D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
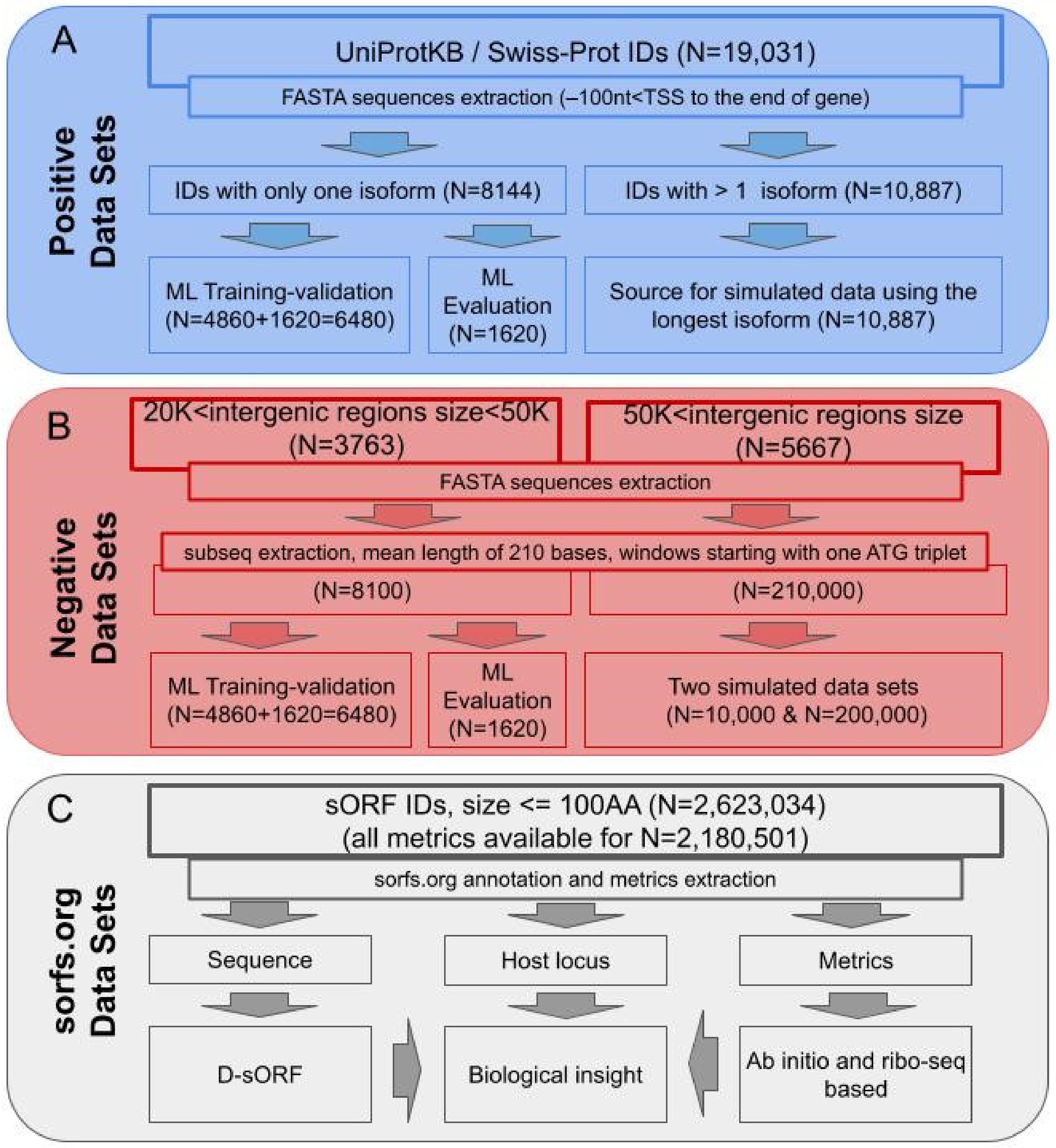
2.1. Positive Datasets
2.2. Negative Datasets
2.3. Small ORF Datasets
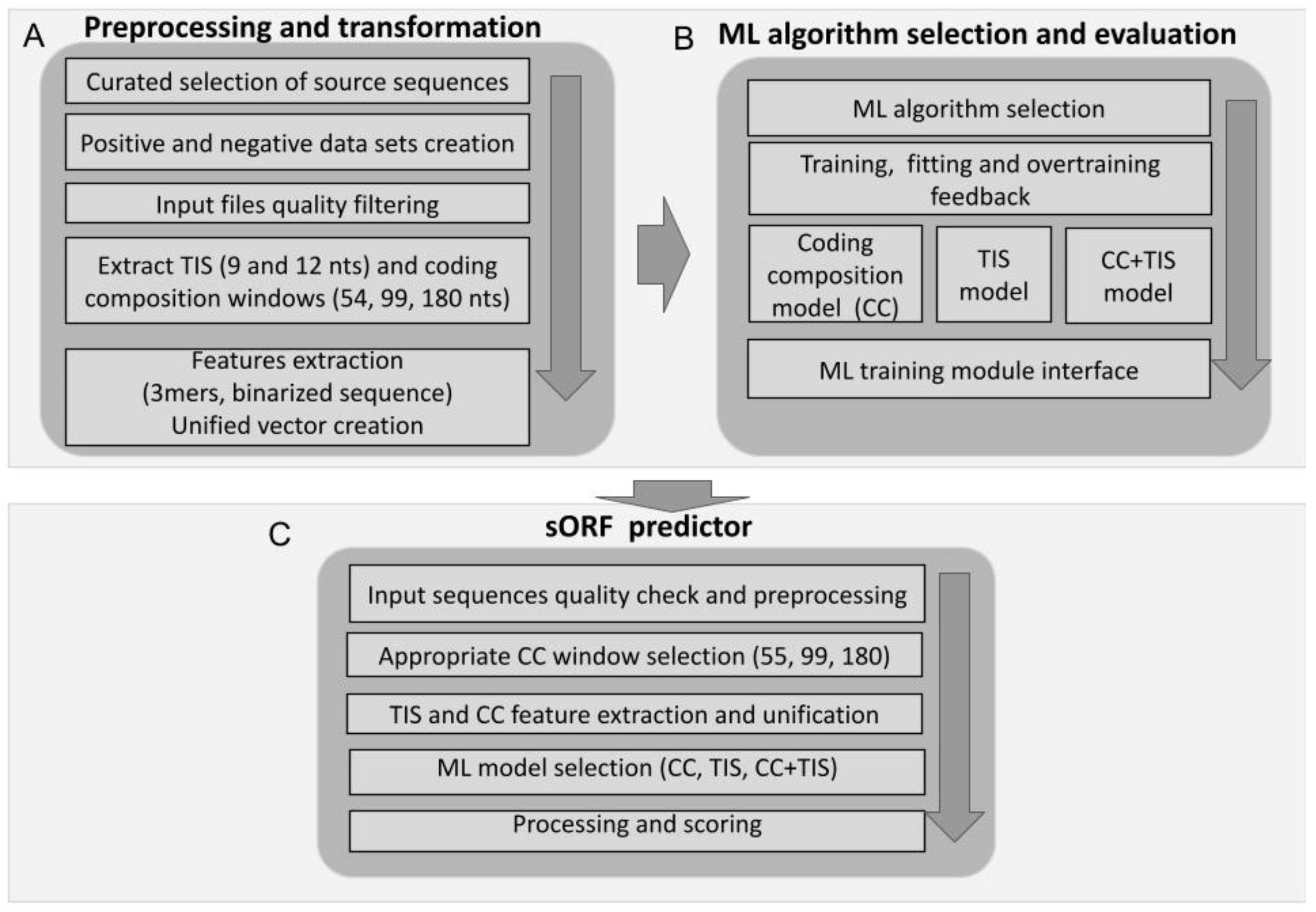
2.4. D-sORF Machine-Learning Framework
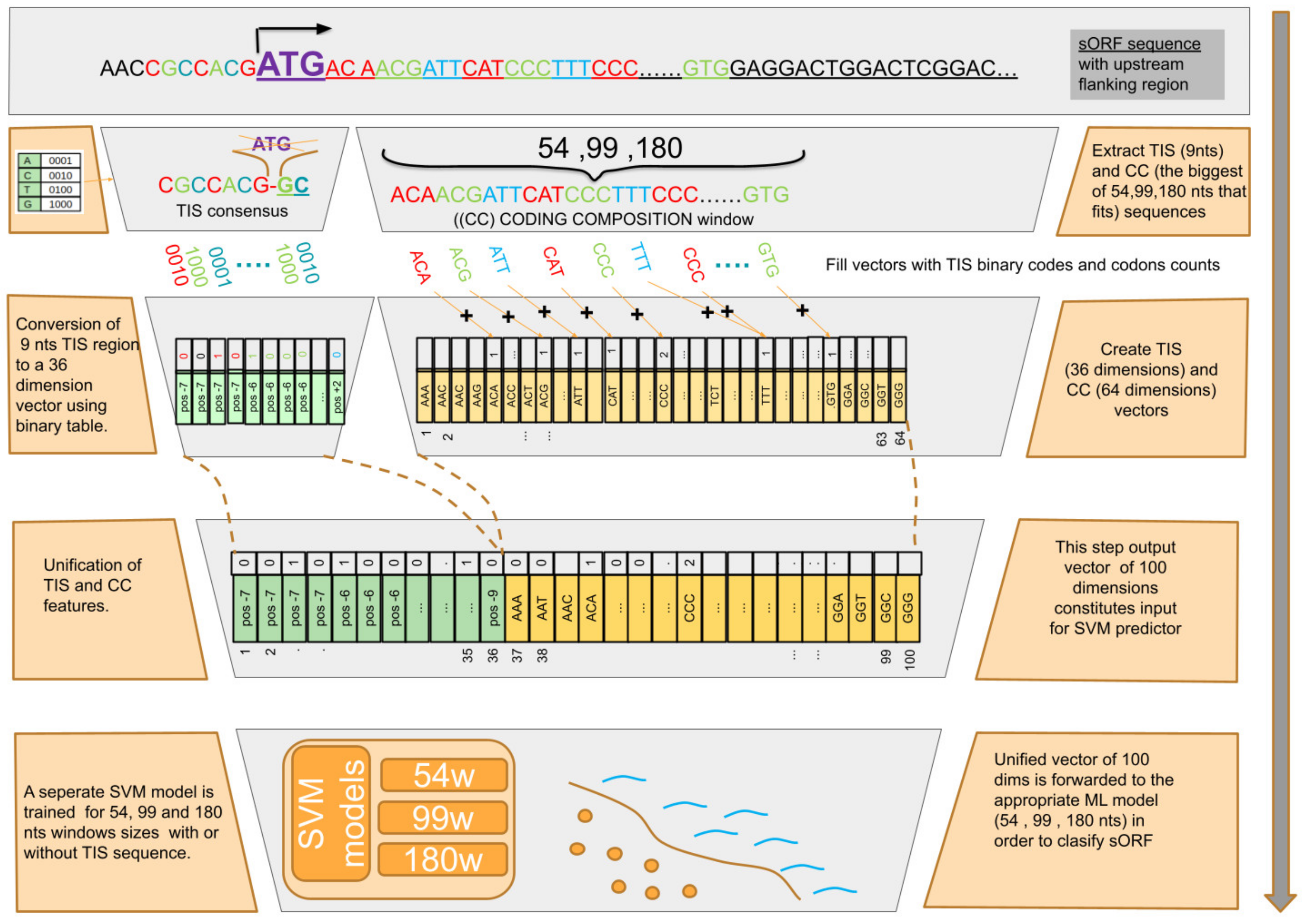
2.5. Data Preprocessing and Transformation
2.6. ML Algorithm Selection and Evaluation
2.7. sORF Predictor
3. Results
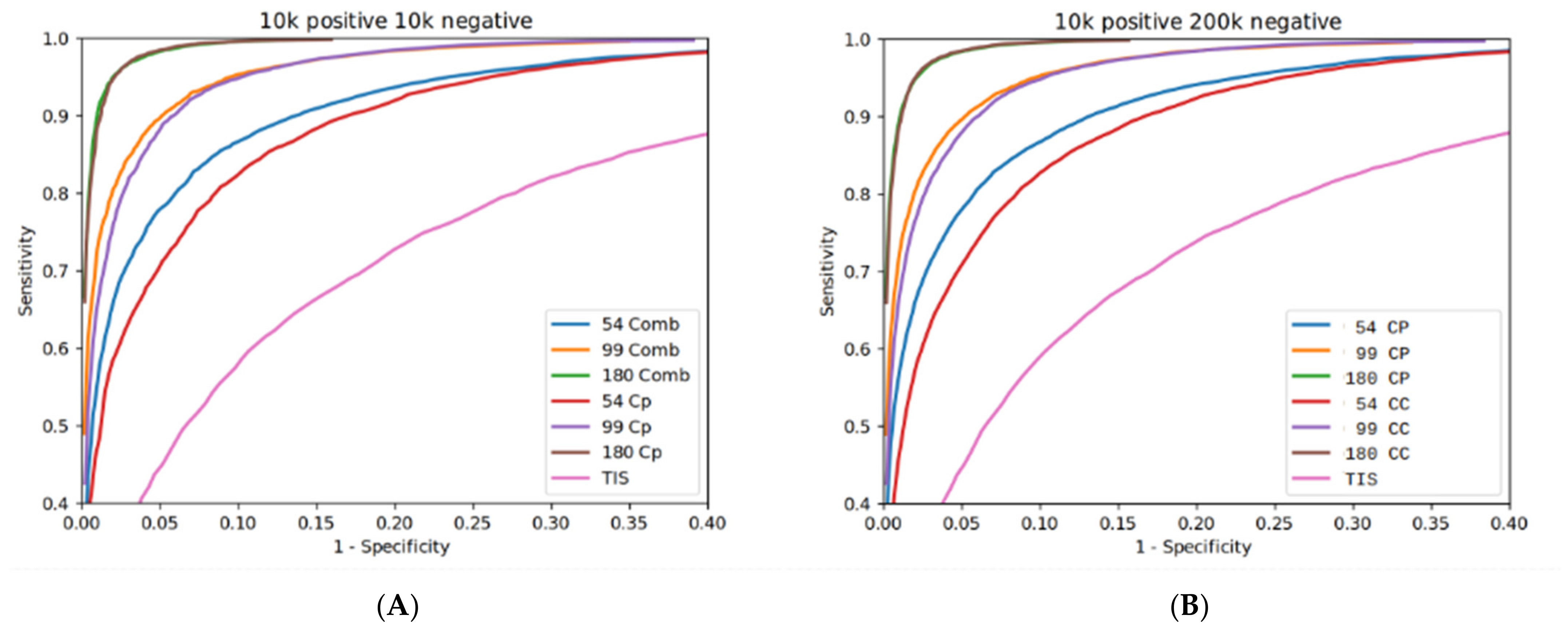
3.1. Algorithm Evaluation
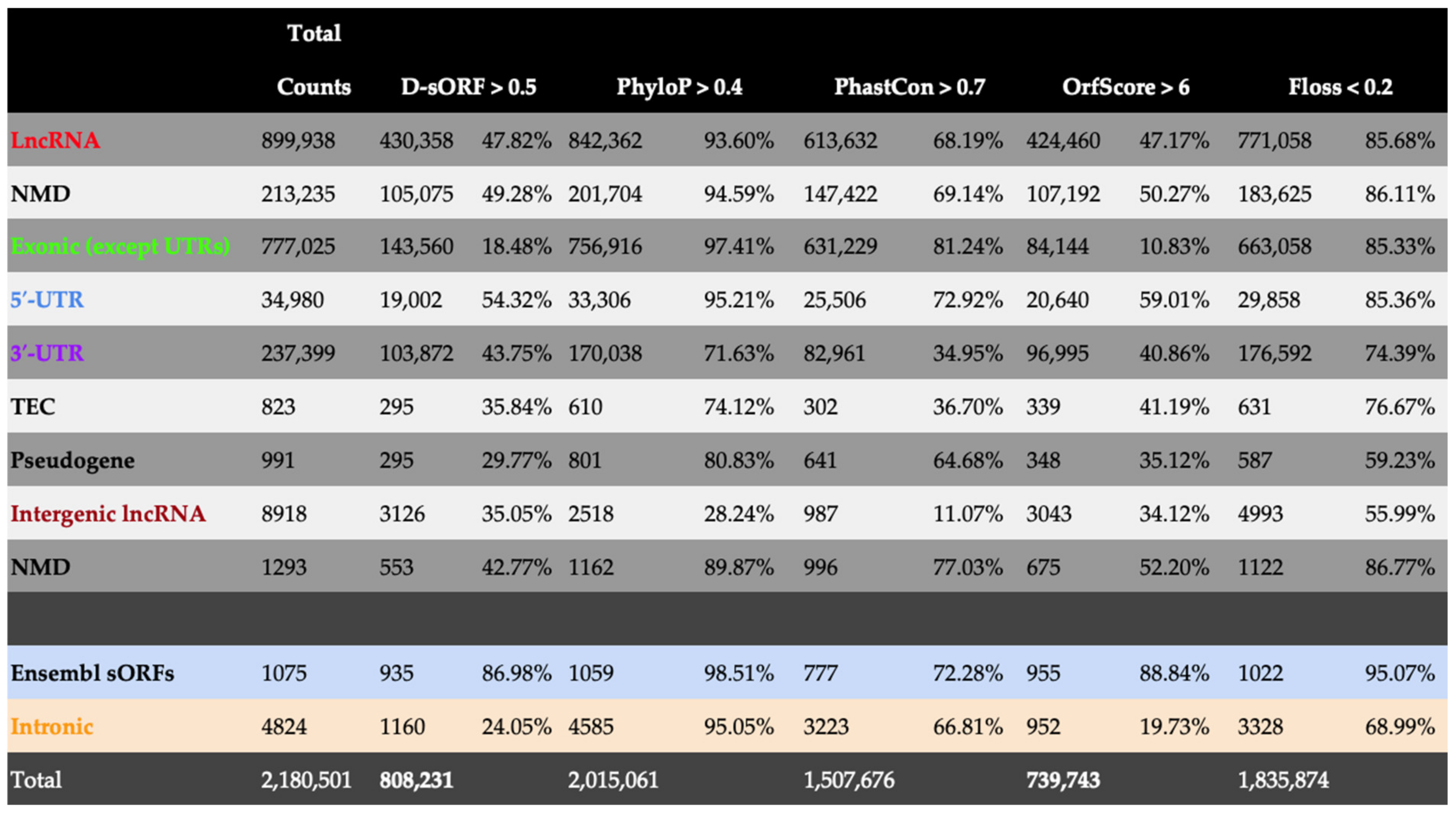
3.2. Application of D-sORF in Annotating Experimentally Detected sORFs in Genomic Regions Enriched with Coding and Non-Coding Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| sORF ID | Length | Sequence |
| SorfId=andreev_2015:538832 | 300 | MVLRRLLAALLHSPQLVERLSESRPIRRAAQLTAFALLQAQLRGQDAARRLQDLAAGPVGSLCRRAERFRDAFTQELRRGLRGRSGPPPGSQRGPGANI |
| SorfId=elkon_2015:50819 | 210 | MLRWLRDFVLPTAACQDAEQPTRYETLFQALDRNGDGVVDIGELQEGLRNLGIPLGQDAEEVGRRRGAA |
| SorfId=fritsch_2012:225652 | 180 | MRQLKGKPKKETSKDKKERKQAMQEARQQITTVVLPTLAVVVLLIVVFVYVATRPTITE |
| SorfId=andreev_2015:496184 | 213 | MAPWSREAVLSLYRALLRQGRQLRYTDRDFYFASIRREFRKNQKLEDAEARERQLEKGLVFLNGKLGRII |
| SorfId=rubio_2014:98637 | 141 | MKESSIDDLMKSLDKNSDQEIDFKEYSVFLTMLCMAYNDFFLEDNK |
| SorfId=andreev_2015:629427 | 261 | MTDRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAIAENESKARVRFNLMEKMLQPCGPPADKPEEN |
| SorfId=crappe_2014:52683 | 249 | MSATWTLSPEPLPPSTGPPVGAGLDAEQRTVFAFVLCLLVVLVLLMVRCVRILLDPYSRMPASSWTDHKEALERGQFDYALV |
| SorfId=calviello_2016:870882 | 252 | MEALGSGHYVGGSIRSMAAAALSGLAVRLSRPQGTRGSYGAFCKTLTRTLLTFFDLAWRLRKNFFYFYILASVILNVHLQVYI |
| SorfId=andreev_2015:491102 | 189 | MAAATLTSKLYSLLFRRTSTFALTIIVGVMFFERAFDQGADAIYDHINEGVRACAIPDLGPA |
| SorfId=calviello_2016:375783 | 189 | MTKAGSKGGNLRDKLDGNELDLSLSDLNEVPVKELVSIVPRGPGAHLRRALAPTWATYFPLL |
| SorfId=liu_HEK_2013:259226 | 204 | MLSRLQELRKEEETLLRLKAALHDQLNRLKVTPRPSPAPPPFLPSDHFPPTWASVVTTCCGQFSATH |
| SorfId=gonzalez_2014:380995 | 267 | MSIMDHSPTTGVVTVIVILIAIAALGALILGCWCYLRLQRISQSEDEESIVGDGETKEPFLLVQYSAKGPCVERKAKLMTPNGPEVHG |
| SorfId=calviello_2016:540138 | 264 | MYCLQWLLPVLLIPKPLNPALWFSHSMFMGFYLLSFLLERKPCTICALVFLAALFLICYSCWGNCFLYHCSDSPLPESAHDPGVVGT |
