A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Isolation of PBMCs
2.3. Construction of Plasmids for the Expression of the Heavy and Light Chains of the Chimeric Antibody against the MT-C34 Peptide
2.4. Production and Purification of Chimeric Antibody
2.5. Size-Exclusion Chromatography
2.6. Polyacrylamide Gel Electrophoresis of Purified chAb
2.7. Western Blotting
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. Calculation of the Equilibrium Dissociation Constant (Kd)
2.10. Flow Cytometry
2.11. Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) Assay
2.12. Statistical Analysis
3. Results
3.1. Cloning of a Mouse Monoclonal Anti-MT-C34 Antibody cDNA and Chimerization with the Human IgG1
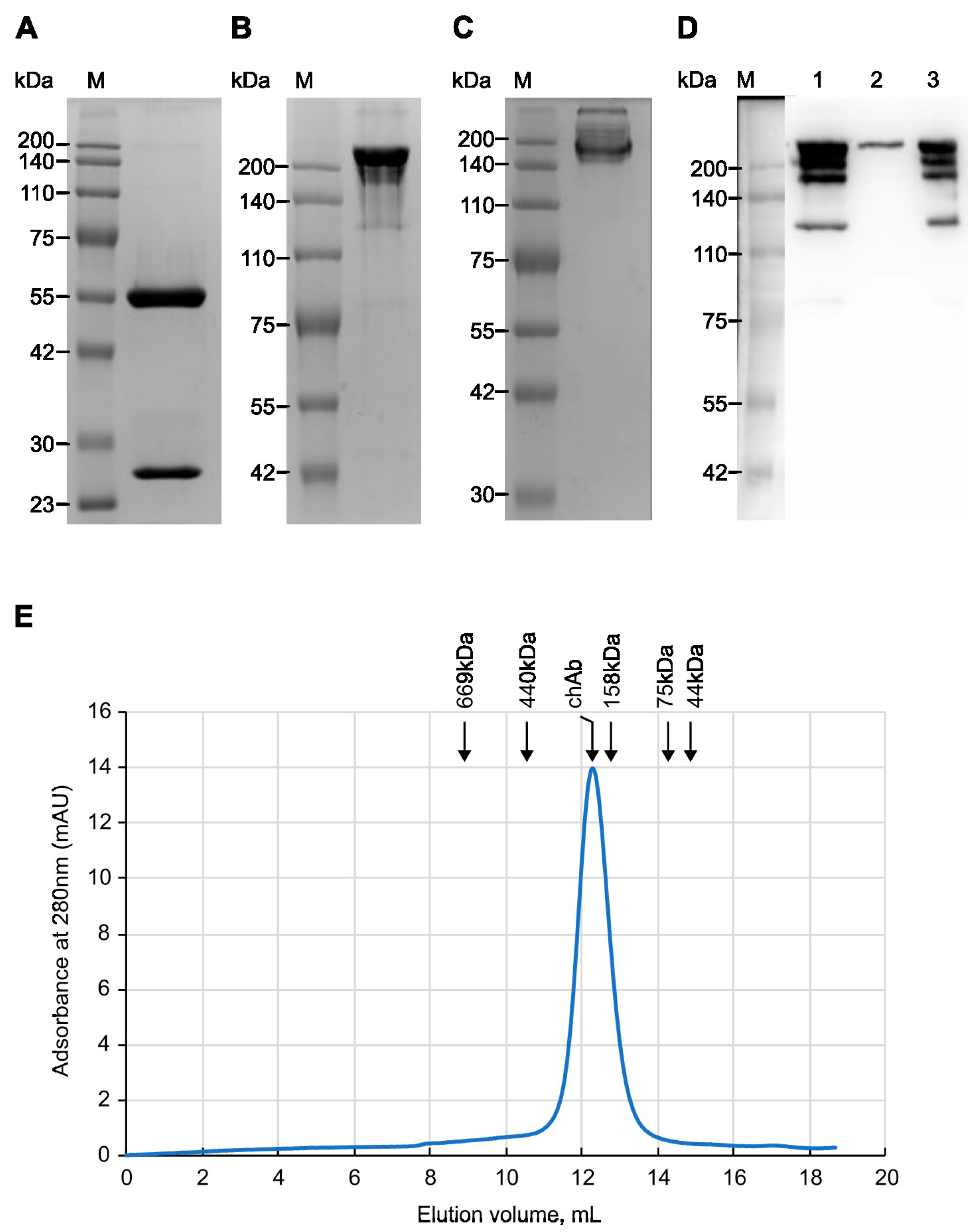
3.2. Analysis of Purified Chimeric Antibody
3.3. Evaluating Binding Affinity of the Mouse and Chimeric Anti-MT-C34 Ab
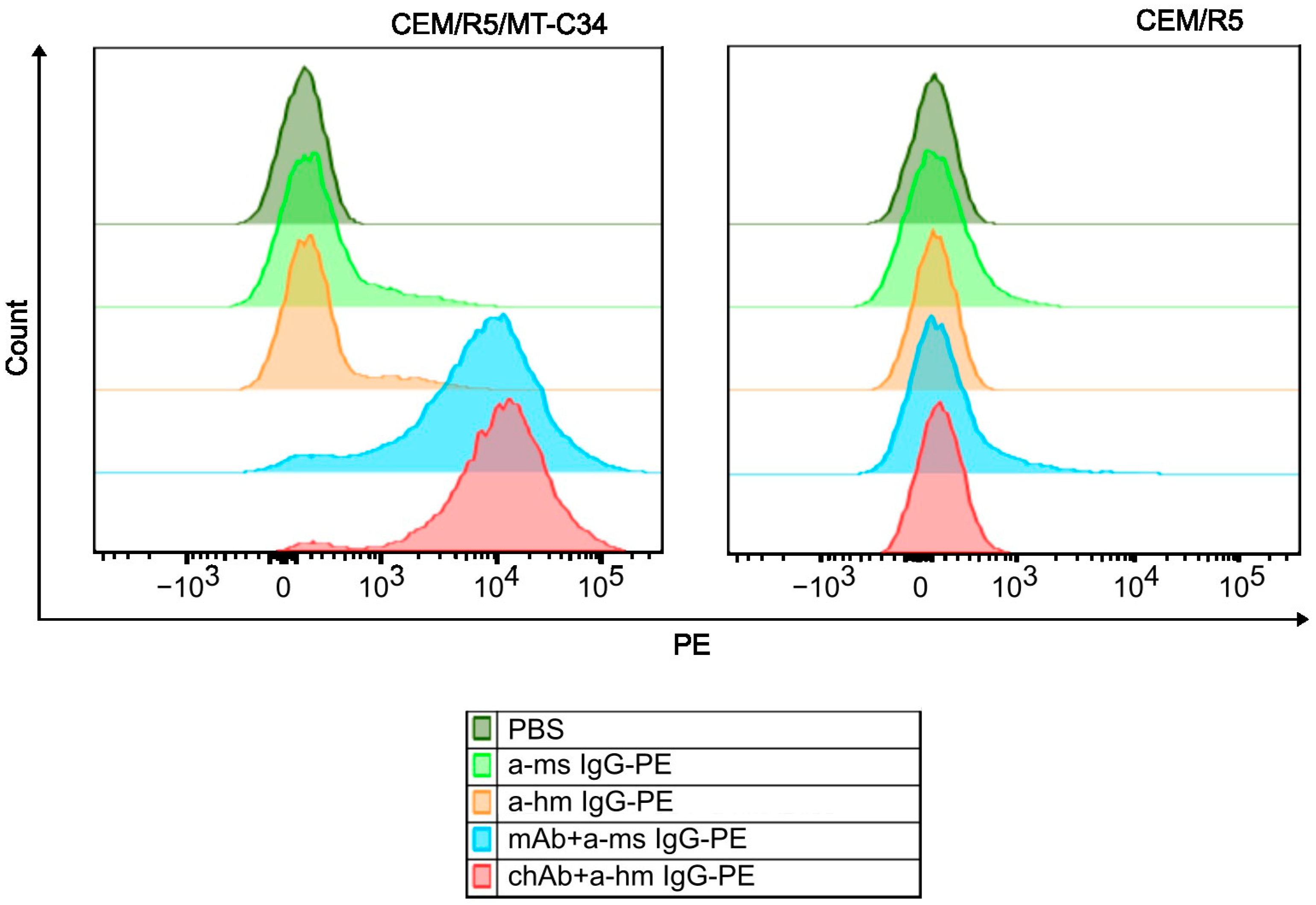
3.4. Measuring Specificity of the chAb Binding Using Flow Cytometry
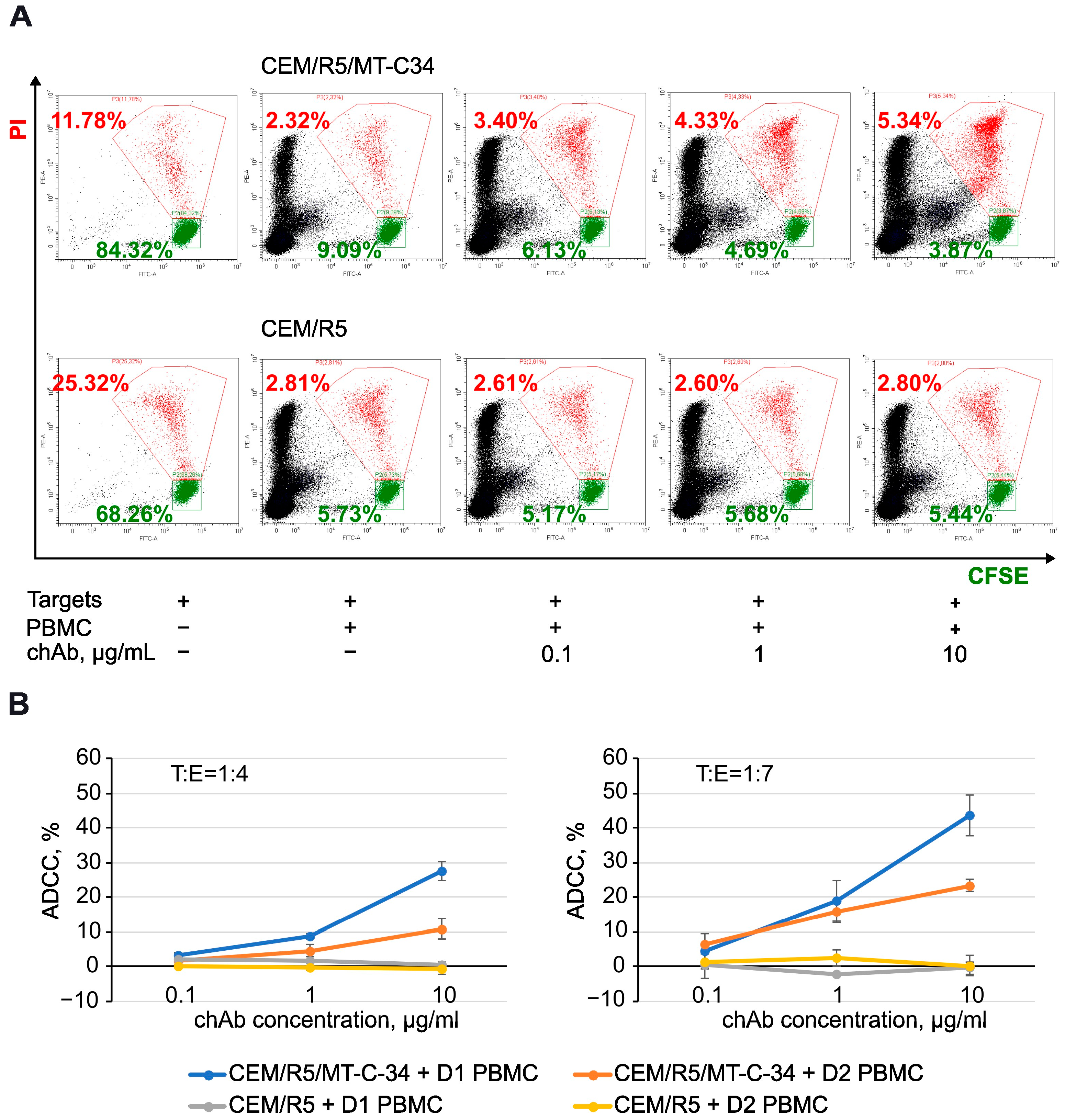
3.5. Antibody-Dependent Cellular Cytotoxic (ADCC) Function of chAb
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.; Farzan, M.; Sun, Y.; Sullivan, N.; Rollins, B.; Ponath, P.D.; Wu, L.; Mackay, C.R.; LaRosa, G.; Newman, W.; et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 1996, 85, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Gerard, N.P.; Wyatt, R.; Choe, H.; Parolin, C.; Ruffing, N.; Borsetti, A.; Cardoso, A.A.; Desjardin, E.; Newman, W.; et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 1996, 384, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Oberlin, E.; Amara, A.; Bachelerie, F.; Bessia, C.; Virelizier, J.L.; Arenzana-Seisdedos, F.; Schwartz, O.; Heard, J.M.; Clark-Lewis, I.; Legler, D.F.; et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 1996, 382, 833–835. [Google Scholar] [CrossRef]
