Hypoxia-Induced Adaptations of Embryonic Fibroblasts: Implications for Developmental Processes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture
2.3. ROS Detection
2.4. Cell Viability Assay
2.5. Wound Healing Assay
2.6. Western Blot Analysis
2.7. Quantitative Real-Time PCR Analysis
2.8. In Vitro Migration Assay
2.9. Procedure for Detection Apoptotic Cells Using Annexin V-FITC Kit
2.10. The RNA-Sequencing and Data Analysis
2.11. The Statistical Analysis
3. Results
3.1. Establishing Hypoxic Models in Mouse Embryo Fibroblasts (MEFs)
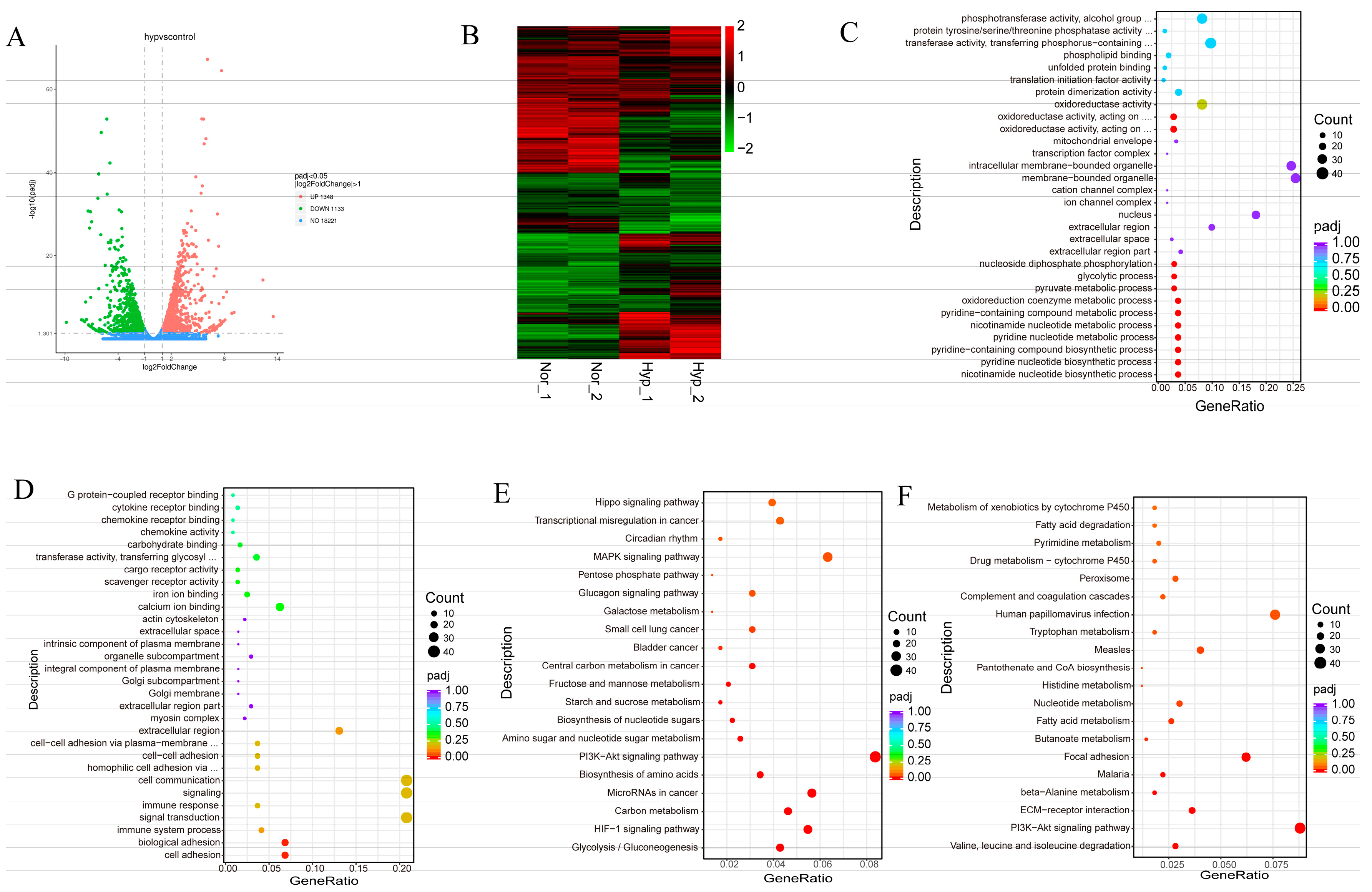
3.2. Transcriptomic Profiles of Normoxic and Hypoxic in MEFs Reveal Common and Divergent Patterns
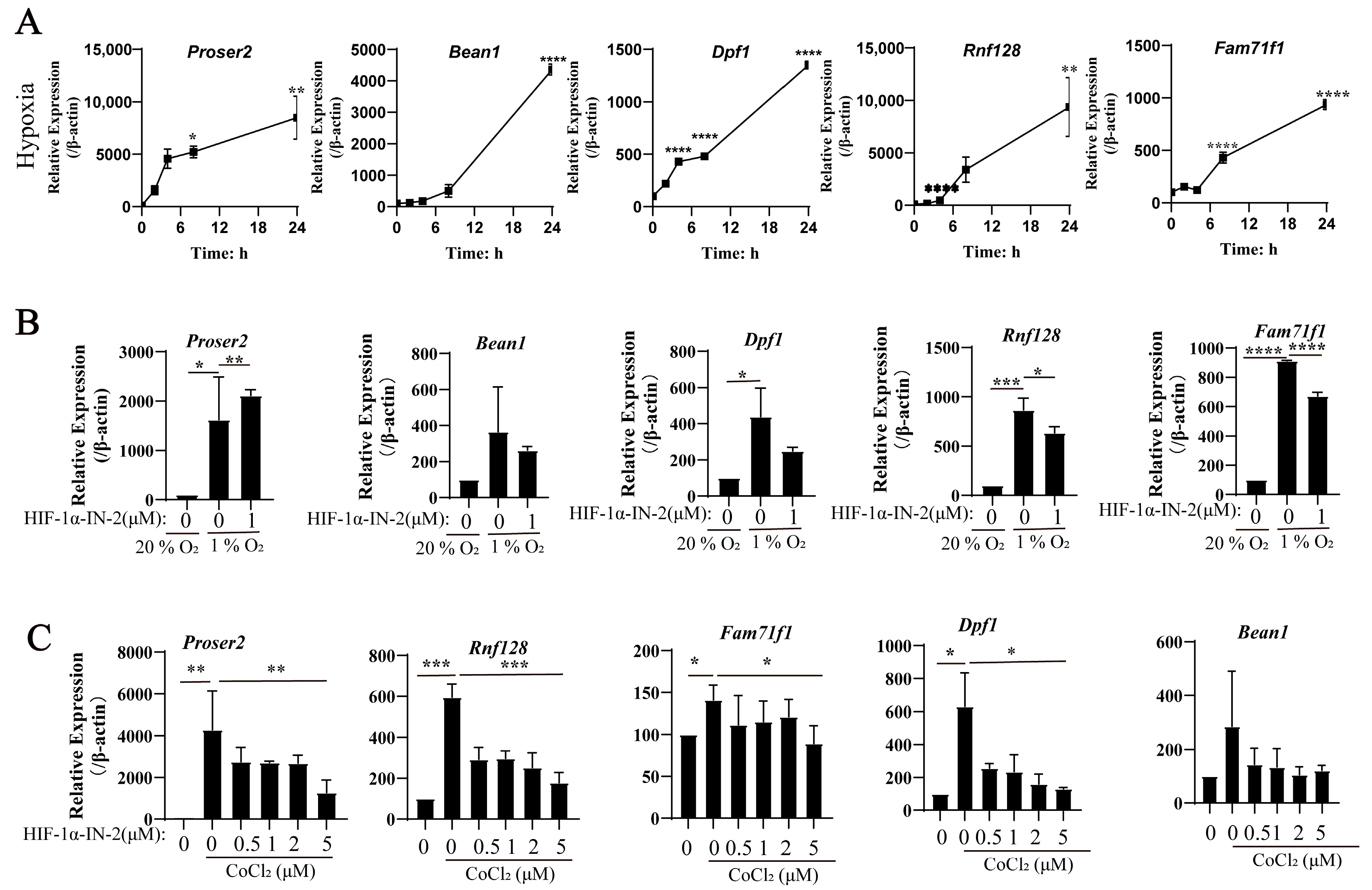
3.3. HIF-1α Is a Key Mediator of the Effects of Hypoxia and CoCl2 on MEFs
3.4. New Genes in Response to Hypoxia in MEFs
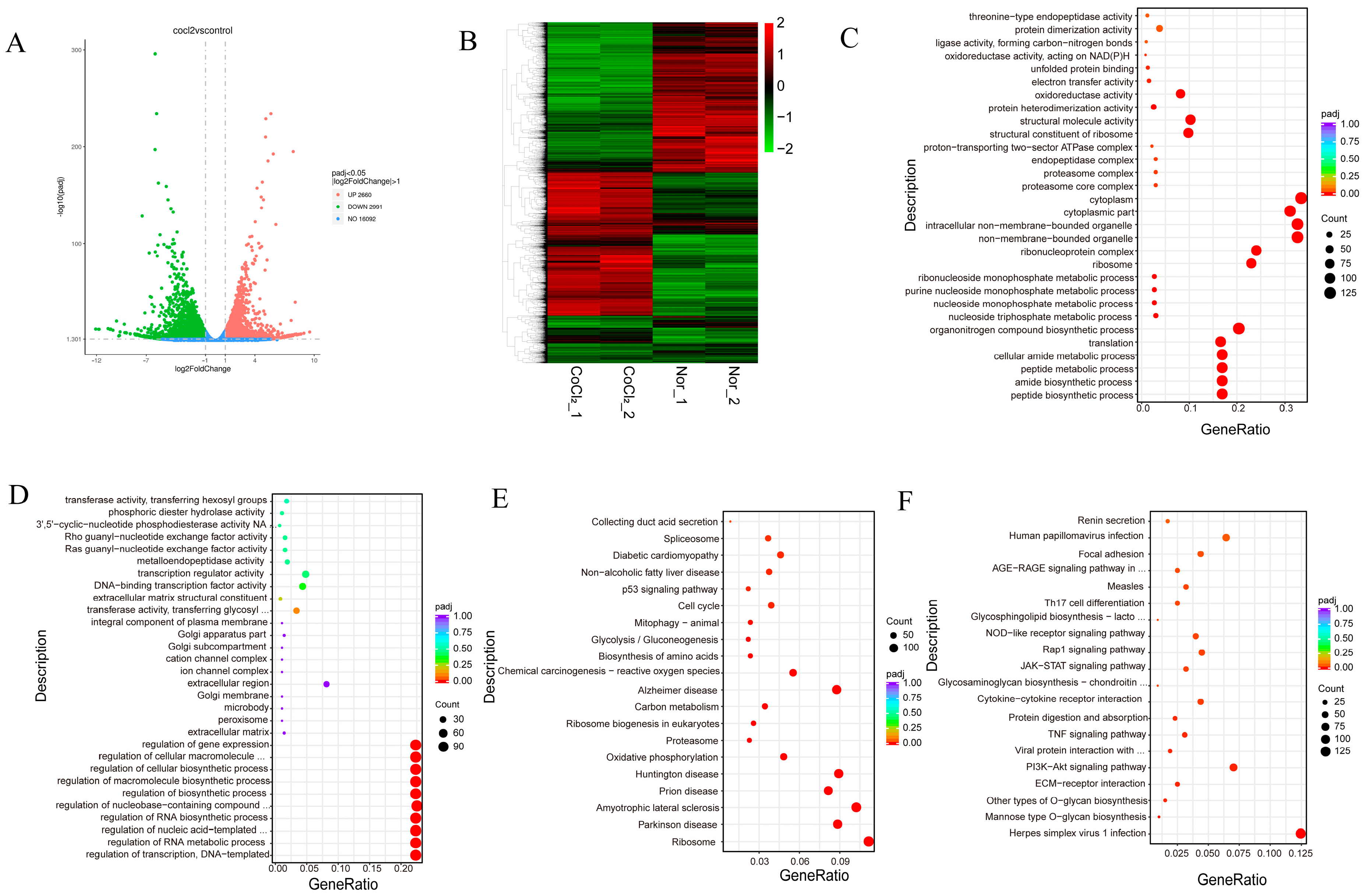
3.5. Transcriptomic Profiles of Normoxic and CoCl2 Treatment in MEFs Reveal Common and Divergent Patterns
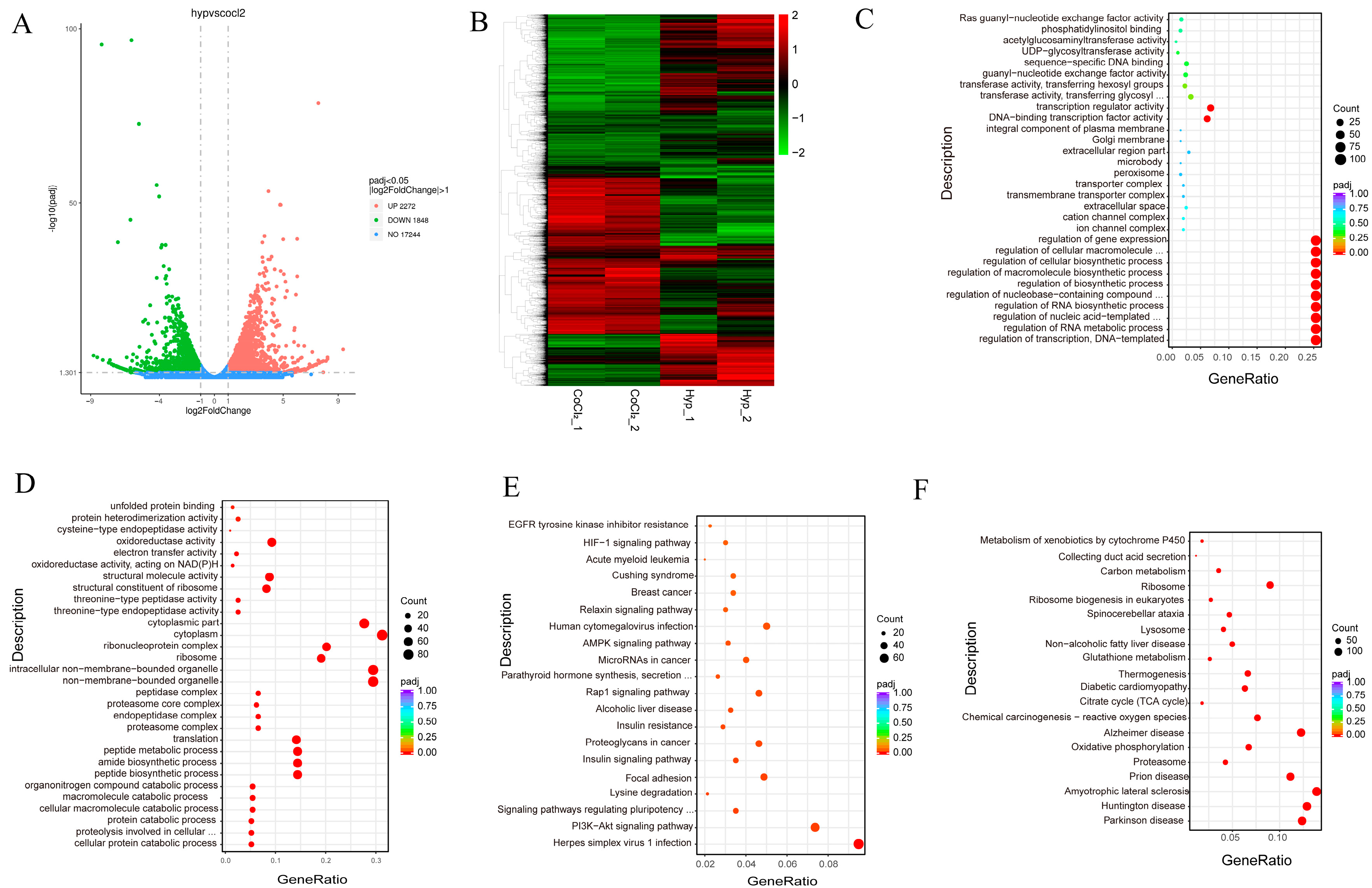
3.6. Transcriptomic Profiles of Hypoxic and CoCl2 Treatment in MEFs Reveal Common and Divergent Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wan, J.J.; Yi, J.; Wang, F.Y.; Zhang, C.; Dai, A.G. Expression and regulation of HIF-1a in hypoxic pulmonary hypertension: Focus on pathological mechanism and Pharmacological Treatment. Int. J. Med. Sci. 2024, 21, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.T.; Ji, C.L.; Yu, L.J.; Song, M.K.; Li, Y.; Liao, Q.; Wei, T.; Olatunji, O.J.; Zuo, J.; Han, J. Resveratrol-induced SIRT1 activation inhibits glycolysis-fueled angiogenesis under rheumatoid arthritis conditions independent of HIF-1alpha. Inflamm. Res. 2023, 72, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ren, Y.; Yang, K.; Wang, W.; Wang, T.; Xiao, W.; Yang, H. The role of hypoxia-inducible factor 1-alpha in inflammatory bowel disease. Cell Biol. Int. 2022, 46, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef]
- Okazaki, K.; Maltepe, E. Oxygen, epigenetics and stem cell fate. Regen. Med. 2006, 1, 71–83. [Google Scholar] [CrossRef]
