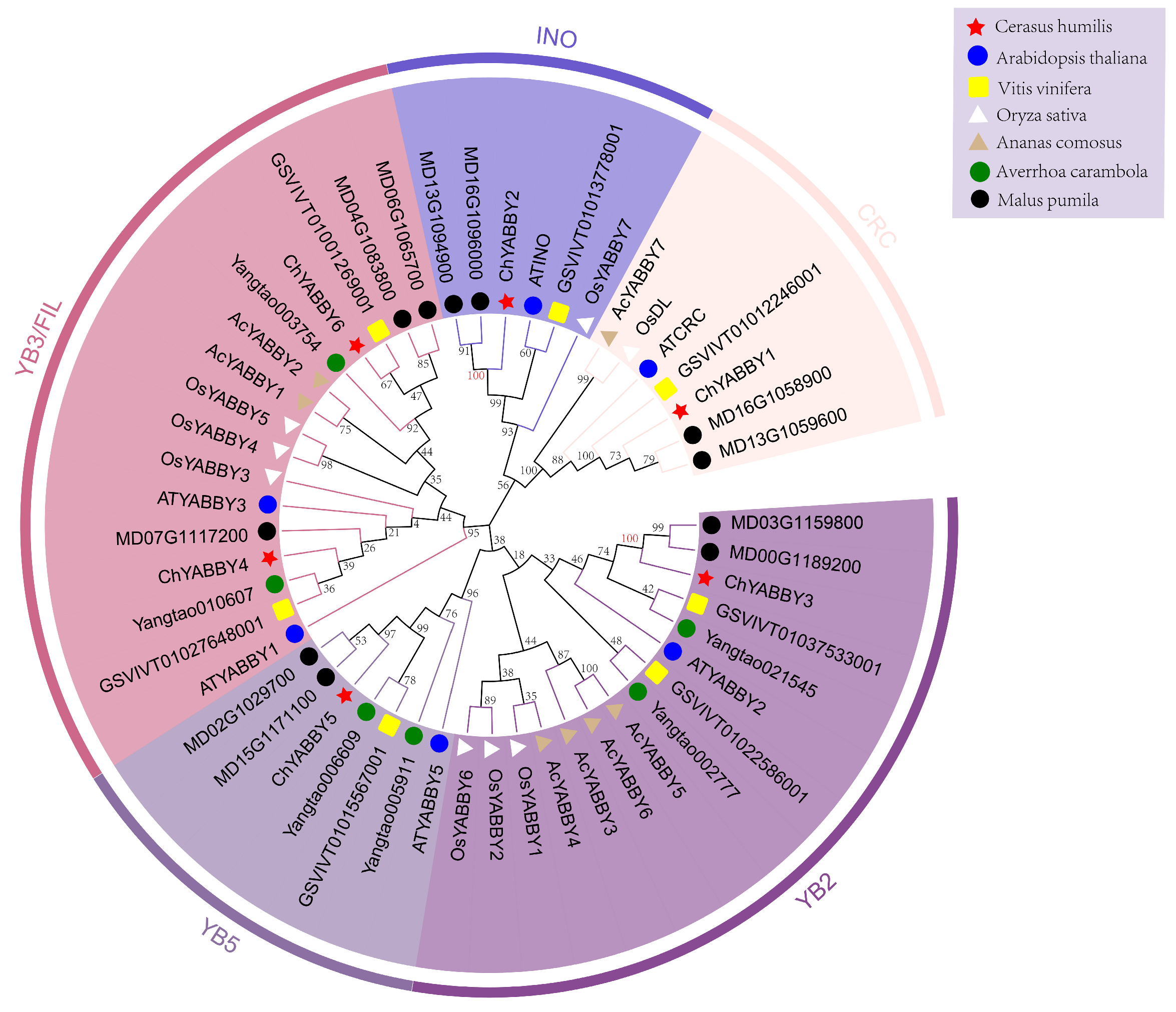
3.3. Phylogenetic Analysis
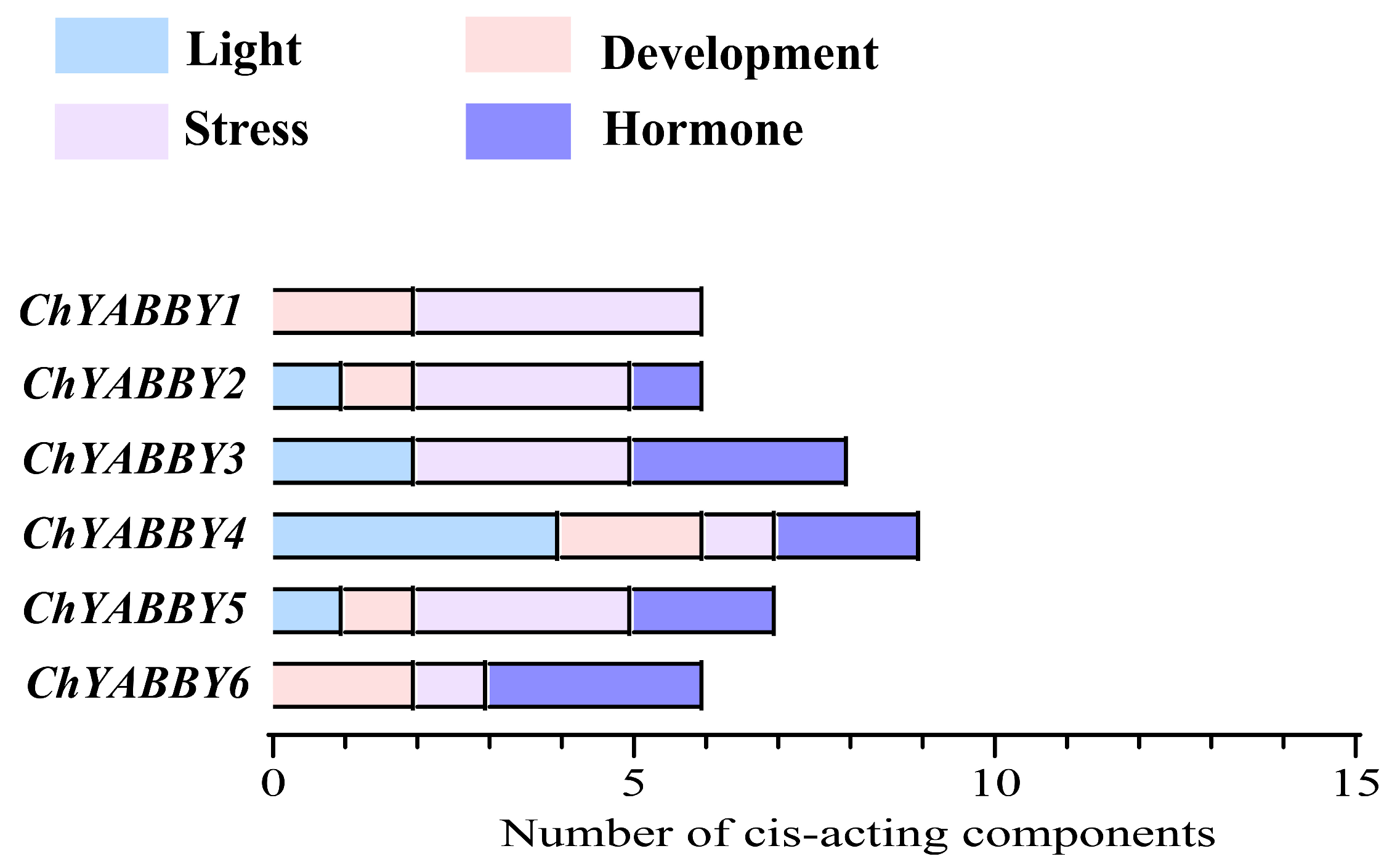
3.4. Promoter Analysis
3.5. Genomic Distribution and Duplication of ChYABBY Genes
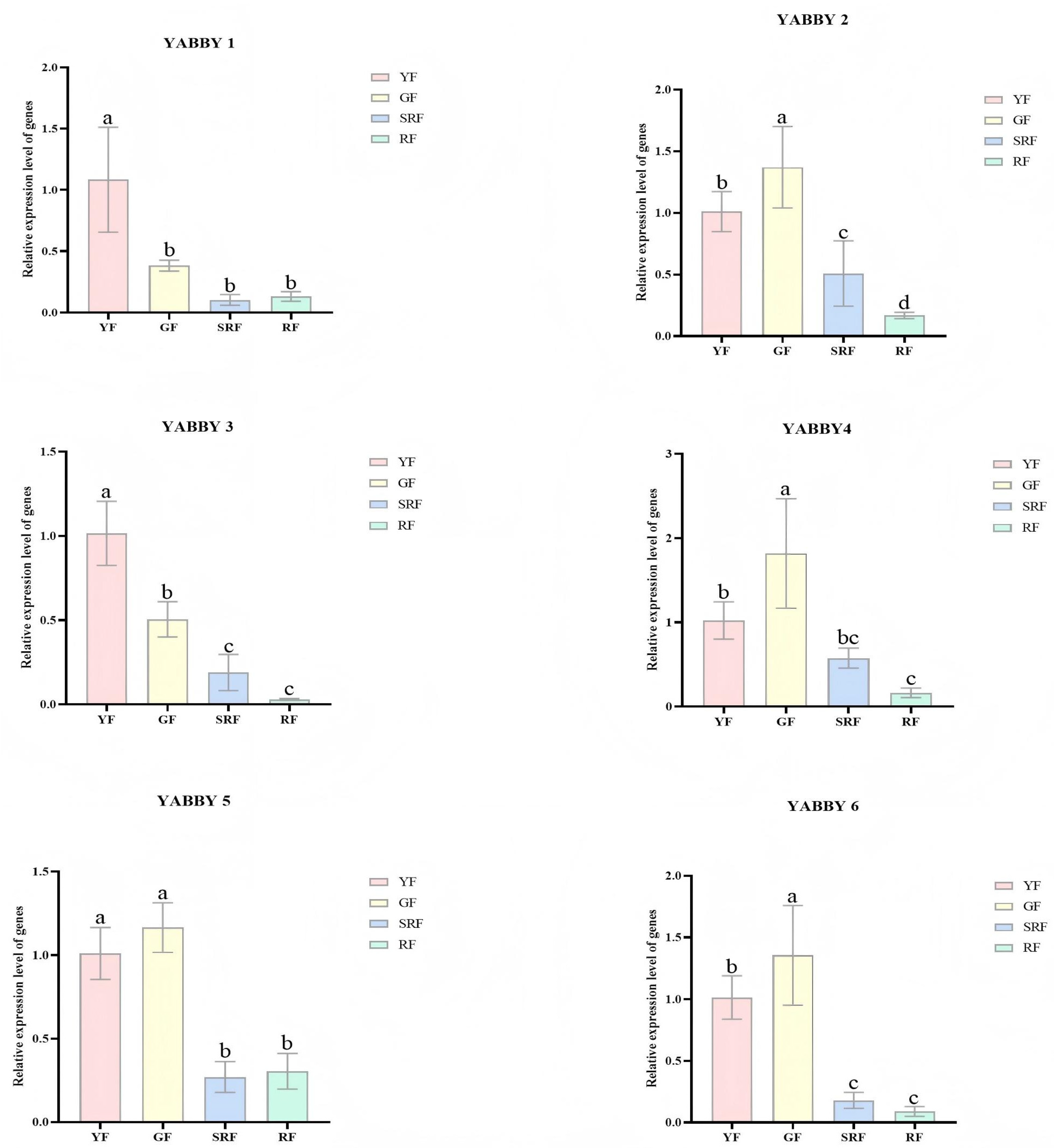
3.6. Targeted Validation of ChYABBY Gene Expression by qRT-PCR
3.7. Real-Time Quantitative Analysis
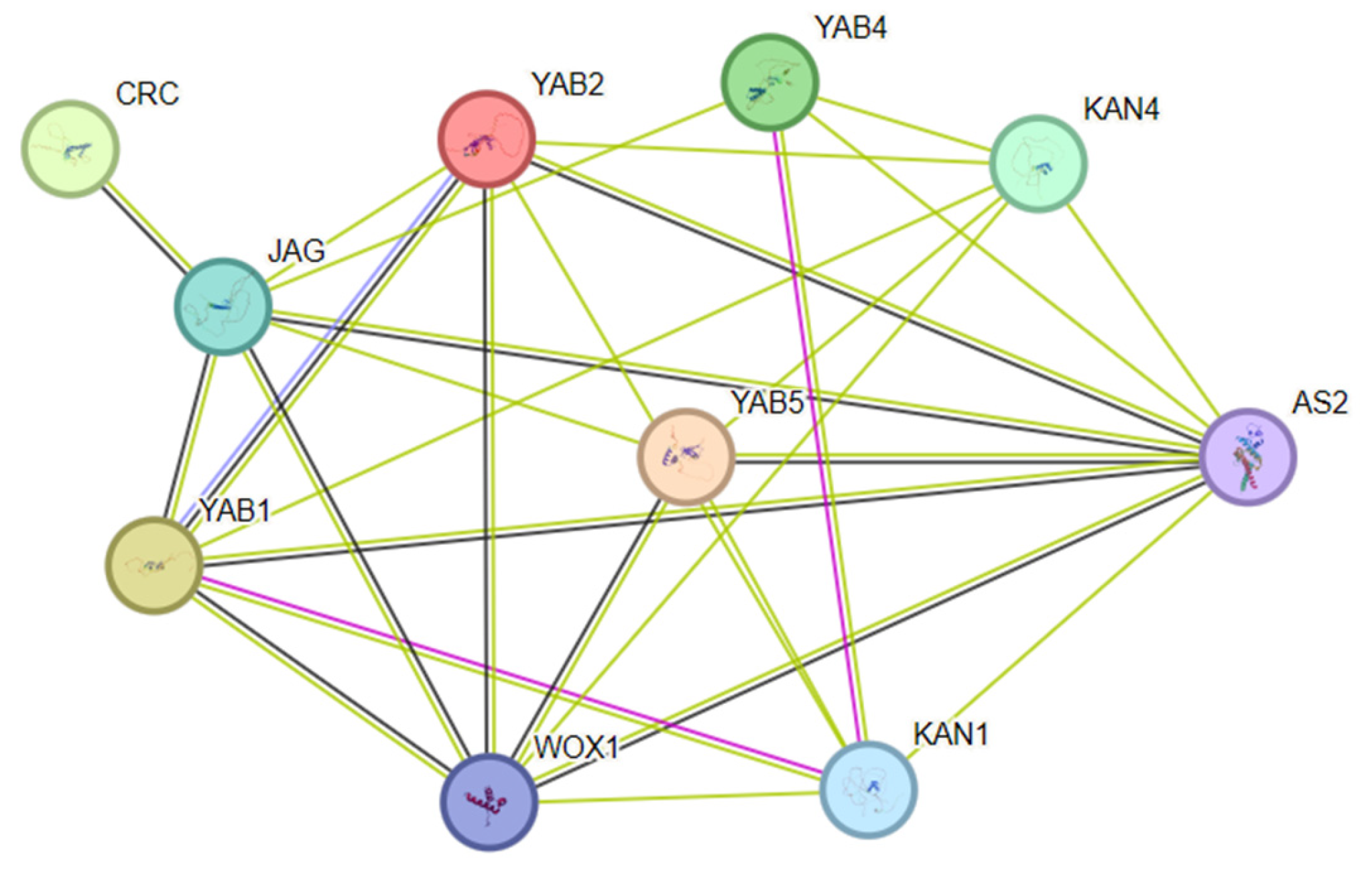
3.8. Inferred Protein Interaction Network Based on Homology
3.9. Determination of Total Sugar and Total Flavone Content
3.10. Determination of Antioxidant Capacity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, G.-Z.; Huang, J.; Zhou, X.-Q.; Hao, Y.; Chen, J.-L.; Zhou, Y.-Z.; Ahmad, S.; Lan, S.; Liu, Z.-J.; Peng, D.-H. Comprehensive analysis for GRF transcription factors in sacred lotus (Nelumbo nucifera). Int. J. Mol. Sci. 2022, 23, 6673. [Google Scholar] [CrossRef]
- Li, C.; Dong, N.; Shen, L.; Lu, M.; Zhai, J.; Zhao, Y.; Chen, L.; Wan, Z.; Liu, Z.; Ren, H.; et al. Genome-wide identification and expression profile of YABBY genes in Averrhoa carambola. PeerJ 2022, 9, e12558. [Google Scholar] [CrossRef]
- Yin, S.; Li, S.; Gao, Y.; Bartholomew, E.S.; Wang, R.; Yang, H.; Liu, C.; Chen, X.; Wang, Y.; Liu, X.; et al. Genome-wide identification of YABBY gene family in cucurbitaceae and expression analysis in cucumber (Cucumis sativus L.). Genes 2022, 13, 467. [Google Scholar] [CrossRef]
- Zhao, S.-P.; Lu, D.; Yu, T.-F.; Ji, Y.-J.; Zheng, W.-J.; Zhang, S.-X.; Chai, S.-C.; Chen, Z.-Y.; Cui, X.-Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017, 119, 132–146. [Google Scholar] [CrossRef]