- Lee, B.; Sharron, M.; Montaner, L.J.; Weissman, D.; Doms, R.W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 1999, 96, 5215–5220. [Google Scholar] [CrossRef]
- Carter, C.C.; Onafuwa-Nuga, A.; McNamara, L.A.; Riddell, J.t.; Bixby, D.; Savona, M.R.; Collins, K.L. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat. Med. 2010, 16, 446–451. [Google Scholar] [CrossRef]
- Carter, C.C.; McNamara, L.A.; Onafuwa-Nuga, A.; Shackleton, M.; Riddell, J.t.; Bixby, D.; Savona, M.R.; Morrison, S.J.; Collins, K.L. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 2011, 9, 223–234. [Google Scholar] [CrossRef]
- Sebastian, N.T.; Zaikos, T.D.; Terry, V.; Taschuk, F.; McNamara, L.A.; Onafuwa-Nuga, A.; Yucha, R.; Signer, R.A.J.; Riddell, J.I.; Bixby, D.; et al. CD4 is expressed on a heterogeneous subset of hematopoietic progenitors, which persistently harbor CXCR4 and CCR5-tropic HIV proviral genomes in vivo. PLoS Pathog. 2017, 13, e1006509. [Google Scholar] [CrossRef]
- Muro-Cacho, C.A.; Pantaleo, G.; Fauci, A.S. Analysis of apoptosis in lymph nodes of HIV-infected persons. Intensity of apoptosis correlates with the general state of activation of the lymphoid tissue and not with stage of disease or viral burden. J. Immunol. 1995, 154, 5555–5566. [Google Scholar] [CrossRef]
- Doitsh, G.; Galloway, N.L.; Geng, X.; Yang, Z.; Monroe, K.M.; Zepeda, O.; Hunt, P.W.; Hatano, H.; Sowinski, S.; Muñoz-Arias, I.; et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 2014, 505, 509–514. [Google Scholar] [CrossRef]
- Katano, H.; Hishima, T.; Mochizuki, M.; Kodama, Y.; Oyaizu, N.; Ota, Y.; Mine, S.; Igari, T.; Ajisawa, A.; Teruya, K.; et al. The prevalence of opportunistic infections and malignancies in autopsied patients with human immunodeficiency virus infection in Japan. BMC Infect. Dis. 2014, 14, 229. [Google Scholar] [CrossRef]