| SorfId=calviello_2016:185382 | 207 | MAPAAAPSSLAVRASSPAATPTSYGVFCKGLSRTLLAFFELAWQLRMNFPYFYVAGSVILNIRLQVHI |
| SorfId=andreev_2015:177588 | 294 | MEEISLANLDTNKLEAIAQEIYVDLIEDSCLGFCFEVHRAVKCGYFYLEFAETGSVKDFGIQPVEDKGACRLPLCSLPGEPGNGPDQQLQRSPPEFQ |
| SorfId=andreev_2015:409703 | 114 | MITDVQLAIFANMLGVSLFLLVVLYHYVAVNNPKKQE |
| SorfId=andreev_2015:37830 | 279 | MTKLAQWLWGLAILGSTWVALTTGALGLELPLSCQEVLWPLPAYLLVSAGCYALGTVGYRVATFHDCEDAARELQSQIQEARADLARRGLRF |
| SorfId=andreev_2015:13871 | 279 | MWPVFWTVVRTYAPYVTFPVAFVVGAVGYHLEWFIRGKDPQPVEEEKSISERREDRKLDELLGKDHTQVVSLKDKLEFAPKAVLNRNRPEKN |
| SorfId=park_2016:1581934 | 237 | MREENKGMPSGGGSDEGLASAAARGLVEKVRQLLEAGADPNGVNRFGRRAIQVAGAPGPRRQGARERGARPRRIGAGT |
| SorfId=gonzalez_2014:153430 | 150 | VRDKKLLNDLNGAVEDAKTARLFNITSSALAASCIILVFIFLRYPLTDY |
| SorfId=fritsch_2012:974998 | 183 | MDAVSQVPMEVVLPKHILDIWVIVLIILATIVIMTSLLLCPATAVIIYRMRTHPILSGAV |
| SorfId=andreev_2015:137699 | 264 | MSTSVPQGHTWTQRVKKDDEEEDPLDQLISRSGCAASHFAVQECMAQHQDWRQCQPQVQAFKDCMSEQQARRQEELQRRQEQAGAHH |
| SorfId=andreev_2015:321927 | 207 | VGVGKNKCLYALEEGIVRYTKEVYVPHPRNTEAVDLITRLPKGAVLYKTFVHVVPAKPEGTFKLVAML |
| SorfId=calviello_2016:206180 | 141 | MPAGVPMSTYLKMFAASLLAMCAGAEVVHRYYRPDLVSAGGLLFYF |
| SorfId=gonzalez_2014:52803 | 162 | VNIDHFTKDITMKNLVEPSLSSFDMAQKRIHALMEKDSLPRFVRSEFYQELIK |
| SorfId=elkon_2015:525198 | 114 | MVVDRLFLWTFIIFTSVGTLVIFLDATYHLPPPDPFP |
| SorfId=andreev_2015:272691 | 150 | MDINRRRYPAHLARSSSRKYTELPHGAISEDQAVGPADIPCDSTGQTST |
| SorfId=calviello_2016:187121 | 222 | LLYLGRDYPKGADYFKKRLKNIFLKNKDVKNPEKIKELIAQGEFVMKELEALYFLRKYRAMKQRYYSDTNKTN |
| SorfId=loayza_puch_2016:211324 | 231 | MDLRRVKEYFSWLYYQYQIISCCAVLEPWERSMFNTILLTIIAMVVYTAYVFIPIHIRLAWEFFSKICGYHSTISN |
| SorfId=gonzalez_2014:438290 | 78 | MLDIFILMFFAIIGLVILSYIIYLL |
| SorfId=cenik_2015:853557 | 171 | MADVSERTLQLSVLVAFASGVLLGWQANRLRRRYLDWRKRRLQDKLAATQKKLDLA |
| SorfId=andreev_2015:169859 | 174 | MPTGKQLADIGYKTFSTSMMLLTVYGGYLCSVRVYHYFQWRRAQRQAAEEQKTSGIM |
| SorfId=crappe_2014:5055 | 144 | DVGSLDEKMKSLDVNQDSELKFNEYWRLIGELAKEIRKKKDLKIRKK |