- Huang, J.; Chen, G.-Z.; Ahmad, S.; Wang, Q.; Tu, S.; Shi, X.-L.; Hao, Y.; Zhou, Y.-Z.; Lan, S.-R.; Liu, Z.-J.; et al. Identification, molecular characteristics, and evolution of YABBY gene family in Melastoma dodecandrum. Int. J. Mol. Sci. 2023, 24, 4174. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Hamidou, A.A.; Zhao, X.; Ouyang, Z.; Lin, H.; Li, J.; Zhang, X.; Luo, K.; Chen, Y. Superoxide dismutase gene family in cassava revealed their involvement in environmental stress via genome-wide analysis. iScience 2023, 26, 107801. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Bartholmes, C.; Hidalgo, O.; Gleissberg, S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. 2012, 14, 11–23. [Google Scholar] [CrossRef]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef]
- Wang, W.; Ma, J.; Liu, H.; Wang, Z.; Nan, R.; Zhong, T.; Sun, M.; Wang, S.; Yao, Y.; Sun, F.; et al. Genome-wide analysis of the switchgrass YABBY family and functional characterization of PvYABBY14 in response to ABA and GA stress in Arabidopsis. BMC Plant Biol. 2024, 24, 114. [Google Scholar] [CrossRef]
- Kayani, S.-I.; Shen, Q.; Ma, Y.; Fu, X.; Xie, L.; Zhong, Y.; Tiantian, C.; Pan, Q.; Li, L.; Rahman, S.-U.; et al. The YABBY family transcription factor AaYABBY5 directly targets cytochrome P450 monooxygenase (CYP71AV1) and double-bond reductase 2 (DBR2) involved in artemisinin biosynthesis in Artemisia Annua. Front. Plant Sci. 2019, 10, 1084. [Google Scholar] [CrossRef] [PubMed]