- Zhou, Y.; Maldini, C.R.; Jadlowsky, J.; Riley, J.L. Challenges and Opportunities of Using Adoptive T-Cell Therapy as Part of an HIV Cure Strategy. J. Infect. Dis. 2021, 223, 38–45. [Google Scholar] [CrossRef] [PubMed]
- De Feo, C.J.; Weiss, C.D. Escape from human immunodeficiency virus type 1 (HIV-1) entry inhibitors. Viruses 2012, 4, 3859–3911. [Google Scholar] [CrossRef]
- Kilby, J.M.; Hopkins, S.; Venetta, T.M.; DiMassimo, B.; Cloud, G.A.; Lee, J.Y.; Alldredge, L.; Hunter, E.; Lambert, D.; Bolognesi, D.; et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998, 4, 1302–1307. [Google Scholar] [CrossRef]
- Matthews, T.; Salgo, M.; Greenberg, M.; Chung, J.; DeMasi, R.; Bolognesi, D. Enfuvirtide: The first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004, 3, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Hildinger, M.; Dittmar, M.T.; Schult-Dietrich, P.; Fehse, B.; Schnierle, B.S.; Thaler, S.; Stiegler, G.; Welker, R.; von Laer, D. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J. Virol. 2001, 75, 3038–3042. [Google Scholar] [CrossRef]
- Ingallinella, P.; Bianchi, E.; Ladwa, N.A.; Wang, Y.J.; Hrin, R.; Veneziano, M.; Bonelli, F.; Ketas, T.J.; Moore, J.P.; Miller, M.D.; et al. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc. Natl. Acad. Sci. USA 2009, 106, 5801–5806. [Google Scholar] [CrossRef]
- Zhu, Y.; Chong, H.; Yu, D.; Guo, Y.; Zhou, Y.; He, Y. Design and Characterization of Cholesterylated Peptide HIV-1/2 Fusion Inhibitors with Extremely Potent and Long-Lasting Antiviral Activity. J. Virol. 2019, 93, e02312-18. [Google Scholar] [CrossRef]
- Lunzen, J.V.; Glaunsinger, T.; Stahmer, I.; Baehr, V.V.; Baum, C.; Schilz, A.; Kuehlcke, K.; Naundorf, S.; Martinius, H.; Hermann, F.; et al. Transfer of Autologous Gene-modified T Cells in HIV-infected Patients with Advanced Immunodeficiency and Drug-resistant Virus. Mol. Ther. 2007, 15, 1024–1033. [Google Scholar] [CrossRef]
