Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks
Simple Summary
Abstract
1. Introduction
2. Discovery of the MCM2-7 Genes
3. Discovery of DNA Helicase Activity in MCM2-7
4. MCM2-7 as a Central Factor for an Active Replicative Helicase Complex
5. Transition of the Inactive MCM2-7 Double Hexamer to an Active Replication Fork
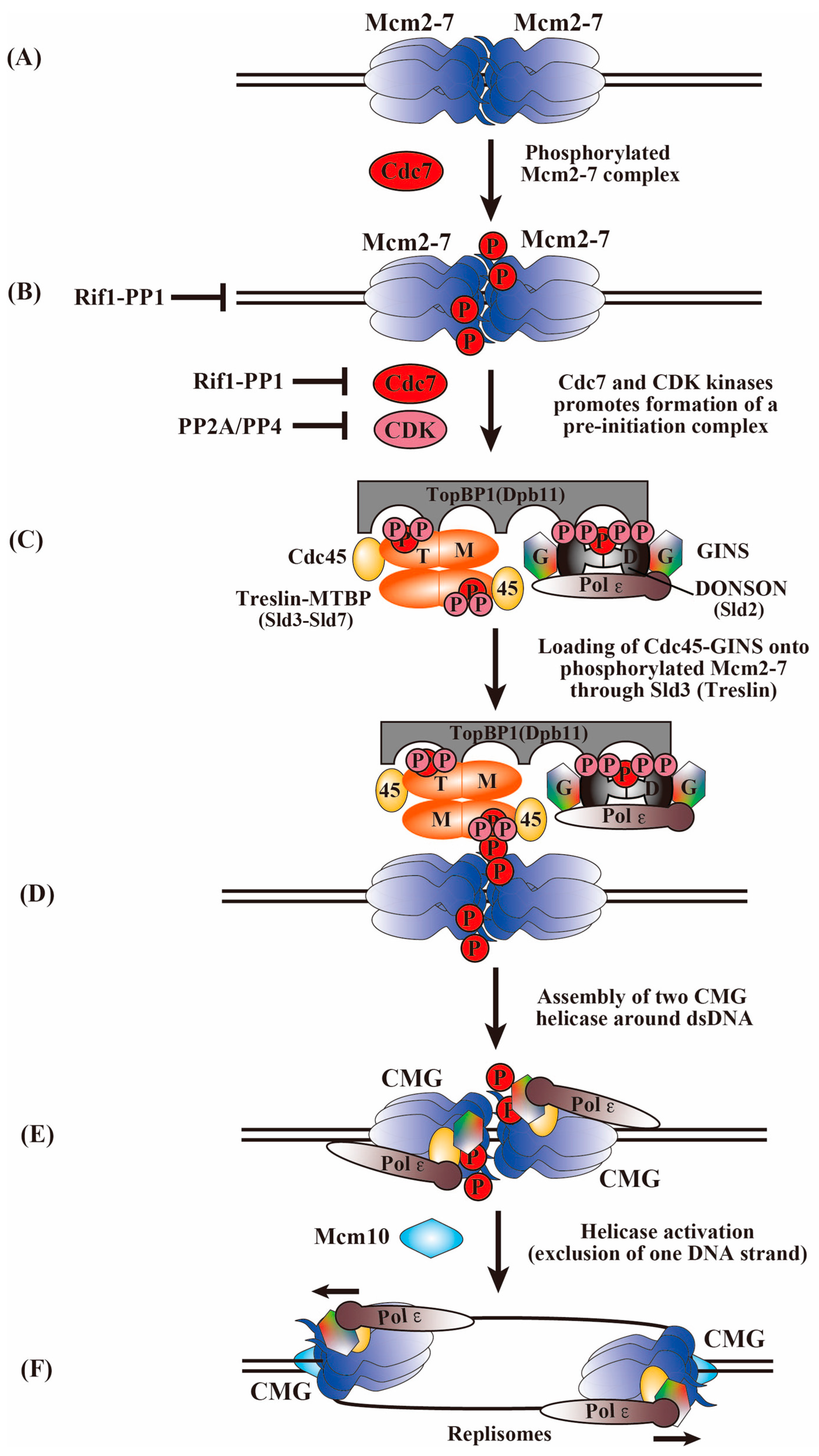
6. Phosphorylation of MCM2-7 by Cdc7 and CDK Kinases for the Activation of the MCM2-7 Helicase and the Initiation of DNA Replication
7. Phosphorylation of Firing Factors by Cdc7 and CDK Kinases for Activation of CMG Assembly
8. Roles of Phosphatases in the Control of Replication Initiation
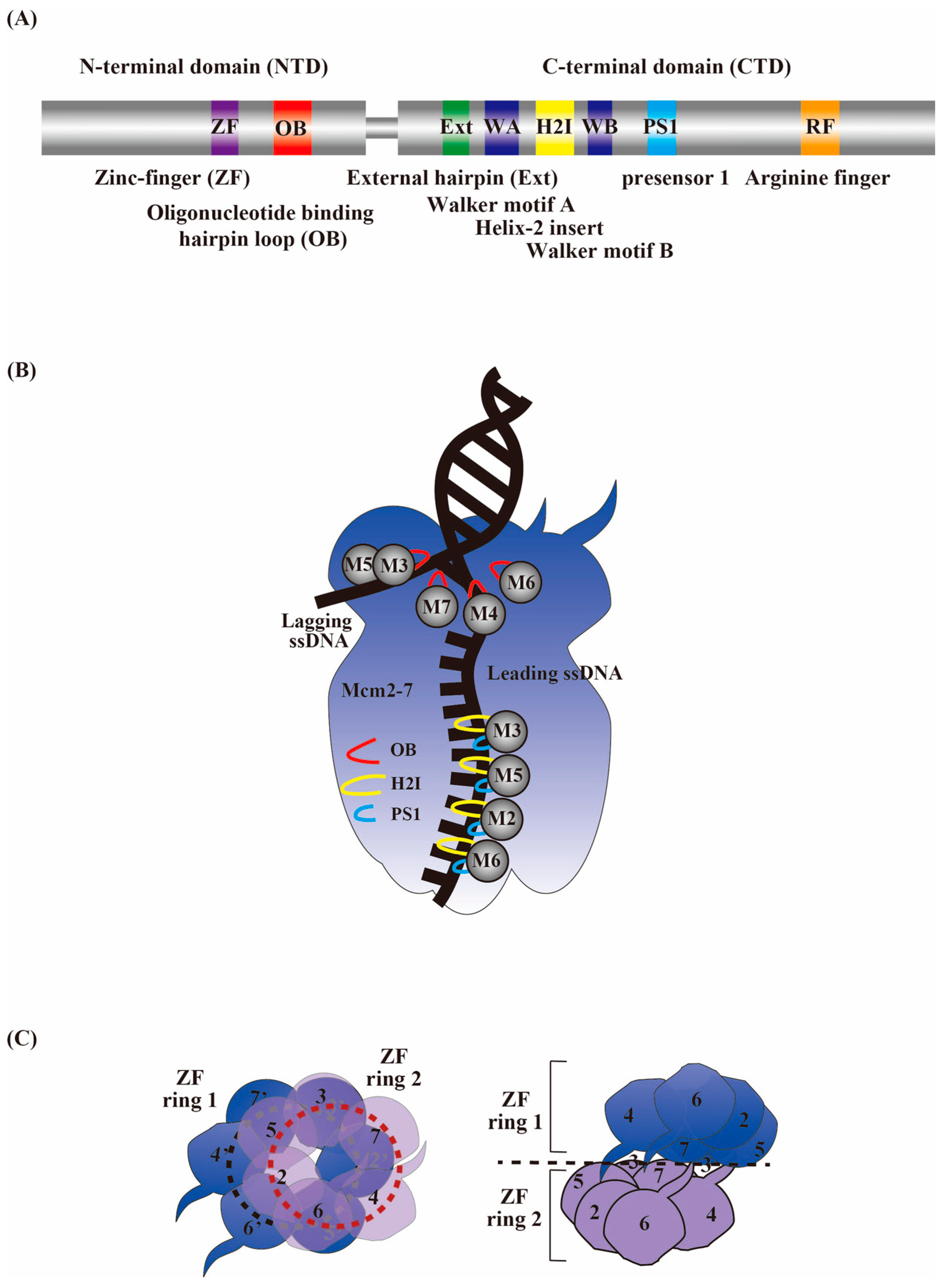
9. Factors Required for CMG Helicase Activation during DNA Replication Initiation
10. A Structural Perspective on the CMG Activation Mechanism
11. A Structural Perspective on the Core Replisome Cooperative Activation Mechanism
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ishimi, Y. Regulation of MCM2-7 function. Genes. Genet. Syst. 2018, 93, 125–133. [Google Scholar] [CrossRef]
- Hu, Y.; Stillman, B. Origins of DNA replication in eukaryotes. Mol. Cell 2023, 83, 352–372. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Blow, J.J. DDK: The Outsourced Kinase of Chromosome Maintenance. Biology 2022, 11, 877. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Ogawa, S. Dimerization of Firing Factors for Replication Origin Activation in Eukaryotes: A Crucial Process for Simultaneous Assembly of Bidirectional Replication Forks? Biology 2022, 11, 928. [Google Scholar] [CrossRef]
- Terui, R.; Berger, S.E.; Sambel, L.A.; Song, D.; Chistol, G. Single-molecule imaging reveals the mechanism of bidirectional replication initiation in metazoa. Cell 2024, 187, 3992–4009.e25. [Google Scholar] [CrossRef]
- Jenkinson, F.; Zegerman, P. Roles of phosphatases in eukaryotic DNA replication initiation control. DNA Repair 2022, 118, 103384. [Google Scholar] [CrossRef] [PubMed]
- Henrikus, S.S.; Gross, M.H.; Willhoft, O.; Puhringer, T.; Lewis, J.S.; McClure, A.W.; Greiwe, J.F.; Palm, G.; Nans, A.; Diffley, J.F.X.; et al. Unwinding of a eukaryotic origin of replication visualized by cryo-EM. Nat. Struct. Mol. Biol. 2024, 8, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lam, W.H.; Zhai, Y. Assembly and activation of replicative helicases at origin DNA for replication initiation. Curr. Opin. Struct. Biol. 2024, 88, 102876. [Google Scholar] [CrossRef] [PubMed]
- Rankin, B.D.; Rankin, S. The MCM2-7 Complex: Roles beyond DNA Unwinding. Biology 2024, 13, 258. [Google Scholar] [CrossRef]
- Maine, G.T.; Sinha, P.; Tye, B.K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 1984, 106, 365–385. [Google Scholar] [CrossRef]
- Gibson, S.I.; Surosky, R.T.; Tye, B.K. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol. Cell. Biol. 1990, 10, 5707–5720. [Google Scholar] [CrossRef]
- Hennessy, K.M.; Lee, A.; Chen, E.; Botstein, D. A group of interacting yeast DNA replication genes. Genes. Dev. 1991, 5, 958–969. [Google Scholar] [CrossRef]
- Coxon, A.; Maundrell, K.; Kearsey, S.E. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992, 20, 5571–5577. [Google Scholar] [CrossRef]
- Nasmyth, K.; Nurse, P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1981, 182, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Okishio, N.; Samejima, I.; Hiraoka, Y.; Toda, T.; Saitoh, I.; Yanagida, M. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol. Biol. Cell 1993, 4, 1003–1015. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Takahashi, K.; Yamada, H.; Yanagida, M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 1994, 5, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Gibson, S.; Tye, B.K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes. Dev. 1991, 5, 944–957. [Google Scholar] [CrossRef]