| SorfId=calviello_2016:673102 | 225 | MFDIKAWAEYVVEWAAKDPYGFLTTVILALTPLFLASAVLSWKLAKMIEAREKEQKKKQKRQENIAKAKRLKKD |
| SorfId=ingolia_2012:368307 | 69 | LGMEEEDVIEVYQEQTGGHSTV |
| SorfId=andreev_2015:611100 | 273 | MPKRKAKGDAKGDKAKVKDEPQRRSARLSAKPAPPKPEPRPKKASAKKGEKLPKGRKGKADAGKDGNNPAKNRDASTLQSQKAEGTGDAK |
| SorfId=elkon_2015:4281 | 108 | MPQTFSGGRGFELDSGSDDMDPGRPRPPRDPDELH |
| SorfId=calviello_2016:40163 | 273 | MRDPVSSQYSSFLFWRMPIPELDLSELEGLGLSDTATYKVKDSSVGKMIGQATAADQEKNPEGDGLLEYSTFNFWRAPIASIHSFELDLL |
| SorfId=elkon_2015:564093 | 213 | MPARRLLLLLTLLLPGLGVSDRGAWGGGQLATAGSGPGQRRGAGAGVRAGSATAAARCPVSPAVGGSGRA |
| SorfId=andreev_2015:217825 | 87 | VERIKERVEEKEGIPPQQQRLIYSGKQM |
| SorfId=calviello_2016:764879 | 126 | VSKCSEEIKNYIEERSGEDPLVKGIPEDKNPFKEKGSCVIS |
| SorfId=calviello_2016:642790 | 204 | VKAAFTRAYNKEAHLTPYSLQAIKASRHSTSPSLDSEYNEELNEDDSQSDEKDQDAIETDAMIKVVF |
| SorfId=andreev_2015:308992 | 210 | MTTLIPILSTFLFEDFSKASGFKGQRPETLHERLTLVSVYAPYLLIPFILLIFMLRSPYYKYEEKRKKK |
| SorfId=gonzalez_2014:172668 | 129 | VSKAAADLMTYCDAHACEDPLITPVPTSENPFREKKFFCALL |
| SorfId=andreev_2015:216730 | 162 | VIPKRTNRPGISTTDRGFPRARYRARTTNYNSSRSRFYSGFNSRPRGRVYRSG |
| SorfId=gonzalez_2014:645009 | 93 | VFRYSLQKLAYTVSRTGRQVLGERRQRAPN |
| SorfId=werner_2015:456667 | 147 | MVITSENDEDRGGQEKESKEESVLAMLGIIGTILNLIVIIFVYIYTTL |
| SorfId=iwasaki_2016:288070 | 114 | MAEGNTLISVDYEIFGKVQGVFFRKHTQVCGLQALGW |
| SorfId=andreev_2015:619791 | 195 | MMNNGLLQQPSALMLLPCRPVLTSVALNANFVSWKSRTKYTITPVKMRKSGGRDHTGGNKDRGI |
| SorfId=werner_2015:555128 | 159 | MGCHSSKSTTVAAESQKLEEEREGREPGLETGTQAADCKDAPLKDGTPEPKS |
References
- Ladoukakis, E.; Pereira, V.; Magny, E.G.; Eyre-Walker, A.; Couso, J.P. Hundreds of putatively functional small open reading frames in Drosophila. Genome Biol. 2011, 12, R118. [Google Scholar] [CrossRef]
- Bliss, M. Banting’s, Best’s, and Collip’s accounts of the discovery of insulin. Bull. Hist. Med. 1982, 56, 554–568. [Google Scholar]
- Wadler, C.S.; Vanderpool, C.K. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 20454–20459. [Google Scholar] [CrossRef] [PubMed]
- Casson, S.A.; Chilley, P.M.; Topping, J.F.; Evans, I.M.; Souter, M.A.; Lindsey, K. The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 2002, 14, 1705–1721. [Google Scholar] [CrossRef]