- Delville, M.; Touzot, F.; Couzin, C.; Hmitou, I.; Djerroudi, L.; Ouedrani, A.; Lefrère, F.; Tuchman-Durand, C.; Mollet, C.; Fabreguettes, J.-R.; et al. Safety of CD34+ Hematopoietic Stem Cells and CD4+ T Lymphocytes Transduced with LVsh5/C46 in HIV-1 Infected Patients with High-Risk Lymphoma. Mol. Ther.—Methods Clin. Dev. 2019, 13, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H.; Keller, M.D.; Hanley, P.J.; Bollard, C.M. Virus-Specific T Cell Therapies for HIV: Lessons Learned from Hematopoietic Stem Cell Transplantation. Front. Cell. Infect. Microbiol. 2020, 10, 298. [Google Scholar] [CrossRef] [PubMed]
- Maslennikova, A.; Kruglova, N.; Kalinichenko, S.; Komkov, D.; Shepelev, M.; Golubev, D.; Siniavin, A.; Vzorov, A.; Filatov, A.; Mazurov, D. Engineering T-Cell Resistance to HIV-1 Infection via Knock-In of Peptides from the Heptad Repeat 2 Domain of gp41. mBio 2022, 13, e0358921. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Yao, X.; Sun, J.; Qiu, Z.; Zhang, M.; Waltersperger, S.; Wang, M.; Cui, S.; He, Y. The M-T hook structure is critical for design of HIV-1 fusion inhibitors. J. Biol. Chem. 2012, 287, 34558–34568. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Borrego, P.; Ding, X.; Zhu, Y.; Martins, A.; Chong, H.; Taveira, N.; He, Y. A Helical Short-Peptide Fusion Inhibitor with Highly Potent Activity against Human Immunodeficiency Virus Type 1 (HIV-1), HIV-2, and Simian Immunodeficiency Virus. J. Virol. 2017, 91, e01839-16. [Google Scholar] [CrossRef]
- Larson, S.M.; Truscott, L.C.; Chiou, T.T.; Patel, A.; Kao, R.; Tu, A.; Tyagi, T.; Lu, X.; Elashoff, D.; De Oliveira, S.N. Pre-clinical development of gene modification of haematopoietic stem cells with chimeric antigen receptors for cancer immunotherapy. Hum. Vaccines Immunother. 2017, 13, 1094–1104. [Google Scholar] [CrossRef]