- Lee, Y.M.; Jeong, C.H.; Koo, S.Y.; Son, M.J.; Song, H.S.; Bae, S.K.; Raleigh, J.A.; Chung, H.Y.; Yoo, M.A.; Kim, K.W. Determination of hypoxic region by hypoxia marker in developing mouse embryos in vivo: A possible signal for vessel development. Dev. Dyn. 2001, 220, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Nanka, O.; Valasek, P.; Dvorakova, M.; Grim, M. Experimental hypoxia and embryonic angiogenesis. Dev. Dyn. 2006, 235, 723–733. [Google Scholar] [CrossRef]
- Patterson, A.J.; Zhang, L. Hypoxia and fetal heart development. Curr. Mol. Med. 2010, 10, 653–666. [Google Scholar] [CrossRef]
- Piesova, M.; Mach, M. Impact of perinatal hypoxia on the developing brain. Physiol. Res. 2020, 69, 199–213. [Google Scholar] [CrossRef]
- Gebb, S.A.; Jones, P.L. Hypoxia and lung branching morphogenesis. Adv. Exp. Med. Biol. 2003, 543, 117–125. [Google Scholar] [CrossRef]
- Yoon, D.; Pastore, Y.D.; Divoky, V.; Liu, E.; Mlodnicka, A.E.; Rainey, K.; Ponka, P.; Semenza, G.L.; Schumacher, A.; Prchal, J.T. Hypoxia-inducible factor-1 deficiency results in dysregulated erythropoiesis signaling and iron homeostasis in mouse development. J. Biol. Chem. 2006, 281, 25703–25711. [Google Scholar] [CrossRef] [PubMed]
- Junker, J.P.; Sommar, P.; Skog, M.; Johnson, H.; Kratz, G. Adipogenic, chondrogenic and osteogenic differentiation of clonally derived human dermal fibroblasts. Cells Tissues Organs 2010, 191, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, J.; Suh, Y.; Cressman, M.; Lee, K. Research Note: All-trans retinoic acids induce adipogenic differentiation of chicken embryonic fibroblasts and preadipocytes. Poult. Sci. 2020, 99, 7142–7146. [Google Scholar] [CrossRef] [PubMed]
- Singhal, P.K.; Sassi, S.; Lan, L.; Au, P.; Halvorsen, S.C.; Fukumura, D.; Jain, R.K.; Seed, B. Mouse embryonic fibroblasts exhibit extensive developmental and phenotypic diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Yun, Z.; Maecker, H.L.; Johnson, R.S.; Giaccia, A.J. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: A mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2002, 2, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Watts, E.R.; Walmsley, S.R. Inflammation and Hypoxia: HIF and PHD Isoform Selectivity. Trends Mol. Med. 2019, 25, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Piret, J.P.; Mottet, D.; Raes, M.; Michiels, C. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y. Acad. Sci. 2002, 973, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ma, R.; Zhang, M. CoCl2 increases the expression of hypoxic markers HIF-1alpha, VEGF and CXCR4 in breast cancer MCF-7 cells. Oncol. Lett. 2018, 15, 1119–1124. [Google Scholar] [CrossRef]
