Genetic Diversity of the Only Natural Population of Corylus avellana L. in Kazakhstan and Prospects for Its In Vitro Conservation
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
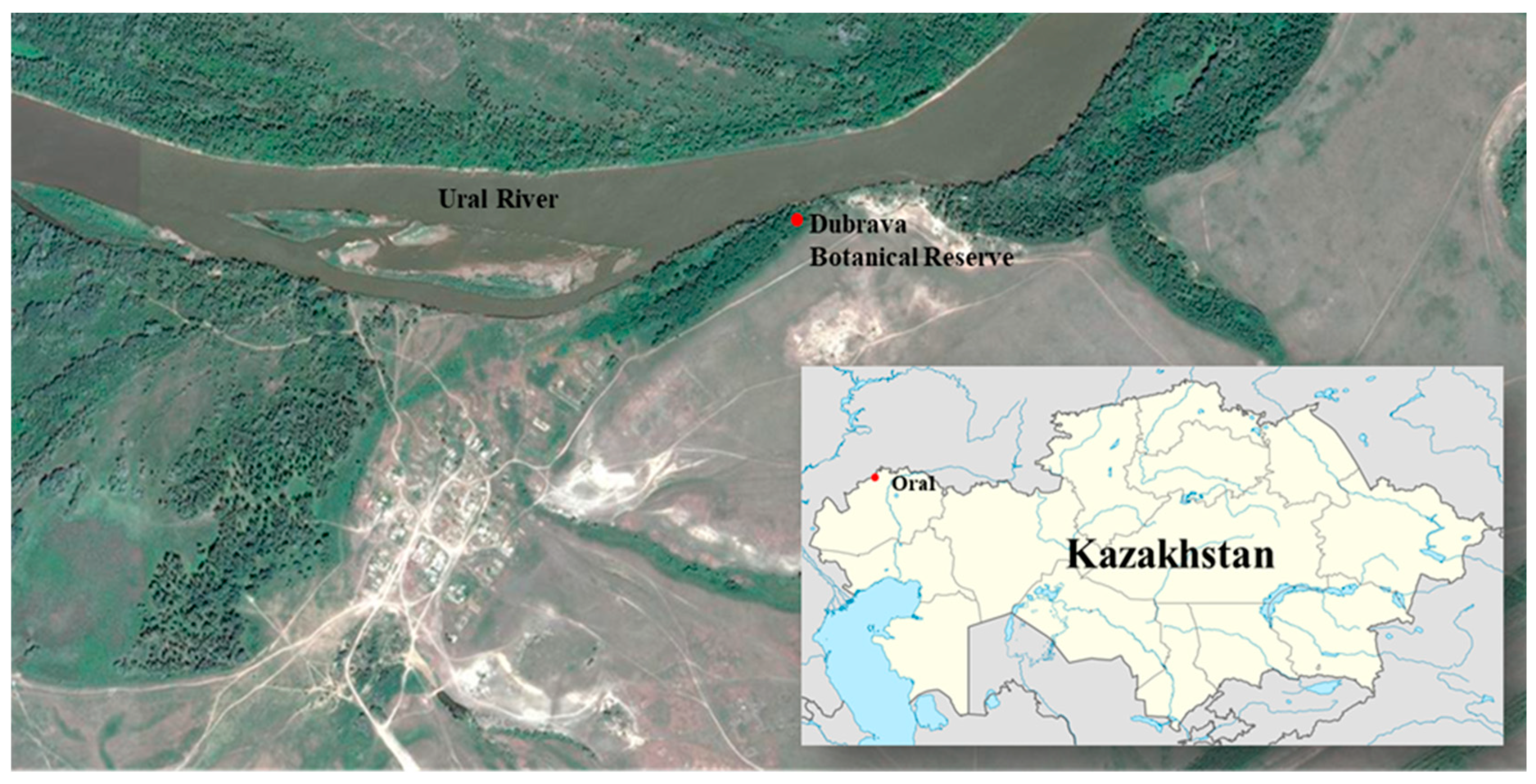
2.1. Plant Material and Collection Site
2.2. Data Collection and Phenotypic Evaluation
2.3. Genomic DNA Extraction and Microsatellite Genotyping
2.4. Genetic Diversity Analysis