- Hoyos, V.; Savoldo, B.; Quintarelli, C.; Mahendravada, A.; Zhang, M.; Vera, J.; Heslop, H.E.; Rooney, C.M.; Brenner, M.K.; Dotti, G. Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia 2010, 24, 1160–1170. [Google Scholar] [CrossRef]
- Budde, L.E.; Berger, C.; Lin, Y.; Wang, J.; Lin, X.; Frayo, S.E.; Brouns, S.A.; Spencer, D.M.; Till, B.G.; Jensen, M.C.; et al. Combining a CD20 chimeric antigen receptor and an inducible caspase 9 suicide switch to improve the efficacy and safety of T cell adoptive immunotherapy for lymphoma. PLoS ONE 2013, 8, e82742. [Google Scholar] [CrossRef]
- Diaconu, I.; Ballard, B.; Zhang, M.; Chen, Y.; West, J.; Dotti, G.; Savoldo, B. Inducible Caspase-9 Selectively Modulates the Toxicities of CD19-Specific Chimeric Antigen Receptor-Modified T Cells. Mol. Ther. 2017, 25, 580–592. [Google Scholar] [CrossRef]
- Wang, X.; Chang, W.C.; Wong, C.W.; Colcher, D.; Sherman, M.; Ostberg, J.R.; Forman, S.J.; Riddell, S.R.; Jensen, M.C. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 2011, 118, 1255–1263. [Google Scholar] [CrossRef]
- Paszkiewicz, P.J.; Fräßle, S.P.; Srivastava, S.; Sommermeyer, D.; Hudecek, M.; Drexler, I.; Sadelain, M.; Liu, L.; Jensen, M.C.; Riddell, S.R.; et al. Targeted antibody-mediated depletion of murine CD19 CAR T cells permanently reverses B cell aplasia. J. Clin. Investig. 2016, 126, 4262–4272. [Google Scholar] [CrossRef]
- Philip, B.; Kokalaki, E.; Mekkaoui, L.; Thomas, S.; Straathof, K.; Flutter, B.; Marin, V.; Marafioti, T.; Chakraverty, R.; Linch, D.; et al. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy. Blood 2014, 124, 1277–1287. [Google Scholar] [CrossRef]