- Kawczyk-Krupka, A.; Czuba, Z.P.; Kwiatek, B.; Kwiatek, S.; Krupka, M.; Sieron, K. The effect of ALA-PDT under normoxia and cobalt chloride (CoCl(2))-induced hypoxia on adhesion molecules (ICAM-1, VCAM-1) secretion by colorectal cancer cells. Photodiagn. Photodyn. Ther. 2017, 19, 103–115. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef] [PubMed]
- Essemine, J.; Li, J.; Chen, G.; Qu, M. Analytical dataset of short-term heat stress induced reshuffling of metabolism and transcriptomes in maize grown under elevated CO(2). Data Brief 2020, 28, 105004. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, D.C.; Gorham, J.M.; Herman, D.S.; Seidman, J.G. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr. Protoc. Mol. Biol. 2011, 94, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, M.C.; McLellan, A.S. Whole-Exome Sequencing (WES) for Illumina Short Read Sequencers Using Solution-Based Capture. Methods Mol. Biol. 2020, 2076, 85–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xue, R.; Qin, W.; Yang, X.; Ye, Q.; Wu, Q. Performance and transcriptome analysis of Salmonella enterica serovar Enteritidis PT 30 under persistent desiccation stress: Cultured by lawn and broth methods. Food Microbiol. 2023, 115, 104323. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Yuan, X.; Song, Y.; Liu, Y.; Wang, X.F. First report of maize yellow mosaic virus (MaYMV) naturally infecting wheat in China. Plant Dis. 2022, 106, 2763. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Im, J.; Yang, Y.; Lee, H.J.; Lee, M.R.; Woo, S.M.; Park, S.J.; Kong, S.Y.; Kim, J.Y.; Hwang, H.; et al. New Function Annotation of PROSER2 in Pancreatic Ductal Adenocarcinoma. J. Proteome Res. 2024, 23, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.R.; Lill, N.L.; Boase, N.; Shi, P.P.; Croucher, D.R.; Shan, H.; Qu, J.; Sweezer, E.M.; Place, T.; Kirby, P.A.; et al. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. 2008, 1, ra5. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Yoshihara, H.; Furuta, H.; Kamei, H.; Hakuno, F.; Luan, J.; Duan, C.; Saeki, Y.; Tanaka, K.; Iemura, S.; et al. Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat. Commun. 2015, 6, 6780. [Google Scholar] [CrossRef] [PubMed]