- Rohrig, H.; Schmidt, J.; Miklashevichs, E.; Schell, J.; John, M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 1915–1920. [Google Scholar] [CrossRef]
- Lee, D.H.; Acharya, S.S.; Kwon, M.; Drane, P.; Guan, Y.; Adelmant, G.; Kalev, P.; Shah, J.; Pellman, D.; Marto, J.A.; et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol. Cell 2014, 54, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, D.; Liu, P.; Li, C.; Zhao, Q.; Zhu, D.; Yu, J. Zm908p11, encoded by a short open reading frame (sORF) gene, functions in pollen tube growth as a profilin ligand in maize. J. Exp. Bot. 2013, 64, 2359–2372. [Google Scholar] [CrossRef]
- Kastenmayer, J.P.; Ni, L.; Chu, A.; Kitchen, L.E.; Au, W.C.; Yang, H.; Carter, C.D.; Wheeler, D.; Davis, R.W.; Boeke, J.D.; et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006, 16, 365–373. [Google Scholar] [CrossRef]
- Gleason, C.A.; Liu, Q.L.; Williamson, V.M. Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol. Plant-Microbe Interact. 2008, 21, 576–585. [Google Scholar] [CrossRef]
- Kondo, T.; Hashimoto, Y.; Kato, K.; Inagaki, S.; Hayashi, S.; Kageyama, Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat. Cell Biol. 2007, 9, 660–665. [Google Scholar] [CrossRef]
- Galindo, M.I.; Pueyo, J.I.; Fouix, S.; Bishop, S.A.; Couso, J.P. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007, 5, e106. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Niikura, T.; Tajima, H.; Yasukawa, T.; Sudo, H.; Ito, Y.; Kita, Y.; Kawasumi, M.; Kouyama, K.; Doyu, M.; et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl. Acad. Sci. USA 2001, 98, 6336–6341. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zeng, J.; Drew, B.G.; Sallam, T.; Martin-Montalvo, A.; Wan, J.; Kim, S.J.; Mehta, H.; Hevener, A.L.; de Cabo, R.; et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015, 21, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Norris, M.L.; Valen, E.; Chew, G.L.; Gagnon, J.A.; Zimmerman, S.; Mitchell, A.; Ma, J.; Dubrulle, J.; Reyon, D.; et al. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. Science 2014, 343, 1248636. [Google Scholar] [CrossRef]