- Hu, B.; Burkhart, R.; Schulte, D.; Musahl, C.; Knippers, R. The P1 family: A new class of nuclear mammalian proteins related to the yeast Mcm replication proteins. Nucleic Acids Res. 1993, 21, 5289–5293. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Nozaki, N.; Sugimoto, K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO J. 1994, 13, 4311–4320. [Google Scholar] [CrossRef]
- Blow, J.J.; Laskey, R.A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 1988, 332, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Mimura, S.; Nishimoto, S.; Takisawa, H.; Nojima, H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell 1995, 81, 601–609. [Google Scholar] [CrossRef]
- Chong, J.P.; Mahbubani, H.M.; Khoo, C.Y.; Blow, J.J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 1995, 375, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Madine, M.A.; Khoo, C.Y.; Mills, A.D.; Laskey, R.A. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature 1995, 375, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993, 21, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Usukura, J.; Yanagida, M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes. Cells 1997, 2, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997, 272, 24508–24513. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Komamura, Y.; Ishimi, Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol. Cell. Biol. 1999, 19, 8003–8015. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Ishimi, Y.; Masai, H.; Hanaoka, F. Roles of Mcm7 and Mcm4 subunits in the DNA helicase activity of the mouse Mcm4/6/7 complex. J. Biol. Chem. 2002, 277, 42471–42479. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Hurwitz, J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 2001, 98, 54–59. [Google Scholar] [CrossRef]
- You, Z.; Ishimi, Y.; Mizuno, T.; Sugasawa, K.; Hanaoka, F.; Masai, H. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J. 2003, 22, 6148–6160. [Google Scholar] [CrossRef]
- Langston, L.D.; Georgescu, R.E.; O’Donnell, M.E. Mechanism of eukaryotic origin unwinding is a dual helicase DNA shearing process. Proc. Natl. Acad. Sci. USA 2023, 120, e2316466120. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Masai, H. DNA binding and helicase actions of mouse MCM4/6/7 helicase. Nucleic Acids Res. 2005, 33, 3033–3047. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Dong, J.; Wang, W.; Yu, D.; Fan, X.; Hui, Y.C.; Lee, C.S.K.; Lam, W.H.; Alary, N.; Yang, Y.; et al. The human pre-replication complex is an open complex. Cell 2023, 186, 98–111.e21. [Google Scholar] [CrossRef]
- Emerson, D.J.; Zhao, P.A.; Cook, A.L.; Barnett, R.J.; Klein, K.N.; Saulebekova, D.; Ge, C.; Zhou, L.; Simandi, Z.; Minsk, M.K.; et al. Cohesin-mediated loop anchors confine the locations of human replication origins. Nature 2022, 606, 812–819. [Google Scholar] [CrossRef]
- Sato, M.; Gotow, T.; You, Z.; Komamura-Kohno, Y.; Uchiyama, Y.; Yabuta, N.; Nojima, H.; Ishimi, Y. Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol. 2000, 300, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Komamura, Y.; You, Z.; Kimura, H. Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 1998, 273, 8369–8375. [Google Scholar] [CrossRef]
- Lee, J.K.; Hurwitz, J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem. 2000, 275, 18871–18878. [Google Scholar] [CrossRef]
- Schwacha, A.; Bell, S.P. Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell 2001, 8, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Prokhorova, T.A.; Blow, J.J. Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem. 2000, 275, 2491–2498. [Google Scholar] [CrossRef] [PubMed]
- Labib, K.; Tercero, J.A.; Diffley, J.F. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 2000, 288, 1643–1647. [Google Scholar] [CrossRef]