- Boter, M.; Golz, J.F.; Gimenez-Ibanez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER Is a Direct Target of JAZ3 and Modulates Responses to Jasmonate. Plant Cell 2015, 27, 3160–3174. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, L.N.; Zhang, Q.Y.; Song, X.S. Drought tolerance is correlated with the activity of antioxidant enzymes in Cerasus humilis seedlings. BioMed Res. Int. 2016, 2016, 9851095. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chang, X.; Yu, J.; Xu, W. Cerasus humilis cherry polyphenol reduces high-fat diet-induced obesity in C57BL/6 mice by mitigating fat deposition, inflammation, and oxidation. J. Agric. Food Chem. 2020, 68, 4424–4436. [Google Scholar] [CrossRef]
- Yang, R.; Yang, Y.; Hu, Y.; Yin, L.; Qu, P.; Wang, P.; Mu, X.; Zhang, S.; Xie, P.; Cheng, C.; et al. Comparison of bioactive compounds and antioxidant activities in differentially pigmented Cerasus humilis fruits. Molecules 2023, 28, 6272. [Google Scholar] [CrossRef]
- Toriba, T.; Harada, K.; Takamura, A.; Nakamura, H.; Ichikawa, H.; Suzaki, T.; Hirano, H.-Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007, 277, 457–468. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Sun, X.; Li, Y.; Yao, J.; van Nocker, S.; Wang, X. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front. Plant Sci. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Li, Z.; Li, G.; Cai, M.; Priyadarshani, S.V.; Aslam, M.; Zhou, Q.; Huang, X.; Wang, X.; Liu, Y.; Qin, Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int. J. Mol. Sci. 2019, 20, 5863. [Google Scholar] [CrossRef]