- Chanut-Delalande, H.; Hashimoto, Y.; Pelissier-Monier, A.; Spokony, R.; Dib, A.; Kondo, T.; Bohère, J.; Niimi, K.; Latapie, Y.; Inagaki, S.; et al. Pri peptides are mediators of ecdysone for the temporal control of development. Nat. Cell Biol. 2014, 16, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Magny, E.G.; Pueyo, J.I.; Pearl, F.M.; Cespedes, M.A.; Niven, J.E.; Bishop, S.A.; Couso, J.P. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013, 341, 1116–1120. [Google Scholar] [CrossRef]
- Tonkin, J.; Rosenthal, N. One small step for muscle: A new micropeptide regulates performance. Cell Metab. 2015, 21, 515–516. [Google Scholar] [CrossRef] [PubMed]
- Crappé, J.; Van Criekinge, W.; Trooskens, G.; Hayakawa, E.; Luyten, W.; Baggerman, G.; Menschaert, G. Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sORFs. BMC Genom. 2013, 14, 648. [Google Scholar] [CrossRef]
- Andrews, S.J.; Rothnagel, J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014, 15, 193–204. [Google Scholar] [CrossRef]
- Slavoff, S.A.; Heo, J.; Budnik, B.A.; Hanakahi, L.A.; Saghatelian, A. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 2014, 289, 10950–10957. [Google Scholar] [CrossRef]
- Yosten, G.L.C.; Liu, J.; Ji, H.; Sandberg, K.; Speth, R.; Samson, W.K. A 5′-upstream short open reading frame encoded peptide regulates angiotensin type 1a receptor production and signalling via the β-arrestin pathway. J. Physiol. 2016, 594, 1601–1605. [Google Scholar] [CrossRef] [PubMed]
- Schwab, S.R.; Li, K.C.; Kang, C.; Shastri, N. Constitutive display of cryptic translation products by MHC class I molecules. Science 2003, 301, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Parkhurst, M.R.; Kawakami, Y.; Robbins, P.F.; Rosenberg, S.A. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J. Exp. Med. 1996, 183, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef] [PubMed]
- McLysaght, A.; Guerzoni, D. New genes from non-coding sequence: The role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140332. [Google Scholar] [CrossRef] [PubMed]