- Kanter, D.M.; Bruck, I.; Kaplan, D.L. Mcm subunits can assemble into two different active unwinding complexes. J. Biol. Chem. 2008, 283, 31172–31182. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 2007, 282, 33795–33804. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Schwacha, A. The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 2008, 31, 287–293. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Masai, H. Potent DNA strand annealing activity associated with mouse Mcm2 approximately 7 heterohexameric complex. Nucleic Acids Res. 2017, 45, 6494–6506. [Google Scholar] [CrossRef] [PubMed]
- Burnham, D.R.; Kose, H.B.; Hoyle, R.B.; Yardimci, H. The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat. Commun. 2019, 10, 2159. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed]
- Calzada, A.; Hodgson, B.; Kanemaki, M.; Bueno, A.; Labib, K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes. Dev. 2005, 19, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Kamimura, Y.; Okawa, M.; Muramatsu, S.; Sugino, A.; Araki, H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes. Dev. 2003, 17, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Kubota, Y.; Takase, Y.; Komori, Y.; Hashimoto, Y.; Arata, T.; Kamimura, Y.; Araki, H.; Takisawa, H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes. Dev. 2003, 17, 1141–1152. [Google Scholar] [CrossRef]
- Kang, Y.H.; Galal, W.C.; Farina, A.; Tappin, I.; Hurwitz, J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 6042–6047. [Google Scholar] [CrossRef]
- Masai, H.; Matsumoto, S.; You, Z.; Yoshizawa-Sugata, N.; Oda, M. Eukaryotic chromosome DNA replication: Where, when, and how? Annu. Rev. Biochem. 2010, 79, 89–130. [Google Scholar] [CrossRef]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022, 91, 107–131. [Google Scholar] [CrossRef]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F. Concerted Loading of Mcm2-7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef]
- Yardimci, H.; Walter, J.C. Prereplication-complex formation: A molecular double take? Nat. Struct. Mol. Biol. 2014, 21, 20–25. [Google Scholar] [CrossRef]
- Labib, K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes. Dev. 2010, 24, 1208–1219. [Google Scholar] [CrossRef]
- Bochman, M.L.; Schwacha, A. The Mcm complex: Unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009, 73, 652–683. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Looke, M.; Maloney, M.F.; Bell, S.P. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes. Dev. 2017, 31, 291–305. [Google Scholar] [CrossRef]
- Miyazawa-Onami, M.; Araki, H.; Tanaka, S. Pre-initiation complex assembly functions as a molecular switch that splits the Mcm2-7 double hexamer. EMBO Rep. 2017, 18, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Gross, M.H.; Sousa, J.; Henrikus, S.S.; Greiwe, J.F.; Nans, A.; Diffley, J.F.X.; Costa, A. Mechanism of replication origin melting nucleated by CMG helicase assembly. Nature 2022, 606, 1007–1014. [Google Scholar] [CrossRef]
- Sato, N.; Arai, K.; Masai, H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: In vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997, 16, 4340–4351. [Google Scholar] [CrossRef]
- Jackson, A.L.; Pahl, P.M.; Harrison, K.; Rosamond, J.; Sclafani, R.A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 1993, 13, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Masai, H. Regulation of chromosome dynamics by Hsk1/Cdc7 kinase. Biochem. Soc. Trans. 2013, 41, 1712–1719. [Google Scholar] [CrossRef]
- Masai, H.; Matsui, E.; You, Z.; Ishimi, Y.; Tamai, K.; Arai, K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J. Biol. Chem. 2000, 275, 29042–29052. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Komamura-Kohno, Y.; Arai, K.; Masai, H. Biochemical activities associated with mouse Mcm2 protein. J. Biol. Chem. 2001, 276, 42744–42752. [Google Scholar] [CrossRef]