2.5. Population Structure Analysis
2.6. In Vitro Culture Initiation
2.7. Statistical Analysis
3. Results
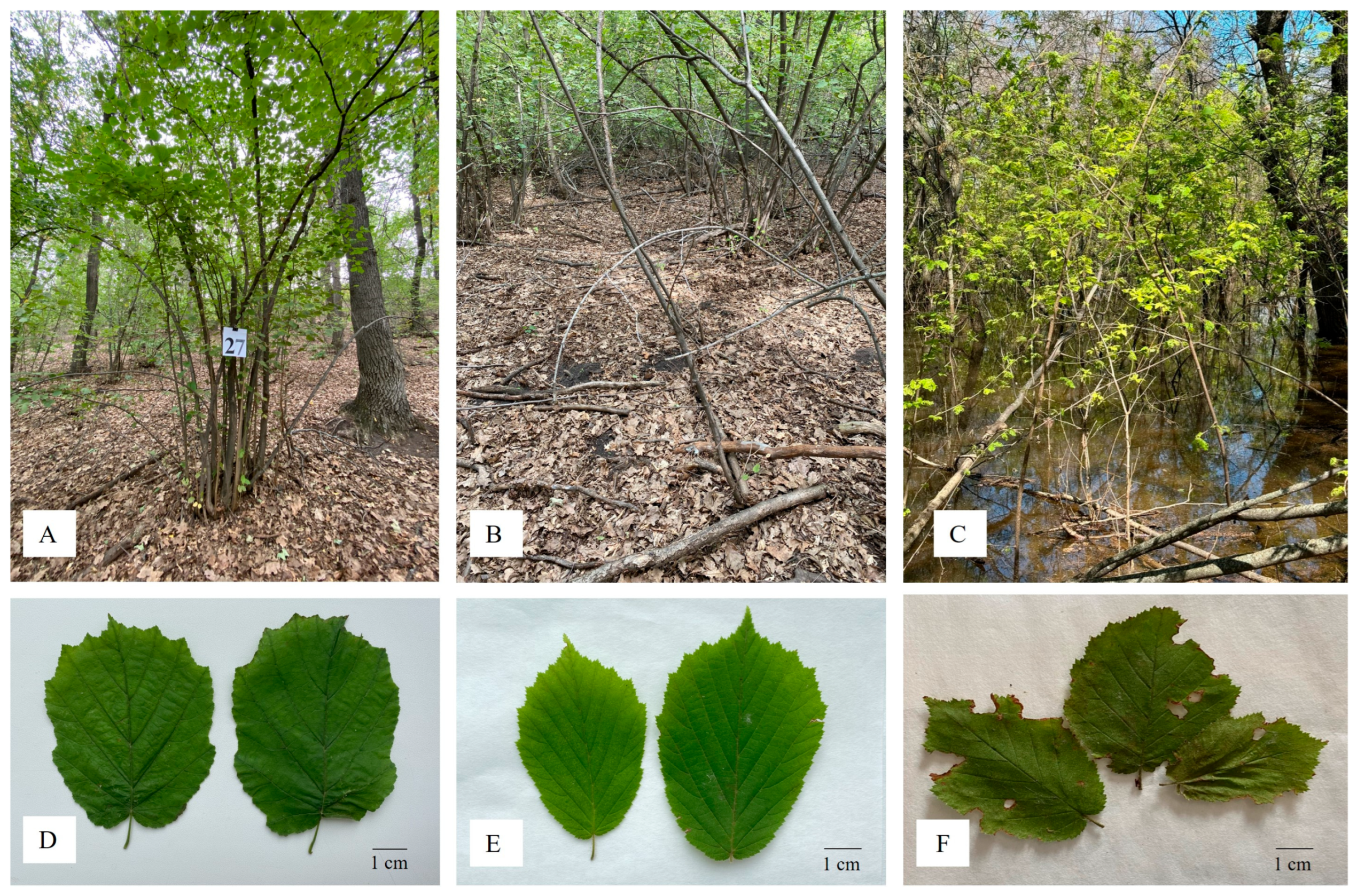
3.1. Phenotypic Evaluation of Corylus avellana
3.2. Genetic Analysis
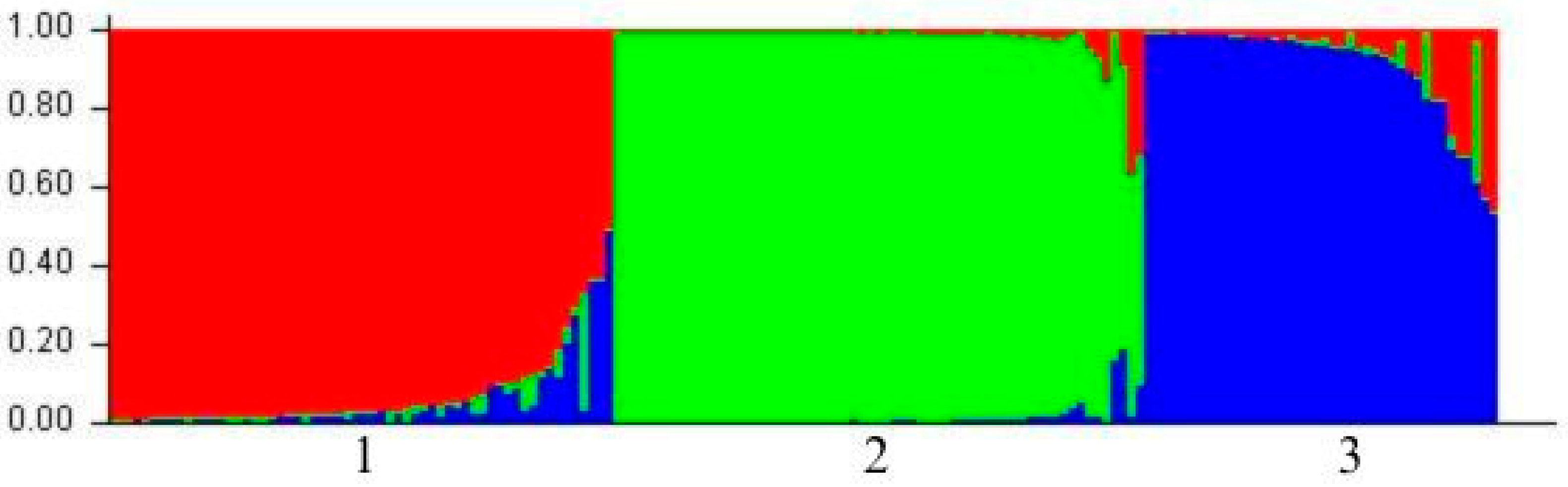
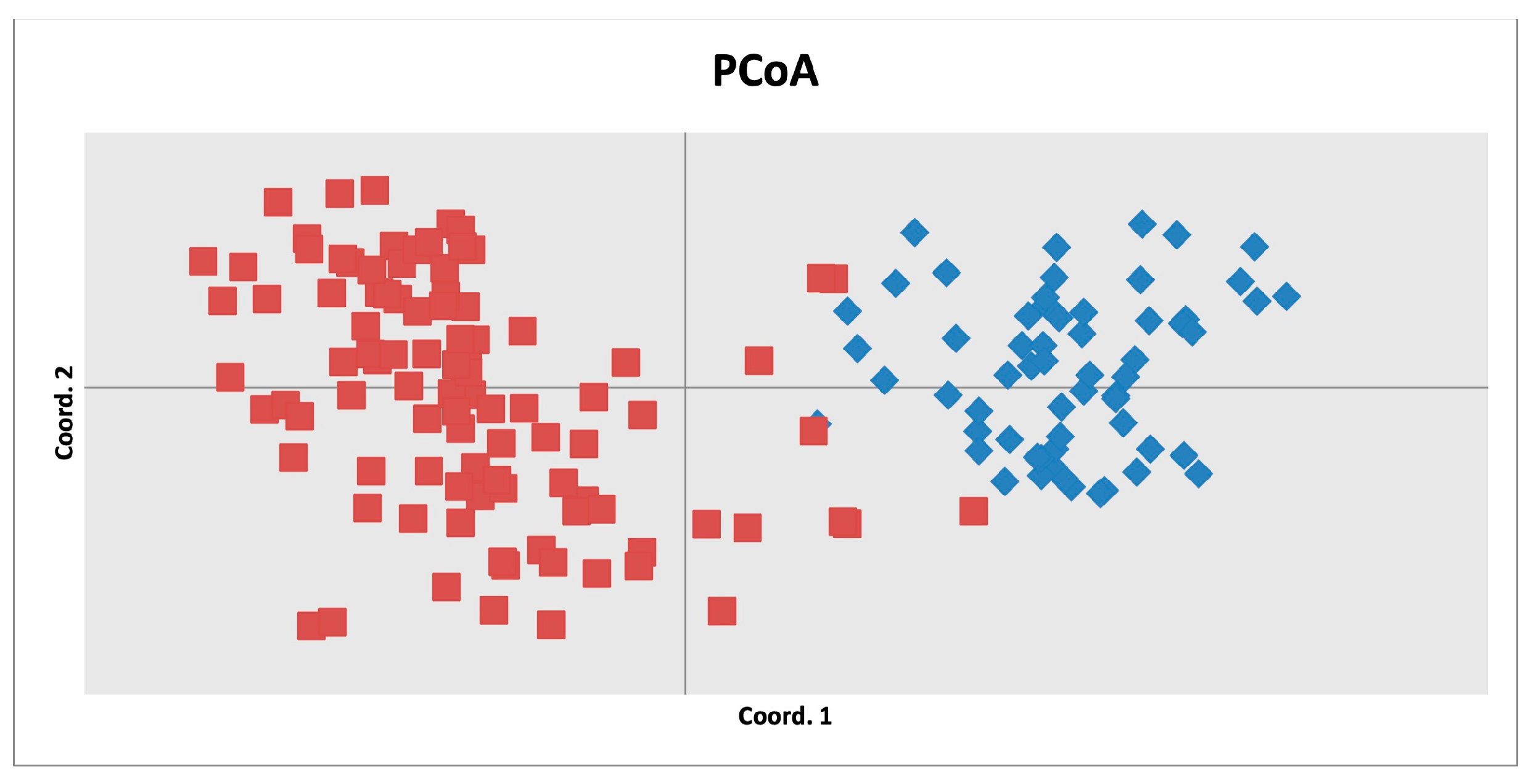
3.3. Structure and Phylogenetic Analysis
3.4. In Vitro Conservation of Corylus avellana L.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Z.; Zhao, T.-T.; Ma, Q.-H.; Liang, L.-S.; Wang, G.-X. Resolving the Speciation Patterns and Evolutionary History of the Intercontinental Disjunct Genus Corylus (Betulaceae) Using Genome-Wide SNPs. Front. Plant Sci. 2018, 9, 1386. [Google Scholar] [CrossRef]
- Enescu, C.M.; Houston Durrant, T.; de Rigo, D.; Caudullo, G. Corylus avellana in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Public Office of the European Union: Luxembourg, 2016; pp. 86–87. [Google Scholar]
- Pavlov, N.V. (Ed.) Flora of Kazakhstan, 3rd ed.; Alma-Ata, AN KazSSR: Almaty, Kazakhstan, 1961; pp. 510–516. [Google Scholar]
- Baitenov, M.S. Flora of Kazakhstan. Ancestral Complex of Flora; Gylym: Almaty, Kazakhstan, 2001; p. 60. [Google Scholar]
- Red Book of Kazakhstan; Volume 2—Plants; “ArtPrintXXI” LLP: Astana, Kazakhstan, 2014; p. 94.