- Zhao, Y.; Fu, G.; Jiang, J.; Li, Y.; Liu, X.; Li, J.; Sun, J.; Wang, Q.; Liu, D.; Luo, Z.; et al. Chromosome-scale Cerasus humilis genome assembly reveals gene family evolution and possible genomic basis of calcium accumulation in fruits. Sci. Hortic. 2022, 299, 111012. [Google Scholar] [CrossRef]
- Kong, L.; Sun, J.; Jiang, Z.; Ren, W.; Wang, Z.; Zhang, M.; Liu, X.; Wang, L.; Ma, W.; Xu, J. Identification and expression analysis of YABBY family genes in Platycodon grandiflorus. Plant Signal. Behav. 2023, 18, 2163069. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, A.; Hu, X.; Deng, Q.; Ma, Z.; Su, L. Comprehensive study of rice YABBY gene family: Evolution, expression and interacting proteins analysis. PeerJ 2023, 11, e14783. [Google Scholar] [CrossRef]
- Fu, W.C. Genome-wide identification and characterization of the AP2/ERF gene family in quinoa (Chenopodium quinoa) and their expression profiling during abiotic stress conditions. J. Plant Growth Regul. 2023, 43, 1118–1136. [Google Scholar] [CrossRef]
- Ma, R.; Huang, B.; Huang, Z.; Zhang, Z. Genome-wide identification and analysis of the YABBY gene family in Moso Bamboo (Phyllostachys edulis (Carriere) J. Houz). PeerJ 2021, 9, e11780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Sami, A.; Haider, M.Z.; Shafiq, M.; Sadiq, S.; Ahmad, F. Genome-wide identification and in-silico expression analysis of CCO gene family in sunflower (Helianthus annuus) against abiotic stress. Plant Mol. Biol. 2024, 114, 34. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.Y.; Wang, R.Y.; Huang, Z.W.; Cui, R.F.; Zhu, H.Q.; Yang, Y.M.; Zhang, D. Genome-wide identification, evolution, and expression analysis of carbonic anhydrases genes in soybean (Glycine max). Funct. Integr. Genom. 2023, 23, 37. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ji, X.; Ren, J.; Zhang, Y.; Lang, S.; Wang, D.; Song, X. Integrated analysis of the metabolome and transcriptome on anthocyanin biosynthesis in four developmental stages of Cerasus humilis peel coloration. Int. J. Mol. Sci. 2021, 22, 11880. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Bartley, G.E.; Ishida, B.K. Developmental gene regulation during tomato fruit ripening and in-vitro sepal morphogenesis. BMC Plant Biol. 2003, 3, 4. [Google Scholar] [CrossRef][Green Version]
- Bartley, G.E.; Ishida, B.K. Ethylene-sensitive and insensitive regulation of transcription factor expression during in vitro tomato sepal ripening. J. Exp. Bot. 2007, 58, 2043–2051. [Google Scholar] [CrossRef]
- Yu, T.; Shen, S.; Xu, Y.; Wang, X.; Yu, Y.; Ma, B.; Chen, X. Identification and expression analysis of the YABBY gene family in strawberry. Sheng Wu Gong Cheng Xue Bao 2024, 40, 104–121. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, C.; Ge, D.; Yan, M.; Ren, Y.; Huang, X.; Yuan, Z. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 2020, 752, 144784. [Google Scholar] [CrossRef]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, A.; Wei, H.; Hao, P.; Zhang, Q.; Tian, M.; Yang, X.; Cheng, S.; Fu, X.; Ma, L.; et al. Genome-wide identification and expression patterns analysis of the RPD3/HDA1 gene family in cotton. BMC Genom. 2020, 21, 643. [Google Scholar] [CrossRef]