- Lei, M.; Kawasaki, Y.; Young, M.R.; Kihara, M.; Sugino, A.; Tye, B.K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes. Dev. 1997, 11, 3365–3374. [Google Scholar] [CrossRef]
- Jiang, W.; McDonald, D.; Hope, T.J.; Hunter, T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999, 18, 5703–5713. [Google Scholar] [CrossRef] [PubMed]
- Takeda, T.; Ogino, K.; Tatebayashi, K.; Ikeda, H.; Arai, K.; Masai, H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell 2001, 12, 1257–1274. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sheu, Y.J.; Stillman, B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010, 463, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hardy, C.F.; Dryga, O.; Seematter, S.; Pahl, P.M.; Sclafani, R.A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA 1997, 94, 3151–3155. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.L.; Leon, R.P.; Pessoa-Brandao, L.; Hunt, S.; Raghuraman, M.K.; Fangman, W.L.; Brewer, B.J.; Sclafani, R.A. Structural changes in Mcm5 protein bypass Cdc7-Dbf4 function and reduce replication origin efficiency in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 7594–7602. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.J.; Bishop, B.E.; Leon, R.P.; Sclafani, R.A.; Ogata, C.M.; Chen, X.S. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003, 10, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Masai, H.; Taniyama, C.; Ogino, K.; Matsui, E.; Kakusho, N.; Matsumoto, S.; Kim, J.M.; Ishii, A.; Tanaka, T.; Kobayashi, T.; et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 2006, 281, 39249–39261. [Google Scholar] [CrossRef]
- Jares, P.; Blow, J.J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes. Dev. 2000, 14, 1528–1540. [Google Scholar] [CrossRef]
- Alver, R.C.; Chadha, G.S.; Gillespie, P.J.; Blow, J.J. Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cell Rep. 2017, 18, 2508–2520. [Google Scholar] [CrossRef]
- Ishimi, Y.; Komamura-Kohno, Y.; You, Z.; Omori, A.; Kitagawa, M. Inhibition of Mcm4,6,7 helicase activity by phosphorylation with cyclin A/Cdk2. J. Biol. Chem. 2000, 275, 16235–16241. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, L.W.; Lents, N.H.; Baldassare, J.J. Cyclin A-CDK activity during G1 phase impairs MCM chromatin loading and inhibits DNA synthesis in mammalian cells. Cell Cycle 2008, 7, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wu, R.A.; Sonneville, R.; Kochenova, O.V.; Labib, K.; Pellman, D.; Walter, J.C. Mitotic CDK Promotes Replisome Disassembly, Fork Breakage, and Complex DNA Rearrangements. Mol. Cell 2019, 73, 915–929.e6. [Google Scholar] [CrossRef]
- Chen, S.; Bell, S.P. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes. Dev. 2011, 25, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Li, N.; Huo, Y.; Dang, S.; Tye, B.K.; Gao, N.; Zhai, Y. Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase. Nat. Commun. 2022, 13, 1396. [Google Scholar] [CrossRef] [PubMed]
- Greiwe, J.F.; Miller, T.C.R.; Locke, J.; Martino, F.; Howell, S.; Schreiber, A.; Nans, A.; Diffley, J.F.X.; Costa, A. Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase. Nat. Struct. Mol. Biol. 2022, 29, 10–20. [Google Scholar] [CrossRef]
- Saleh, A.; Noguchi, Y.; Aramayo, R.; Ivanova, M.E.; Stevens, K.M.; Montoya, A.; Sunidhi, S.; Carranza, N.L.; Skwark, M.J.; Speck, C. The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer. Nat. Commun. 2022, 13, 2915. [Google Scholar] [CrossRef] [PubMed]
- Abd Wahab, S.; Remus, D. Antagonistic control of DDK binding to licensed replication origins by Mcm2 and Rad53. eLife 2020, 9, e58571. [Google Scholar] [CrossRef]
- Masumoto, H.; Muramatsu, S.; Kamimura, Y.; Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 2002, 415, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.S.; Tanaka, Y.; Endo, S.; Kamimura, Y.; Araki, H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006, 25, 1987–1996. [Google Scholar] [CrossRef]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef]