- Norioka, R.; Sugaya, K.; Murayama, A.; Kawazoe, T.; Tobisawa, S.; Kawata, A.; Takahashi, K. Midbrain atrophy related to parkinsonism in a non-coding repeat expansion disorder: Five cases of spinocerebellar ataxia type 31 with nigrostriatal dopaminergic dysfunction. Cerebellum Ataxias 2021, 8, 11. [Google Scholar] [CrossRef]
- Liu, P.Y.; Chen, C.Y.; Lin, Y.L.; Lin, C.M.; Tsai, W.C.; Tsai, Y.L.; Lin, G.J.; Chen, Y.G.; Wang, S.Y.; Sun, R.N.; et al. RNF128 regulates neutrophil infiltration and myeloperoxidase functions to prevent acute lung injury. Cell Death Dis. 2023, 14, 369. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.S.; Zhang, C.; Peng, R.; Jiang, G.Q.; Jin, S.J.; Wang, Q.; Ke, A.W.; Bai, D.S. RNF128 Promotes Malignant Behaviors via EGFR/MEK/ERK Pathway in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 10129–10141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Gan, Y.; Zou, R.; Sha, H.; Lu, Y.; Zhang, Y.; Feng, J. RNF128 suppresses the malignancy of colorectal cancer cells via inhibition of Wnt/beta-catenin signaling. Am. J. Transl. Res. 2021, 13, 13567–13578. [Google Scholar] [PubMed]
- Gupta, A.; Lutolf, M.P.; Hughes, A.J.; Sonnen, K.F. Bioengineering in vitro models of embryonic development. Stem Cell Rep. 2021, 16, 1104–1116. [Google Scholar] [CrossRef]
- Copp, A.J.; Greene, N.D. Genetics and development of neural tube defects. J. Pathol. 2010, 220, 217–230. [Google Scholar] [CrossRef]
- Houyel, L.; Meilhac, S.M. Heart Development and Congenital Structural Heart Defects. Annu. Rev. Genom. Hum. Genet. 2021, 22, 257–284. [Google Scholar] [CrossRef] [PubMed]
- Nimonkar, S.V.; Belkhode, V.M. Bilateral cleft lip, alveolus, and palate deformity in an infant. Pan. Afr. Med. J. 2022, 42, 316. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A.; Zeller, R.; Probst, S. The molecular basis of human congenital limb malformations. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 803–822. [Google Scholar] [CrossRef] [PubMed]