- Strullu-Derrien, C.; Selosse, M.-A.; Kenrick, P.; Martin, F.M. The origin and evolution of mycorrhizal symbioses: From palaeomycology to phylogenomics. New Phytol. 2018, 220, 1012–1030. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Kutter, C.; Watt, S.; Stefflova, K.; Wilson, M.D.; Goncalves, A.; Ponting, C.P.; Odom, D.T.; Marques, A.C. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012, 8, e1002841. [Google Scholar] [CrossRef] [PubMed]
- Laing, W.A.; Martínez-Sánchez, M.; Wright, M.A.; Bulley, S.M.; Brewster, D.; Dare, A.P.; Rassam, M.; Wang, D.; Storey, R.; Macknight, R.C.; et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 2015, 27, 772–786. [Google Scholar] [CrossRef]
- Lauressergues, D.; Couzigou, J.M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef]
- Couso, J.-P.; Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017, 18, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Andreev, D.E.; O’Connor, P.B.; Fahey, C.; Kenny, E.M.; Terenin, I.M.; Dmitriev, S.E.; Cormican, P.; Morris, D.W.; Shatsky, I.N.; Baranov, P.V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 2015, 4, e03971. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef]
- Olexiouk, V.; Van Criekinge, W.; Menschaert, G. An update on sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018, 46, D497–D502. [Google Scholar] [CrossRef]
- Olexiouk, V.; Crappé, J.; Verbruggen, S.; Verhegen, K.; Martens, L.; Menschaert, G. sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016, 44, D324–D329. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.M. Everything old is new again: (linc)RNAs make proteins! EMBO J. 2014, 33, 937–938. [Google Scholar] [CrossRef]
- Raj, A.; Wang, S.H.; Shim, H.; Harpak, A.; Li, Y.I.; Engelmann, B.; Stephens, M.; Gilad, Y.; Pritchard, J.K. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife 2016, 5, e13328. [Google Scholar] [CrossRef]
- Kozak, M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- Michel, A.M.; Kiniry, S.J.; O’Connor, P.B.F.; Mullan, J.P.; Baranov, P.V. GWIPS-viz: 2018 update. Nucleic Acids Res. 2018, 46, D823–D830. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Chen, X.; Zheng, Y.; Kang, Q.; Hao, D.; Zhang, L.; Song, T.; Luo, H.; Hao, Y.; et al. SmProt: A Reliable Repository with Comprehensive Annotation of Small Proteins Identified from Ribosome Profiling. Genom. Proteom. Bioinform. 