- Zegerman, P.; Diffley, J.F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Muramatsu, S.; Hirai, K.; Tak, Y.S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon, and GINS in budding yeast. Genes. Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J. Cell Biol. 2011, 193, 995–1007. [Google Scholar] [CrossRef]
- Deegan, T.D.; Yeeles, J.T.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016, 35, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Umemori, T.; Endo, S.; Muramatsu, S.; Kanemaki, M.; Kamimura, Y.; Obuse, C.; Araki, H. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J. 2011, 30, 2019–2030. [Google Scholar] [CrossRef]
- Kamimura, Y.; Tak, Y.S.; Sugino, A.; Araki, H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001, 20, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef]
- Yuan, Z.; Bai, L.; Sun, J.; Georgescu, R.; Liu, J.; O’Donnell, M.E.; Li, H. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 2016, 23, 217–224. [Google Scholar] [CrossRef]
- Goswami, P.; Abid Ali, F.; Douglas, M.E.; Locke, J.; Purkiss, A.; Janska, A.; Eickhoff, P.; Early, A.; Nans, A.; Cheung, A.M.C.; et al. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat. Commun. 2018, 9, 5061. [Google Scholar] [CrossRef]
- De Jesus-Kim, L.; Friedman, L.J.; Looke, M.; Ramsoomair, C.K.; Gelles, J.; Bell, S.P. DDK regulates replication initiation by controlling the multiplicity of Cdc45-GINS binding to Mcm2-7. eLife 2021, 10, e65471. [Google Scholar] [CrossRef]
- Nougarede, R.; Della Seta, F.; Zarzov, P.; Schwob, E. Hierarchy of S-phase-promoting factors: Yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 2000, 20, 3795–3806. [Google Scholar] [CrossRef]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic Origin-Dependent DNA Replication In Vitro Reveals Sequential Action of DDK and S-CDK Kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Hayano, M.; Kanoh, Y.; Matsumoto, S.; Renard-Guillet, C.; Shirahige, K.; Masai, H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes. Dev. 2012, 26, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Hayano, M.; Kanoh, Y.; Masai, H. Multiple pathways can bypass the essential role of fission yeast Hsk1 kinase in DNA replication initiation. J. Cell Biol. 2011, 195, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Suski, J.M.; Ratnayeke, N.; Braun, M.; Zhang, T.; Strmiska, V.; Michowski, W.; Can, G.; Simoneau, A.; Snioch, K.; Cup, M.; et al. CDC7-independent G1/S transition revealed by targeted protein degradation. Nature 2022, 605, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.; Alvino, G.M.; Chang, F.; Lian, H.Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes. Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.I.; Ly, T.; Garzon, J.; Horejsi, Z.; Ohkubo, Y.N.; Endo, A.; Obuse, C.; Boulton, S.J.; Lamond, A.I.; Donaldson, A.D. Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 2017, 18, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Ishii, A.; Kanoh, Y.; Oda, M.; Nishito, Y.; Masai, H. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012, 31, 3667–3677. [Google Scholar] [CrossRef]
- Volpi, I.; Gillespie, P.J.; Chadha, G.S.; Blow, J.J. The role of DDK and Treslin-MTBP in coordinating replication licensing and pre-initiation complex formation. Open Biol. 2021, 11, 210121. [Google Scholar] [CrossRef]
- Jenkinson, F.; Tan, K.W.; Schopf, B.; Santos, M.M.; Zegerman, P. Dephosphorylation of the pre-initiation complex is critical for origin firing. Mol. Cell 2023, 83, 12–25.e10. [Google Scholar] [CrossRef]
- Godfrey, M.; Touati, S.A.; Kataria, M.; Jones, A.; Snijders, A.P.; Uhlmann, F. PP2A(Cdc55) Phosphatase Imposes Ordered Cell-Cycle Phosphorylation by Opposing Threonine Phosphorylation. Mol. Cell 2017, 65, 393–402.e3. [Google Scholar] [CrossRef]