- Vogler, I.; Newrzela, S.; Hartmann, S.; Schneider, N.; von Laer, D.; Koehl, U.; Grez, M. An improved bicistronic CD20/tCD34 vector for efficient purification and in vivo depletion of gene-modified T cells for adoptive immunotherapy. Mol. Ther. 2010, 18, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.; López, T.; Espinosa, R.; Arias, C.F.; Vollmers, C.; DuBois, R.M. A simplified workflow for monoclonal antibody sequencing. PLoS ONE 2019, 14, e0218717. [Google Scholar] [CrossRef] [PubMed]
- Rybchenko, V.S.; Panina, A.A.; Aliev, T.K.; Solopova, O.N.; Balabashin, D.S.; Novoseletsky, V.N.; Dolgikh, D.A.; Sveshnikov, P.G.; Kirpichnikov, M.P. Bispecific Antibodies for IFN-β Delivery to ErbB2(+) Tumors. Biomolecules 2021, 11, 1915. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Friguet, B.; Chaffotte, A.F.; Djavadi-Ohaniance, L.; Goldberg, M.E. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J. Immunol. Methods 1985, 77, 305–319. [Google Scholar] [CrossRef]
- Beaudoin-Bussières, G.; Richard, J.; Prévost, J.; Goyette, G.; Finzi, A. A new flow cytometry assay to measure antibody-dependent cellular cytotoxicity against SARS-CoV-2 Spike-expressing cells. STAR Protoc. 2021, 2, 100851. [Google Scholar] [CrossRef] [PubMed]
- Petricevic, B.; Laengle, J.; Singer, J.; Sachet, M.; Fazekas, J.; Steger, G.; Bartsch, R.; Jensen-Jarolim, E.; Bergmann, M. Trastuzumab mediates antibody-dependent cell-mediated cytotoxicity and phagocytosis to the same extent in both adjuvant and metastatic HER2/neu breast cancer patients. J. Transl. Med. 2013, 11, 307. [Google Scholar] [CrossRef]
- Mimura, K.; Kono, K.; Hanawa, M.; Kanzaki, M.; Nakao, A.; Ooi, A.; Fujii, H. Trastuzumab-mediated antibody-dependent cellular cytotoxicity against esophageal squamous cell carcinoma. Clin. Cancer Res. 2005, 11, 4898–4904. [Google Scholar] [CrossRef][Green Version]
- Kabat, E.A. Sequences of Proteins of Immunological Interest; DIANE Publishing Company: Collingdale, PA, USA, 1992. [Google Scholar]
- IGBLAST. Available online: https://www.ncbi.nlm.nih.gov/igblast/ (accessed on 3 November 2021).