- Hartman, R.J.; Rasmussen, S.A.; Botto, L.D.; Riehle-Colarusso, T.; Martin, C.L.; Cragan, J.D.; Shin, M.; Correa, A. The contribution of chromosomal abnormalities to congenital heart defects: A population-based study. Pediatr. Cardiol. 2011, 32, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Slavotinek, A.M. Single gene disorders associated with congenital diaphragmatic hernia. Am. J. Med. Genet. C Semin. Med. Genet. 2007, 145C, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Shai, S.Y.; Harpf, A.E.; Babbitt, C.J.; Jordan, M.C.; Fishbein, M.C.; Chen, J.; Omura, M.; Leil, T.A.; Becker, K.D.; Jiang, M.; et al. Cardiac myocyte-specific excision of the beta1 integrin gene results in myocardial fibrosis and cardiac failure. Circ. Res. 2002, 90, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Marcelain, K.; Armisen, R.; Aguirre, A.; Ueki, N.; Toro, J.; Colmenares, C.; Hayman, M.J. Chromosomal instability in mouse embryonic fibroblasts null for the transcriptional co-repressor Ski. J. Cell. Physiol. 2012, 227, 278–287. [Google Scholar] [CrossRef]
- Dunwoodie, S.L. The role of hypoxia in development of the Mammalian embryo. Dev. Cell 2009, 17, 755–773. [Google Scholar] [CrossRef]
- Fajersztajn, L.; Veras, M.M. Hypoxia: From Placental Development to Fetal Programming. Birth Defects Res. 2017, 109, 1377–1385. [Google Scholar] [CrossRef]
- Cosin-Roger, J.; Simmen, S.; Melhem, H.; Atrott, K.; Frey-Wagner, I.; Hausmann, M.; de Valliere, C.; Spalinger, M.R.; Spielmann, P.; Wenger, R.H.; et al. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat. Commun. 2017, 8, 98. [Google Scholar] [CrossRef]
- Perkins, N.D. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 2007, 8, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Bowen, M.E.; Attardi, L.D. The role of p53 in developmental syndromes. J. Mol. Cell Biol. 2019, 11, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.; Karis, A.; Visser, P.; Grosveld, F.; Philipsen, S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 1997, 89, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Raz, R.; Lee, C.K.; Cannizzaro, L.A.; d’Eustachio, P.; Levy, D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 1999, 96, 2846–2851. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef] [PubMed]