- Krasinska, L.; Domingo-Sananes, M.R.; Kapuy, O.; Parisis, N.; Harker, B.; Moorhead, G.; Rossignol, M.; Novak, B.; Fisher, D. Protein phosphatase 2A controls the order and dynamics of cell-cycle transitions. Mol. Cell 2011, 44, 437–450. [Google Scholar] [CrossRef]
- Lin, X.H.; Walter, J.; Scheidtmann, K.; Ohst, K.; Newport, J.; Walter, G. Protein phosphatase 2A is required for the initiation of chromosomal DNA replication. Proc. Natl. Acad. Sci. USA 1998, 95, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Boos, D.; Yekezare, M.; Diffley, J.F. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 2013, 340, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010, 140, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Sansam, C.G.; Goins, D.; Siefert, J.C.; Clowdus, E.A.; Sansam, C.L. Cyclin-dependent kinase regulates the length of S phase through TICRR/TRESLIN phosphorylation. Genes. Dev. 2015, 29, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Tamayo-Orrego, L.; Schmid, E.; Tarnauskaite, Z.; Kochenova, O.V.; Gruar, R.; Muramatsu, S.; Lynch, L.; Schlie, A.V.; Carroll, P.L.; et al. In silico protein interaction screening uncovers DONSON’s role in replication initiation. Science 2023, 381, eadi3448. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Sadano, K.; Miyata, N.; Ito, H.; Tanaka, H. Novel role of DONSON in CMG helicase assembly during vertebrate DNA replication initiation. EMBO J. 2023, 42, e114131. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sonneville, R.; Jenkyn-Bedford, M.; Ji, L.; Alabert, C.; Hong, Y.; Yeeles, J.T.P.; Labib, K.P.M. DNSN-1 recruits GINS for CMG helicase assembly during DNA replication initiation in Caenorhabditis elegans. Science 2023, 381, eadi4932. [Google Scholar] [CrossRef]
- Kingsley, G.; Skagia, A.; Passaretti, P.; Fernandez-Cuesta, C.; Reynolds-Winczura, A.; Koscielniak, K.; Gambus, A. DONSON facilitates Cdc45 and GINS chromatin association and is essential for DNA replication initiation. Nucleic Acids Res. 2023, 51, 9748–9763. [Google Scholar] [CrossRef]
- Evrin, C.; Alvarez, V.; Ainsworth, J.; Fujisawa, R.; Alabert, C.; Labib, K.P. DONSON is required for CMG helicase assembly in the mammalian cell cycle. EMBO Rep. 2023, 24, e57677. [Google Scholar] [CrossRef]
- Padayachy, L.; Ntallis, S.G.; Halazonetis, T.D. RECQL4 is not critical for firing of human DNA replication origins. Sci. Rep. 2024, 14, 7708. [Google Scholar] [CrossRef] [PubMed]
- Sangrithi, M.N.; Bernal, J.A.; Madine, M.; Philpott, A.; Lee, J.; Dunphy, W.G.; Venkitaraman, A.R. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005, 121, 887–898. [Google Scholar] [CrossRef]
- Matsuno, K.; Kumano, M.; Kubota, Y.; Hashimoto, Y.; Takisawa, H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 2006, 26, 4843–4852. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, M.A.; Passaretti, P.; Butryn, A.; Reynolds-Winczura, A.; Kingsley, G.; Skagia, A.; Fernandez-Cuesta, C.; Poovathumkadavil, D.; George, R.; Chauhan, A.S.; et al. The structural mechanism of dimeric DONSON in replicative helicase activation. Mol. Cell 2023, 83, 4017–4031.e9. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Tetik, B.; Parlak, M.; Almeida-Hernandez, Y.; Raschle, M.; Kaschani, F.; Siegert, H.; Marko, A.; Sanchez-Garcia, E.; Kaiser, M.; et al. TopBP1 utilises a bipartite GINS binding mode to support genome replication. Nat. Commun. 2024, 15, 1797. [Google Scholar] [CrossRef]
- van Deursen, F.; Sengupta, S.; De Piccoli, G.; Sanchez-Diaz, A.; Labib, K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012, 31, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Kanke, M.; Kodama, Y.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012, 31, 2182–2194. [Google Scholar] [CrossRef]