2021, 19, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Brunet, M.A.; Brunelle, M.; Lucier, J.F.; Delcourt, V.; Levesque, M.; Grenier, F.; Samandi, S.; Leblanc, S.; Aguilar, J.D.; Dufour, P.; et al. OpenProt: A more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res. 2019, 47, D403–D410. [Google Scholar] [CrossRef]
- Chen, Y.; Long, W.; Yang, L.; Zhao, Y.; Wu, X.; Li, M.; Du, F.; Chen, Y.; Yang, Z.; Wen, Q.; et al. Functional Peptides Encoded by Long Non-Coding RNAs in Gastrointestinal Cancer. Front. Oncol. 2021, 11, 777374. [Google Scholar] [CrossRef] [PubMed]
- Leong, H.S.; Dawson, K.; Wirth, C.; Li, Y.; Connolly, Y.; Smith, D.L.; Wilkinson, C.R.; Miller, C.J. A global non-coding RNA system modulates fission yeast protein levels in response to stress. Nat. Commun. 2014, 5, 3947. [Google Scholar] [CrossRef]
- Mazin, P.V.; Fisunov, G.Y.; Gorbachev, A.Y.; Kapitskaya, K.Y.; Altukhov, I.A.; Semashko, T.A.; Alexeev, D.G.; Govorun, V.M. Transcriptome analysis reveals novel regulatory mechanisms in a genome-reduced bacterium. Nucleic Acids Res. 2014, 42, 13254–13268. [Google Scholar] [CrossRef]
- Giannakakis, A.; Zhang, J.; Jenjaroenpun, P.; Nama, S.; Zainolabidin, N.; Aau, M.Y.; Yarmishyn, A.A.; Vaz, C.; Ivshina, A.V.; Grinchuk, O.V.; et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci. Rep. 2015, 5, 9737. [Google Scholar] [CrossRef]
- Pircher, A.; Gebetsberger, J.; Polacek, N. Ribosome-associated ncRNAs: An emerging class of translation regulators. RNA Biol. 2014, 11, 1335–1339. [Google Scholar] [CrossRef]
- Slavoff, S.A.; Mitchell, A.J.; Schwaid, A.G.; Cabili, M.N.; Ma, J.; Levin, J.Z.; Karger, A.D.; Budnik, B.A.; Rinn, J.L.; Saghatelian, A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013, 9, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Hanada, K.; Zhang, X.; Borevitz, J.O.; Li, W.-H.; Shiu, S.-H. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007, 17, 632–640. [Google Scholar] [CrossRef]