- Ruas, J.L.; Poellinger, L. Hypoxia-dependent activation of HIF into a transcriptional regulator. Semin. Cell Dev. Biol. 2005, 16, 514–522. [Google Scholar] [CrossRef]
- Park, S.K.; Dadak, A.M.; Haase, V.H.; Fontana, L.; Giaccia, A.J.; Johnson, R.S. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1alpha (HIF-1alpha): Role of cytoplasmic trapping of HIF-2alpha. Mol. Cell Biol. 2003, 23, 4959–4971. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Sanchez, J.; Chanez-Cardenas, M.E. The use of cobalt chloride as a chemical hypoxia model. J. Appl. Toxicol. 2019, 39, 556–570. [Google Scholar] [CrossRef]
- Goda, N.; Ryan, H.E.; Khadivi, B.; McNulty, W.; Rickert, R.C.; Johnson, R.S. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol. Cell Biol. 2003, 23, 359–369. [Google Scholar] [CrossRef]


| Gene | Sequence (5′-3′) |
|---|---|
| Bean1-F | GCACGATACAACCGTACCAG |
| Bean2-R | ACCACACCTATGACAGCGTTC |
| Proser2-F | GAGGTGGCAGTCTGGAGAG |
| Proser3-R | AGAACAGGATAACGTCCTTTTCC |
| Pdk1-F | GGACTTCGGGTCAGTGAATGC |
| Pdk1-R | TCCTGAGAAGATTGTCGGGGA |
| Ldha-F | CAAAGACTACTGTGTAACTGCGA |
| Ldha-R | TGGACTGTACTTGACAATGTTGG |
| Dpf1-F | TTGCTGGAGTTTCCGCATGAT |
| Dpf2-R | TACGGCTTGTCTCGGTCCT |
| Rnf128-F | ATTCAAAGAGGCATCCAAGTCAC |
| Rnf129-R | TGCATTTCGTAATCTTCGAGCAG |
| Fam71f1-F | ACCTTCCCTTCCTTGAATGCC |
| Fam71f2-R | GCAGAGTAGTCCTTCCTCCAC |
| Glut1-F | GCAGTTCGGCTATAACACTGG |
| Glut1-R | GCGGTGGTTCCATGTTTGATTG |
| Vegf-F | CTGCCGTCCGATTGAGACC |
| Vegf-R | CCCCTCCTTGTACCACTGTC |
| Actin-F | GGCTGTATTCCCCTCCATCG |
| Actin-R | CCAGTTGGTAACAATGCCATGT |
| Key TF | # of Overlapped Genes | p Value | Q Value | List of Overlapped Genes |
|---|---|---|---|---|
| p53 | 44 | 1.97 × 10−48 | 3.11 × 10−46 | Polk, E2f1, Gdf15, Ptgs2, Nupr1, Klf4, Fos, Mdm2, Rps6ka1, Lif, Elf4, Rrm2b, Pcna, Btg2, Foxo3, Slc19a2, Mcm7, Cdkn1a, Rela, Atf3, Hmox1, Dusp4, Pmaip1, Krt19, Klf2, Mt1, Ankrd1, Myc, Gabre, Srebf1, E4f1, Dusp1, Casp6, Plk3, Bbc3, Adgrb1, Cdkn1b, Ddit4, Serpine1, Egr1, Igf1r, Ndrg1, Mxi1, Id2 |
| Sp1 | 38 | 8.07 × 10−31 | 6.38 × 10−29 | Smad7, Myc, Dck, Pld2, Fos, Mgarp, Bhlhe40, Serpine1, Acsbg1, Srebf1, Slc7a5, Plaur, Egr1, Ccnd3, Timp3, Slc2a1, Rnf141, Tec, Klf16, Epor, Tnfaip3, Cdkn2d, Ndrg1, Slc2a3, Hinfp, Ptgs2, Ets1, Rest, Ccnd2, Cdkn1a, Jun, Ecm1, Jund, Cdh5, Igfbp3, Mrc1, Bsg, Itga6 |