- Watase, G.; Takisawa, H.; Kanemaki, M.T. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr. Biol. 2012, 22, 343–349. [Google Scholar] [CrossRef]
- Langston, L.D.; Mayle, R.; Schauer, G.D.; Yurieva, O.; Zhang, D.; Yao, N.Y.; Georgescu, R.E.; O’Donnell, M.E. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. eLife 2017, 6, e29118. [Google Scholar] [CrossRef]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Cook, C.R.; Kung, G.; Peterson, F.C.; Volkman, B.F.; Lei, M. A novel zinc finger is required for Mcm10 homocomplex assembly. J. Biol. Chem. 2003, 278, 36051–36058. [Google Scholar] [CrossRef]
- Mayle, R.; Langston, L.; Molloy, K.R.; Zhang, D.; Chait, B.T.; O’Donnell, M.E. Mcm10 has potent strand-annealing activity and limits translocase-mediated fork regression. Proc. Natl. Acad. Sci. USA 2019, 116, 798–803. [Google Scholar] [CrossRef]
- Kurat, C.F.; Yeeles, J.T.P.; Patel, H.; Early, A.; Diffley, J.F.X. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol. Cell 2017, 65, 117–130. [Google Scholar] [CrossRef]
- Kose, H.B.; Xie, S.; Cameron, G.; Strycharska, M.S.; Yardimci, H. Duplex DNA engagement and RPA oppositely regulate the DNA-unwinding rate of CMG helicase. Nat. Commun. 2020, 11, 3713. [Google Scholar] [CrossRef]
- Spinks, R.R.; Spenkelink, L.M.; Dixon, N.E.; van Oijen, A.M. Single-Molecule Insights Into the Dynamics of Replicative Helicases. Front. Mol. Biosci. 2021, 8, 741718. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Baris, Y.; Taylor, M.R.G.; Yeeles, J.T.P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 2021, 40, e108819. [Google Scholar] [CrossRef] [PubMed]
- Jenkyn-Bedford, M.; Jones, M.L.; Baris, Y.; Labib, K.P.M.; Cannone, G.; Yeeles, J.T.P.; Deegan, T.D. A conserved mechanism for regulating replisome disassembly in eukaryotes. Nature 2021, 600, 743–747. [Google Scholar] [CrossRef]
- Baretic, D.; Jenkyn-Bedford, M.; Aria, V.; Cannone, G.; Skehel, M.; Yeeles, J.T.P. Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork. Mol. Cell 2020, 78, 926–940.e13. [Google Scholar] [CrossRef] [PubMed]
- Abid Ali, F.; Renault, L.; Gannon, J.; Gahlon, H.L.; Kotecha, A.; Zhou, J.C.; Rueda, D.; Costa, A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun. 2016, 7, 10708. [Google Scholar] [CrossRef] [PubMed]
- Eickhoff, P.; Kose, H.B.; Martino, F.; Petojevic, T.; Abid Ali, F.; Locke, J.; Tamberg, N.; Nans, A.; Berger, J.M.; Botchan, M.R.; et al. Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome. Cell Rep. 2019, 28, 2673–2688.e8. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Georgescu, R.; Schauer, G.D.; O’Donnell, M.E.; Li, H. Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome. Nat. Commun. 2020, 11, 3156. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.; O’Donnell, M. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. eLife 2017, 6, e23449. [Google Scholar] [CrossRef]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Scharer, O.D.; Walter, J.C. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Yuan, Z.; Bai, L.; Schneider, S.; Zhao, G.; Stillman, B.; Speck, C.; Li, H. Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc. Natl. Acad. Sci. USA 2017, 114, E9529–E9538. [Google Scholar] [CrossRef]
- Li, N.; Zhai, Y.; Zhang, Y.; Li, W.; Yang, M.; Lei, J.; Tye, B.K.; Gao, N. Structure of the eukaryotic MCM complex at 3.8 A. Nature 2015, 524, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.C.R.; Locke, J.; Greiwe, J.F.; Diffley, J.F.X.; Costa, A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 2019, 575, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Abid Ali, F.; Douglas, M.E.; Locke, J.; Pye, V.E.; Nans, A.; Diffley, J.F.X.; Costa, A. Cryo-EM structure of a licensed DNA replication origin. Nat. Commun. 2017, 8, 2241. [Google Scholar] [CrossRef] [PubMed]