- IMGT/V-QUEST. Available online: https://www.imgt.org/IMGT_vquest/input (accessed on 3 November 2021).
- Torres, M.; Casadevall, A. The immunoglobulin constant region contributes to affinity and specificity. Trends Immunol. 2008, 29, 91–97. [Google Scholar] [CrossRef]
- Casadevall, A.; Janda, A. Immunoglobulin isotype influences affinity and specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 12272–12273. [Google Scholar] [CrossRef] [PubMed]
- Janda, A.; Bowen, A.; Greenspan, N.S.; Casadevall, A. Ig Constant Region Effects on Variable Region Structure and Function. Front. Microbiol. 2016, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, D.; Yan, H.; Chong, H.; He, Y. Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J. Virol. 2020, 94, e00635-20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, D.; Hu, Y.; Wu, T.; Chong, H.; He, Y. SARS-CoV-2-derived fusion inhibitor lipopeptides exhibit highly potent and broad-spectrum activity against divergent human coronaviruses. Signal Transduct. Target. Ther. 2021, 6, 294. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Yu, Y.; Liu, N.; Ju, X.; Ding, Q.; He, Y. Design and characterization of novel SARS-CoV-2 fusion inhibitors with N-terminally extended HR2 peptides. Antivir. Res. 2023, 212, 105571. [Google Scholar] [CrossRef]
- Chong, H.; Xue, J.; Xiong, S.; Cong, Z.; Ding, X.; Zhu, Y.; Liu, Z.; Chen, T.; Feng, Y.; He, L.; et al. A Lipopeptide HIV-1/2 Fusion Inhibitor with Highly Potent In Vitro, Ex Vivo, and In Vivo Antiviral Activity. J. Virol. 2017, 91, e00288-17. [Google Scholar] [CrossRef]
- Harman, S.; Herrera, C.; Armanasco, N.; Nuttall, J.; Shattock, R.J. Preclinical Evaluation of the HIV-1 Fusion Inhibitor L’644 as a Potential Candidate Microbicide. Antimicrob. Agents Chemother. 2012, 56, 2347–2356. [Google Scholar] [CrossRef]
- Zotova, A.; Pichugin, A.; Atemasova, A.; Knyazhanskaya, E.; Lopatukhina, E.; Mitkin, N.; Holmuhamedov, E.; Gottikh, M.; Kuprash, D.; Filatov, A.; et al. Isolation of gene-edited cells via knock-in of short glycophosphatidylinositol-anchored epitope tags. Sci. Rep. 2019, 9, 3132. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef]
- Tebas, P.; Jadlowsky, J.K.; Shaw, P.A.; Tian, L.; Esparza, E.; Brennan, A.L.; Kim, S.; Naing, S.Y.; Richardson, M.W.; Vogel, A.N.; et al. CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. J. Clin. Investig. 2024, 131, e144486. [Google Scholar] [CrossRef]
- Xu, L.; Wang, J.; Liu, Y.; Xie, L.; Su, B.; Mou, D.; Wang, L.; Liu, T.; Wang, X.; Zhang, B.; et al. CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N. Engl. J. Med. 2019, 381, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Foote, J. Immunogenicity of engineered antibodies. Methods 2005, 36, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Verdun, N.; Marks, P. Secondary Cancers after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med. 2024, 390, 584–586. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.J.; Nguyen, T.; Rahman, M.; Er, J.; Li, J.; Li, K.; Lendvai, N.; Schecter, J.M.; Banerjee, A.; Roccia, T.; et al. CAR+ T-Cell Lymphoma Post Ciltacabtagene Autoleucel Therapy for Relapsed Refractory Multiple Myeloma. Blood 2023, 142, 6939. [Google Scholar] [CrossRef]