| Nfkb1 | 34 | 1.81 × 10−30 | 9.53 × 10−29 | Ntn1, Nfkbia, Pmaip1, Ptgs2, Nos2, Tgfb1, Myoz2, Ppargc1a, Cdkn1a, Adora2b, Ccnd3, Stk39, Slc2a1, Gadd45b, Lif, Myc, Klf5, Fos, Per1, Tnfaip3, Msx1, Dusp1, Wt1, Timp3, Sox9, Banp, Plin2, Kdm2a, Yy1, Plaur, Junb, Hmox1, Ppp1r13l, Tec |
| Stat3 | 16 | 3.85 × 10−16 | 1.52 × 10−14 | Klf4, Epor, Tgfb1, Cdkn2d, Nos2, Ccnd3, Mt1, Ccnd2, Cdkn1b, Eed, Srebf1, Egr1, Rela, Fos, Myc, Lif |
| Hif1a | 10 | 7.71 × 10−15 | 2.44 × 10−13 | Fam162a, Mt1, Hey1, Hey2, Hmox1, Serpine1, Ptgs2, Bsg, Pfkfb3, Ctgf |
| Rela | 17 | 2.73 × 10−14 | 7.19 × 10−13 | Tnfaip3, Myc, Klf5, Stk39, Fos, Tec, Pecam1, Ptgs2, Wt1, Cxcr4, Gabre, Cdkn1b, Pgf, Yy1, Nos2, Sox9, Kdm2a |
| Egr1 | 14 | 4.19 × 10−14 | 9.46 × 10−13 | Cacna1h, Cdkn2d, Smad7, Itga7, Rcan1, Serpine1, Jun, Bnip3, Ndrg1, Ccnd2, Ltb, Jund, Igf1r, Gadd45b |
| Myc | 12 | 1.77 × 10−13 | 3.49 × 10−12 | Myc, Rhoa, Ndrg1, Hnrnpa1, Ccnd2, Suz12, Mxi1, Zfp36, Timp3, Cdkn1a, Bcat1, Eed |
| Cebpb | 11 | 8.2 × 10−13 | 1.44 × 10−11 | Cdkn1a, Ptgs2, Myc, Atf3, Btg2, Cxcr4, Cyr61, Id2, Serpine1, Ppargc1a, Fos |
| Jun | 16 | 1.48 × 10−12 | 2.34 × 10−11 | Plin2, Plaur, Ccnd2, Jun, Ccnd3, Ptgs2, Fos, Eno2, Slc2a1, Pdk1, Nos2, Serpine1, Tgfb1, Fosl1, Hmox1, Ppp1r3b |
| Trp73 | 8 | 1.44 × 10−11 | 2.07 × 10−10 | Hey2, Pmaip1, Gls2, Mdm2, Cdkn1a, Bbc3, E2f1, Foxo3 |
| Ep300 | 11 | 5.5 × 10−11 | 7.25 × 10−10 | Klf5, Ldha, Txnip, Fos, Ptgs2, Cryab, Mt1, Hmox1, Nos2, Cdkn1a, Hinfp |
| Clock | 7 | 1.17 × 10−10 | 1.42 × 10−9 | Cry1, Serpine1, Nampt, Bhlhe41, Bhlhe40, Per1, Cry2 |
| Crebbp | 9 | 5.84 × 10−10 | 6.6 × 10−9 | Fos, Nfyb, Cryab, Fosb, Hmox1, Hinfp, Mt1, Per1, Ldha |
| Creb1 | 8 | 1.34 × 10−9 | 1.41 × 10−8 | Fos, Noct, Ppargc1a, Ptgs2, Slc2a3, Ldha, Hspa5, Nmnat2 |
| Foxo1 | 9 | 3.17 × 10−9 | 3.13 × 10−8 | Ccng2, Cdkn1a, Cdkn1b, Ppargc1a, Trib3, Egr1, Srebf1, Ccnd2, Klf2 |
| Nfe2l2 | 9 | 6.58 × 10−9 | 6.12 × 10−8 | Mcm7, Txnip, Areg, Fos, Sqstm1, Mdm2, Hmox1, Mt1, Tgfb1 |
| Dmtf1 | 5 | 9.01 × 10−9 | 7.91 × 10−8 | Junb, Ccnd2, Ets1, Egr1, Areg |
| Fos | 9 | 1.09 × 10−8 | 9.09 × 10−8 | Jun, Tgfb1, Bdnf, Ppp1r3b, Tinagl1, Fos, Mt1, Egr1, Nos2 |