- Shan, D.P.; Huang, J.G.; Yang, Y.T.; Guo, Y.H.; Wu, C.A.; Yang, G.D.; Gao, Z.; Zheng, C.C. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 2007, 176, 70–81. [Google Scholar] [CrossRef]
- Li, S.; Wu, P.; Yu, X.; Cao, J.; Chen, X.; Gao, L.; Chen, K.; Grierson, D. Contrasting roles of ethylene response factors in pathogen response and ripening in fleshy fruit. Cells 2022, 11, 2484. [Google Scholar] [CrossRef]
- Mazhar, H.S.-U.; Shafiq, M.; Ali, H.; Ashfaq, M.; Anwar, A.; Tabassum, J.; Ali, Q.; Jilani, G.; Awais, M.; Sahu, R.; et al. Genome-wide identification, and in-silico expression analysis of YABBY gene family in response to biotic and abiotic stresses in potato (Solanum tuberosum). Genes 2023, 14, 824. [Google Scholar] [CrossRef]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, P.; Won, S.Y.; Park, D.S.; Lee, Y.-H.; Kim, J.S. Comparative analysis of the YABBY gene family of Bienertia sinuspersici, a single-cell C4 plant. Plants 2019, 8, 536. [Google Scholar] [CrossRef] [PubMed]
- Kayani, S.-I.; Shen, Q.; Rahman, S.-U.; Fu, X.; Li, Y.; Wang, C.; Hassani, D.; Tang, K. Transcriptional regulation of flavonoid biosynthesis in Artemisia annua by AaYABBY5. Hortic. Res. 2022, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Huang, X.; Li, M.; Li, M.; Gu, L.; Gao, L.; Li, C.; Qin, S.; Liu, D.; Zhang, Z. Integrated multi-omics analysis reveals genes involved in flavonoid biosynthesis and trichome development of Artemisia argyi. Plant Sci. 2024, 346, 112158. [Google Scholar] [CrossRef]