- Brewster, A.S.; Wang, G.; Yu, X.; Greenleaf, W.B.; Carazo, J.M.; Tjajadia, M.; Klein, M.G.; Chen, X.S. Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA 2008, 105, 20191–20196. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Georgescu, R.; Bai, L.; Zhang, D.; Li, H.; O’Donnell, M.E. DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nat. Commun. 2020, 11, 688. [Google Scholar] [CrossRef]
- Georgescu, R.; Yuan, Z.; Bai, L.; de Luna Almeida Santos, R.; Sun, J.; Zhang, D.; Yurieva, O.; Li, H.; O’Donnell, M.E. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. USA 2017, 114, E697–E706. [Google Scholar] [CrossRef]
- Poplawski, A.; Grabowski, B.; Long, S.E.; Kelman, Z. The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem. 2001, 276, 49371–49377. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.J.; Shen, J.; Gomez-Llorente, Y.; Martin, C.S.; Carazo, J.M.; Chen, X.S. Double hexamer disruption and biochemical activities of Methanobacterium thermoautotrophicum MCM. J. Biol. Chem. 2005, 280, 42405–42410. [Google Scholar] [CrossRef]
- Evrin, C.; Fernandez-Cid, A.; Riera, A.; Zech, J.; Clarke, P.; Herrera, M.C.; Tognetti, S.; Lurz, R.; Speck, C. The ORC/Cdc6/MCM2-7 complex facilitates MCM2-7 dimerization during prereplicative complex formation. Nucleic Acids Res. 2014, 42, 2257–2269. [Google Scholar] [CrossRef] [PubMed]
- Langston, L.D.; O’Donnell, M.E. An explanation for origin unwinding in eukaryotes. eLife 2019, 8, e46515. [Google Scholar] [CrossRef] [PubMed]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Feng, J.; Yu, D.; Huo, Y.; Ma, X.; Lam, W.H.; Liu, Z.; Li, X.D.; Ishibashi, T.; Dang, S.; et al. Synergism between CMG helicase and leading strand DNA polymerase at replication fork. Nat. Commun. 2023, 14, 5849. [Google Scholar] [CrossRef] [PubMed]
- Charlton, S.J.; Flury, V.; Kanoh, Y.; Genzor, A.V.; Kollenstart, L.; Ao, W.; Brogger, P.; Weisser, M.B.; Adamus, M.; Alcaraz, N.; et al. The fork protection complex promotes parental histone recycling and epigenetic memory. Cell 2024, 187, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Gao, Y.; Zhang, Y.; Yu, D.; Lin, J.; Feng, J.; Li, J.; Xu, Z.; Zhang, Y.; Dang, S.; et al. Parental histone transfer caught at the replication fork. Nature 2024, 627, 890–897. [Google Scholar] [CrossRef]
- Yang, C.C.; Kato, H.; Shindo, M.; Masai, H. Cdc7 activates replication checkpoint by phosphorylating the Chk1-binding domain of Claspin in human cells. eLife 2019, 8, e50796. [Google Scholar] [CrossRef]
- Yang, C.C.; Suzuki, M.; Yamakawa, S.; Uno, S.; Ishii, A.; Yamazaki, S.; Fukatsu, R.; Fujisawa, R.; Sakimura, K.; Tsurimoto, T.; et al. Claspin recruits Cdc7 kinase for initiation of DNA replication in human cells. Nat. Commun. 2016, 7, 12135. [Google Scholar] [CrossRef]
- Lou, H.; Komata, M.; Katou, Y.; Guan, Z.; Reis, C.C.; Budd, M.; Shirahige, K.; Campbell, J.L. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 2008, 32, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.C.; Janska, A.; Goswami, P.; Renault, L.; Abid Ali, F.; Kotecha, A.; Diffley, J.F.X.; Costa, A. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc. Natl. Acad. Sci. USA 2017, 114, 4141–4146. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, Z.; Masai, H. Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks. Biology 2024, 13, 629. https://doi.org/10.3390/biology13080629
You Z, Masai H. Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks. Biology. 2024; 13(8):629. https://doi.org/10.3390/biology13080629
Chicago/Turabian StyleYou, Zhiying, and Hisao Masai. 2024. "Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks" Biology 13, no. 8: 629. https://doi.org/10.3390/biology13080629
APA StyleYou, Z., & Masai, H. (2024). Assembly, Activation, and Helicase Actions of MCM2-7: Transition from Inactive MCM2-7 Double Hexamers to Active Replication Forks. Biology, 13(8), 629. https://doi.org/10.3390/biology13080629