- Kushnarenko, S.; Romadanova, N.; Aralbaeva, M. Current state and in vitro conservation of the only endangered population of Corylus avellana L. in Kazakhstan. Res. Crops 2020, 21, 681–686. [Google Scholar] [CrossRef]
- Pérez, C.; Rodríguez, R.; Tamés, R.S. “In vitro” filbert (Corylus avellana L.) micropropagation from shoot and cotyledonary node segments. Plant Cell Rep. 1985, 4, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Damiano, C.; Catenaro, E.; Giovinazzi, J.; Frattarelli, A.; Caboni, E. Micropropagation of hazelnut (Corylus avellana L.). Acta Hortic. 2005, 686, 221–226. [Google Scholar] [CrossRef]
- Ellena, M.; González, A.; Matamala, L. The effect of cytokinins on micropropagation of hazelnut (Corylus avellana L.) rootstock selection ‘Selfe J’. Acta Hortic. 2023, 1379, 327–332. [Google Scholar] [CrossRef]
- Clapa, D.; Harta, M.; Borsai, O.; Pamfil, D. Micropropagation of Vaccinium corymbosum L. and Corylus avellana L. using a temporary immersion bioreactor system. Agricultura 2019, 3–4, 102–109. [Google Scholar] [CrossRef]
- Kıvrak Kiran, S.; Galatali, S.; Yeniocak, S.; Ozkaya, D.E.; Mercan, T.; Guldag, S.; Celik, O.; Abdul Ghafoor, N.; Kaya, E. Investigation of modified WPM medium for the best meristem proliferation of Corylus avellana L. Adv. Hort. Sci. 2021, 35, 285–292. [Google Scholar] [CrossRef]
- Yu, X.; Reed, B.M. Improved shoot multiplication of mature hazelnut (Corylus avellana L.) in vitro using glucose as a carbon source. Plant Cell Rep. 1993, 12, 256–259. [Google Scholar] [CrossRef]
- Boccacci, P.; Botta, R.; Rovira, M. Genetic diversity of hazelnut (Corylus avellana L.) germplasm in Northeastern Spain. HortScience 2008, 43, 667–672. [Google Scholar] [CrossRef]
- Pop, I.F.; Pamfil, D.; Raica, P.A.; Petricele, I.V.; Botu, M.; Vicol, A.C.; Harta, M.; Sisea, C.R. Evaluation of the Genetic Diversity of several Corylus avellana Accessions from the Romanian National Hazelnut Collection. Not. Bot. Horti Agrobot. Cluj-Napoca 2010, 38, 61–67. [Google Scholar] [CrossRef]
- Boccacci, P.; Aramini, M.; Valentini, N.; Bacchetta, L.; Rovira, M.; Drogoudi, P.; Silva, A.P.; Solar, A.; Calizzano, F.; Erdoğan, V.; et al. Molecular and morphological diversity of on-farm hazelnut (Corylus avellana L.) landraces from southern Europe and their role in the origin and diffusion of cultivated germplasm. Tree Genet. Genomes 2013, 9, 1465–1480. [Google Scholar] [CrossRef]
- Akin, M.; Nyberg, A.; Postman, J.; Mehlenbacher, S.; Bassil, N.V. A multiplexed microsatellite fingerprinting set for hazelnut cultivar identification. Eur. J. Hortic. Sci. 2016, 81, 327–338. [Google Scholar] [CrossRef]
- Ozturk, S.C.; Ozturk, S.E.; Celik, I.; Stampar, F.; Veberic, R.; Doganlar, S.; Solar, A.; Frary, A. Molecular genetic diversity and association mapping of nut and kernel traits in Slovenian hazelnut (Corylus avellana) germplasm. Tree Genet. Genomes 2017, 13, 16. [Google Scholar] [CrossRef]
- Tanhuanpää, P.; Heinonen, M.; Bitz, L.; Rokka, V.M. Genetic diversity and structure in the northern populations of European hazelnut (Corylus avellana L.). Genome 2019, 62, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Simões, F.; Matos, J.; Silva, A.P.; Carnide, V. Genetic relationship among wild, landraces and cultivars of hazelnut (Corylus avellana) from Portugal revealed through ISSR and AFLP markers. Plant Syst. Evol. 2014, 300, 1035–1046. [Google Scholar] [CrossRef]
- Karakaya, O.; Balta, M.F.; Uzun, A.; Balta, F.; Uzun, S. Genetic diversity and marker-trait associations among Çakıldak hazelnut (Corylus avellana L.) clones in the North Eastern Anatolia of Türkiye based on morphological and molecular markers. Genet. Resour. Crop. Evol. 2025, 72, 5921–5937. [Google Scholar] [CrossRef]