| Rb1 | 6 | 6.13 × 10−8 | 0.000000484 | Fosl1, Cdkn1a, Mybl2, Fos, Jun, E2f1 |
| Myod1 | 8 | 7.59 × 10−8 | 0.000000571 | Smad7, Fosl1, Cdkn1a, Itga7, Ppargc1a, Rb1, Ccnd3, Myh7b |
| Key TF | # of Overlapped Genes | p Value | Q Value | List of Overlapped Genes |
|---|---|---|---|---|
| Trp53 | 18 | 0.000000132 | 0.00000818 | E2f1, Gdf15, Nupr1, Chek1, Klf4, Mdm2, Gtse1, Pcna, Bax, Slc19a2, Mcm7, Hspa1b, Afp, Mt1, Serpine1, Ndrg1, Id2, Birc5 |
| Sp1 | 21 | 0.00000288 | 0.0000893 | Serpine1, Sirt1, Slc7a5, Slc2a1, Slc3a2, Tec, Prdx6, Klf16, Apoe, Mertk, Cebpa, Cdk5r1, Ndrg1, Hspa1b, Nrgn, Jun, Tspo, Tgm1, Pemt, Bsg, Itga6 |
| Egr1 | 9 | 0.0000523 | 0.00108 | Wnt4, Serpine1, Cdk5r1, Bax, Jun, Birc5, Nr4a1, Hsd11b2, Ndrg1 |
| Nfe2l2 | 7 | 0.000276 | 0.00427 | Mcm7, Gclc, Slc7a11, Abcc4, Srxn1, Mdm2, Mt1 |
| Nfkb1 | 14 | 0.000385 | 0.00478 | Ntn1, Mmp3, Birc5, Phex, Cebpa, Apoe, Gclm, Slc2a1, Klf5, Per1, Amh, Plin2, Hsd11b2, Tec |
| Trp73 | 4 | 0.000988 | 0.01 | Gls2, Mdm2, E2f1, Bax |
| Myc | 6 | 0.00113 | 0.01 | Bax, Wnt4, Ndrg1, Nop56, Zfp36, Nrgn |
| Jun | 10 | 0.00134 | 0.0103 | Plin2, Gclc, Jun, Tgm1, Mmp3, Slc2a1, Serpine1, Phex, Sirt1, Srxn1 |
| Rbl2 | 3 | 0.00149 | 0.0103 | Birc5, Stmn1, Pcna |
| Nr1i2 | 3 | 0.00264 | 0.0164 | Insig1, Cyp24a1, E2f1 |
| Sp3 | 7 | 0.00399 | 0.0216 | Nrgn, Bsg, Itga6, Tspo, Mertk, Cdk5r1, Cebpa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Han, D.; Li, Z.; Luo, L. Hypoxia-Induced Adaptations of Embryonic Fibroblasts: Implications for Developmental Processes. Biology 2024, 13, 598. https://doi.org/10.3390/biology13080598
Li Z, Han D, Li Z, Luo L. Hypoxia-Induced Adaptations of Embryonic Fibroblasts: Implications for Developmental Processes. Biology. 2024; 13(8):598. https://doi.org/10.3390/biology13080598
Chicago/Turabian StyleLi, Zeyu, Delong Han, Zhenchi Li, and Lingjie Luo. 2024. "Hypoxia-Induced Adaptations of Embryonic Fibroblasts: Implications for Developmental Processes" Biology 13, no. 8: 598. https://doi.org/10.3390/biology13080598
APA StyleLi, Z., Han, D., Li, Z., & Luo, L. (2024). Hypoxia-Induced Adaptations of Embryonic Fibroblasts: Implications for Developmental Processes. Biology, 13(8), 598. https://doi.org/10.3390/biology13080598