- Micklethwaite, K.P.; Gowrishankar, K.; Gloss, B.S.; Li, Z.; Street, J.A.; Moezzi, L.; Mach, M.A.; Sutrave, G.; Clancy, L.E.; Bishop, D.C.; et al. Investigation of product-derived lymphoma following infusion of piggyBac-modified CD19 chimeric antigen receptor T cells. Blood 2021, 138, 1391–1405. [Google Scholar] [CrossRef]


| T:E = 1:4 | ||||||
|---|---|---|---|---|---|---|
| chAb Concentration, µg/mL | 0.1 | 1 | 10 | |||
| ADCC, % | p-value | ADCC, % | p-value | ADCC, % | p-value | |
| CEM/R5/MT-C34 + D1 PBMCs | 3.28 ± 0.32 | 0.2285 | 8.88 ± 0.9 | 0.001 * | 27.88 ± 2.76 | <0.0001 * |
| CEM/R5/MT-C-34 + D2 PBMCs | 1.79 ± 2.11 | 0.2423 | 4.3 ± 2.04 | 0.027 * | 10.9 ± 3.00 | 0.0047 * |
| CEM/R5 + D1 PBMCs | 1.99 ± 1.54 | - | 1.91 ± 1.08 | - | 0.71 ± 0.41 | - |
| CEM/R5 + D2 PBMCs | 0.11 ± 0.23 | - | −0.06 ± 0.86 | - | −0.4 ± 1.68 | - |
| T:E = 1:7 | ||||||
| chAb concentration, µg/mL | 0.1 | 1 | 10 | |||
| ADCC, % | p-value | ADCC, % | p-value | ADCC, % | p-value | |
| CEM/R5/MT-C34 + D1 PBMC | 4.39 ± 5.14 | 0.2607 | 18.83 ± 5.88 | 0.0034 * | 43.62 ± 5.93 | 0.0003 * |
| CEM/R5/MT-C-34 + D2 PBMC | 6.57 ± 2.93 | 0.1795 | 15.85 ± 3.19 | 0.004 * | 23.27 ± 1.78 | 0.0003 * |
| CEM/R5 + D1 PBMC | 0.49 ± 0.46 | - | −2.29 ± 0.15 | - | −0.28 ± 1.77 | - |
| CEM/R5 + D2 PBMC | 1.39 ± 4.68 | - | 2.30 ± 2.34 | - | 0.22 ± 2.99 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinichenko, S.V.; Ramadan, L.; Kruglova, N.A.; Balagurov, K.I.; Lukashina, M.I.; Mazurov, D.V.; Shepelev, M.V. A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity. Biology 2024, 13, 675. https://doi.org/10.3390/biology13090675
Kalinichenko SV, Ramadan L, Kruglova NA, Balagurov KI, Lukashina MI, Mazurov DV, Shepelev MV. A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity. Biology. 2024; 13(9):675. https://doi.org/10.3390/biology13090675
Chicago/Turabian StyleKalinichenko, Svetlana V., Lama Ramadan, Natalia A. Kruglova, Konstantin I. Balagurov, Marina I. Lukashina, Dmitriy V. Mazurov, and Mikhail V. Shepelev. 2024. "A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity" Biology 13, no. 9: 675. https://doi.org/10.3390/biology13090675
APA StyleKalinichenko, S. V., Ramadan, L., Kruglova, N. A., Balagurov, K. I., Lukashina, M. I., Mazurov, D. V., & Shepelev, M. V. (2024). A New Chimeric Antibody against the HIV-1 Fusion Inhibitory Peptide MT-C34 with a High Affinity and Fc-Mediated Cellular Cytotoxicity. Biology, 13(9), 675. https://doi.org/10.3390/biology13090675