- Lombardoni, J.J.; Honig, J.A.; Vaiciunas, J.N.; Kubik, C.; Capik, J.M.; Mehlenbacher, S.A.; Molnar, T.J. Genotyping-by-sequencing shows high genetic diversity in Corylus avellana germplasm resistant to eastern filbert blight. Tree Genet. Genomes 2024, 20, 45. [Google Scholar] [CrossRef]
- Helmstetter, A.J.; Oztolan-Erol, N.; Lucas, S.J.; Buggs, R.J.A. Genetic diversity and domestication of hazelnut (Corylus avellana L.) in Turkey. Plants People Planet 2020, 2, 326–339. [Google Scholar] [CrossRef]
- Oztolan-Erol, N.; Helmstetter, A.J.; İnan, A.; Buggs, R.J.A.; Lucas, S.J. Unraveling Genetic Diversity Amongst European Hazelnut (Corylus avellana L.) Varieties in Turkey. Front. Plant Sci. 2021, 12, 661274. [Google Scholar] [CrossRef]
- Bioversity International. Descriptors for Hazelnut (Corylus avellana L.); Bioversity International: Rome, Italy; Food and Agricultural Organization of the United Nations: Rome, Italy; International Centre for Advanced Mediterranean Agronomic Studies: Zagaroza, Spain, 2008; p. 55. [Google Scholar]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Boccacci, P.; Akkak, A.; Bassil, N.V.; Mehlenbacher, S.A.; Botta, R. Characterization and evaluation of microsatellite loci in European hazelnut (Corylus avellana L.) and their transferability to other Corylus species. Mol. Ecol. Notes 2005, 5, 934–937. [Google Scholar] [CrossRef]
- Bassil, N.V.; Botta, R.; Mehlenbacher, S.A. Additional microsatellites of the European hazelnut. Acta Hort. 2005, 686, 105–110. [Google Scholar] [CrossRef]
- Boccacci, P.; Akkak, A.; Botta, R. DNA typing and genetic relations among European hazelnut (Corylus avellana L.) cultivars using microsatellite markers. Genome 2006, 49, 598–611. [Google Scholar] [CrossRef]
- Matschiner, M.; Salzburger, W. TANDEM: Integrating automated allele binning into genetics and genomics workflows. Bioinformatics 2009, 25, 1982–1983. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in excel: Population genetic software for teaching and research—An update. Bioinformatics 2012, 19, 2537–2539. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Kalinowski, S.T. HP-Rare: A computer program for performing rarefaction on measures of allelic diversity. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Cornille, A.; Feurtey, A.; Gelin, U.; Ropars, J.; Misvanderbrugge, K.; Gladieux, P.; Giraud, T. Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: A basis for conservation and breeding programs in apples. Evol. Appl. 2015, 8, 373–384. [Google Scholar] [CrossRef]
- Gascuel, O.; Mirkin, B.; McMorris, F.; Roberts, F.; Rzhetsky, A. Concerning the NJ algorithm and its unweighted version, UNJ. In Mathematical Hierarchies and Biology; DIMACS Series in Discrete Mathematics and Theoretical Computer Science; RI American Mathematical Society: Providence, RI, USA, 1997; pp. 149–170. [Google Scholar]
- Huson, D.H.; Richter, D.C.; Rausch, C.; Dezulian, T.; Franz, M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinform. 2007, 8, 460. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Akin, M.; Eyduran, E.; Niedz, R.P.; Reed, B.M. Developing hazelnut tissue culture medium free of ion confounding. Plant Cell Tissue Organ Cult. 2017, 130, 483–494. [Google Scholar] [CrossRef]
- Viss, P.R.; Brooks, E.M.; Driver, J.A. A simplifed method for the control of bacterial contamination in woody plant tissue culture. In Vitr. Cell. Dev. Biol.-Plant 1991, 27, 42. [Google Scholar] [CrossRef]
- SYSTAT. Statistics Software, SYSTAT 13.0; SYSTAT Software, Inc.: San Jose, CA, USA, 2009; Available online: https://systatsoftware.com/ (accessed on 15 July 2025).