- Huang, X.; Wu, Y.; Zhang, S.; Yang, H.; Wu, W.; Lyu, L.; Li, W. Changes in antioxidant substances and antioxidant enzyme activities in raspberry fruits at different developmental stages. Sci. Hortic. 2023, 321, 112314. [Google Scholar] [CrossRef]
- Li, L.; Qin, Y.; Xin, X.; Wang, S.; Liu, Z.; Feng, X. The great potential of flavonoids as candidate drugs for NAFLD. Biomed. Pharmacother. 2023, 164, 114991. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Jiang, S.; Kong, L.; Yue, C.; Ma, L.; Ma, J.; Ma, W.; Liu, X. Identification of the YABBY Gene Family in Cerasus humilis and Analysis of Expression Patterns During Different Growth Stages. Biology 2025, 14, 1511. https://doi.org/10.3390/biology14111511
Ren W, Jiang S, Kong L, Yue C, Ma L, Ma J, Ma W, Liu X. Identification of the YABBY Gene Family in Cerasus humilis and Analysis of Expression Patterns During Different Growth Stages. Biology. 2025; 14(11):1511. https://doi.org/10.3390/biology14111511
Chicago/Turabian StyleRen, Weichao, Shan Jiang, Lingyang Kong, Chenzhuo Yue, Lengleng Ma, Junbai Ma, Wei Ma, and Xiubo Liu. 2025. "Identification of the YABBY Gene Family in Cerasus humilis and Analysis of Expression Patterns During Different Growth Stages" Biology 14, no. 11: 1511. https://doi.org/10.3390/biology14111511
APA StyleRen, W., Jiang, S., Kong, L., Yue, C., Ma, L., Ma, J., Ma, W., & Liu, X. (2025). Identification of the YABBY Gene Family in Cerasus humilis and Analysis of Expression Patterns During Different Growth Stages. Biology, 14(11), 1511. https://doi.org/10.3390/biology14111511