- Martins, S.; Simões, F.; Mendonça, D.; Matos, J.; Silva, A.P.; Carnide, V. Western European wild and landraces hazelnuts evaluated by SSR markers. Plant Mol. Biol. Rep. 2015, 33, 1712–1720. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, Y.; Gou, X.; Du, J.; Wang, M. Genetic Diversity and Population Structure of Corylus yunnanensis (Franch.) A. Camus Using Microsatellite Markers in Sichuan Province. Forests 2023, 14, 932. [Google Scholar] [CrossRef]
- Yang, W.; Yang, B.; Lu, L.; Zhang, X.; Sun, J.; Wang, L.; Zhen, Z.; Liang, D.; Wang, K.; Yan, X.; et al. Utilizing SSR-based core collection development to improve conservation and utilization of Corylus L. genetic resources. PLoS ONE 2024, 19, e0312116. [Google Scholar] [CrossRef]
- Bassil, N.; Boccacci, P.; Botta, R.; Postman, J.; Mehlenbacher, S. Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Genet. Resour. Crop Evol. 2013, 60, 543–568. [Google Scholar] [CrossRef]
- Gökirmak, T.; Mehlenbacher, S.A.; Bassil, N.V. Characterization of European hazelnut (Corylus avellana) cultivars using SSR markers. Genet. Resour. Crop Evol. 2009, 56, 147–172. [Google Scholar] [CrossRef]
- Palmé, A.E.; Vendramin, G.G. Chloroplast DNA variation, postglacial recolonization and hybridization in hazel, Corylus avellana. Mol. Ecol. 2002, 11, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Campa, A.; Trabanco, N.; Pérez-Vega, E.; Rovira, M.; Ferreira, J.J. Genetic relationship between cultivated and wild hazelnuts (Corylus avellana L.) collected in northern Spain. Plant Breed. 2011, 130, 360–366. [Google Scholar] [CrossRef]
- Ferreira, J.J.; Garcia-González, C.; Tous, J.; Rovira, M. Genetic diversity revealed by morphological traits and ISSR markers in hazelnut germplasm from northern Spain. Plant Breed. 2010, 129, 435–441. [Google Scholar] [CrossRef]
- Boccacci, P.; Botta, R. Investigating the origin of hazelnut (Coryllus avellana L.) cultivars using chloroplast microsatellites. Genet. Resour. Crop Evol. 2009, 56, 851–859. [Google Scholar] [CrossRef]
- Reed, B.M.; Mentzer, J.; Tanprasert, P.; Yu, X. Internal bacterial contamination of micropropagated hazelnut: Identification and antibiotic treatment. Plant Cell Tissue Organ Cult. 1998, 52, 67–70. [Google Scholar] [CrossRef]
- Hand, C.R.; Wada, N.; Stockwell, V.; Reed, B.M. Node position infuences viability and contamination in hazelnut shoot explants. In Vitr. Cell. Dev. Biol.-Plant 2016, 52, 580–589. [Google Scholar] [CrossRef]
- Sekerli, M. Season, thermotherapy and surface sterilization play important roles in microbial contaminsation of hazelnut in vitro cultures. Plant Cell Tissue Organ Cult. 2024, 157, 70. [Google Scholar] [CrossRef]
- Kushnarenko, S.; Aralbayeva, M.; Rymkhanova, N.; Reed, B.M. Initiation pretreatment with Plant Preservative MixtureTM increases the percentage of aseptic walnut shoots. Vitr. Cell. Dev. Biol.-Plant 2022, 58, 964–971. [Google Scholar] [CrossRef]
- George, M.W.; Tripepi, R.R. Plant Preservative Mixture™ can affect shoot regeneration from leaf explants of chrysanthemum, European birch, and rhododendron. HortScience 2001, 36, 768–769. [Google Scholar] [CrossRef]
- Ho, W.J.; Huang, Y.K.; Huang, W.W.; Huang, Y.C.; Chung, J.P. Effective in vitro culture using dormant bud of nodal sections from a mature Acacia tree. Vitr. Cell. Dev. Biol.-Plant 2021, 58, 437–446. [Google Scholar] [CrossRef]
- Romadanova, N.V.; Tolegen, A.B.; Kushnarenko, S.V.; Zholdybayeva, E.V.; Bettoni, J.C. Effect of Plant Preservative MixtureTM on endophytic bacteria eradication from in vitro-grown apple shoots. Plants 2022, 11, 2624. [Google Scholar] [CrossRef]
- Tolegen, A.B.; Romadanova, N.V.; Aralbaeva, M.M.; Ukhatova, Y.V.; Agakhanov, M.M.; Kushnarenko, S.V. Using Plant Preservative MixtureTM to eliminate endophytic bacterial contamination and establishing an in vitro collection of commercially valuable blackberry varieties. Int. J. Biol. Chem. 2025, 18, 89–99. [Google Scholar] [CrossRef]



| CaT-B505 | CaT- B503 | CaT- B507 | CaC- B020 | CaT- B501 | CaT- B107 | CaT- B504 | CaT- B502 | CaC- B028 | CaT- B508 | Mean | SE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na | 9 | 12 | 10 | 14 | 10 | 16 | 12 | 12 | 13 | 12 | 12 | - |
| Ne | 3.09 | 5.02 | 4.14 | 3.06 | 3.81 | 3.46 | 5.84 | 3.68 | 2.9 | 3.61 | 3.86 | 0.09 |
| Ho | 0.70 | 0.85 | 0.81 | 0.57 | 0.76 | 0.78 | 0.87 | 0.68 | 0.46 | 0.52 | 0.7 | 0.04 |
| He | 0.68 | 0.80 | 0.76 | 0.67 | 0.74 | 0.71 | 0.83 | 0.73 | 0.65 | 0.72 | 0.73 | 0.02 |
| PIC | 0.62 | 0.77 | 0.72 | 0.62 | 0.71 | 0.66 | 0.81 | 0.7 | 0.61 | 0.69 | 0.7 | - |
| PI | 0.16 | 0.07 | 0.1 | 0.16 | 0.1 | 0.13 | 0.05 | 0.1 | 0.17 | 0.11 | 0.26 | - |
| I | 1.3 | 1.68 | 1.51 | 1.35 | 1.64 | 1.51 | 1.84 | 1.59 | 1.29 | 1.56 | 1.53 | 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushnarenko, S.V.; Omasheva, M.; Romadanova, N.; Aralbayeva, M.; Rymkhanova, N.; Manapkanova, U.; Botta, R.; Ruffa, P.; Valentini, N.; Marinoni, D.T. Genetic Diversity of the Only Natural Population of Corylus avellana L. in Kazakhstan and Prospects for Its In Vitro Conservation. Biology 2025, 14, 1472. https://doi.org/10.3390/biology14111472
Kushnarenko SV, Omasheva M, Romadanova N, Aralbayeva M, Rymkhanova N, Manapkanova U, Botta R, Ruffa P, Valentini N, Marinoni DT. Genetic Diversity of the Only Natural Population of Corylus avellana L. in Kazakhstan and Prospects for Its In Vitro Conservation. Biology. 2025; 14(11):1472. https://doi.org/10.3390/biology14111472
Chicago/Turabian StyleKushnarenko, Svetlana V., Madina Omasheva, Natalya Romadanova, Moldir Aralbayeva, Nazgul Rymkhanova, Ulzhan Manapkanova, Roberto Botta, Paola Ruffa, Nadia Valentini, and Daniela Torello Marinoni. 2025. "Genetic Diversity of the Only Natural Population of Corylus avellana L. in Kazakhstan and Prospects for Its In Vitro Conservation" Biology 14, no. 11: 1472. https://doi.org/10.3390/biology14111472
APA StyleKushnarenko, S. V., Omasheva, M., Romadanova, N., Aralbayeva, M., Rymkhanova, N., Manapkanova, U., Botta, R., Ruffa, P., Valentini, N., & Marinoni, D. T. (2025). Genetic Diversity of the Only Natural Population of Corylus avellana L. in Kazakhstan and Prospects for Its In Vitro Conservation. Biology, 14(11), 1472. https://doi.org/10.3390/biology14111472