- Hanada, K.; Akiyama, K.; Sakurai, T.; Toyoda, T.; Shinozaki, K.; Shiu, S.-H. sORF finder: A program package to identify small open reading frames with high coding potential. Bioinformatics 2010, 26, 399–400. [Google Scholar] [CrossRef]
- Chugunova, A.; Navalayeu, T.; Dontsova, O.; Sergiev, P. Mining for Small Translated ORFs. J. Proteome Res. 2018, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef]
- McGeoch, D.J. On the predictive recognition of signal peptide sequences. Virus Res. 1985, 3, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Fix, E.; Hodges, J.L. Discriminatory Analysis: Nonparametric Discrimination: Consistency Properties; USAF School of Aviation Medicine: Randolf Fiend, TX, USA, 1951. [Google Scholar]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Ivanov, I.P.; Hinnebusch, A.G. Translational regulation by uORFs and start codon selection stringency. Genes Dev. 2023, 37, 474–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hao, W.; Yang, J.; Zhang, Y.; Wang, X.; Zhang, C. Emerging roles and potential clinical applications of translatable circular RNAs in cancer and other human diseases. Genes Dis. 2023, 10, 1994–2012. [Google Scholar] [CrossRef]
- Mudge, J.M.; Ruiz-Orera, J.; Prensner, J.R.; Brunet, M.A.; Calvet, F.; Jungreis, I.; Gonzalez, J.M.; Magrane, M.; Martinez, T.F.; Schulz, J.F.; et al. Standardized annotation of translated open reading frames. Nat. Biotechnol. 2022, 40, 994–999. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perdikopanis, N.; Giannakakis, A.; Kavakiotis, I.; Hatzigeorgiou, A.G. D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery. Biology 2024, 13, 563. https://doi.org/10.3390/biology13080563
Perdikopanis N, Giannakakis A, Kavakiotis I, Hatzigeorgiou AG. D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery. Biology. 2024; 13(8):563. https://doi.org/10.3390/biology13080563
Chicago/Turabian StylePerdikopanis, Nikos, Antonis Giannakakis, Ioannis Kavakiotis, and Artemis G. Hatzigeorgiou. 2024. "D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery" Biology 13, no. 8: 563. https://doi.org/10.3390/biology13080563
APA StylePerdikopanis, N., Giannakakis, A., Kavakiotis, I., & Hatzigeorgiou, A. G. (2024). D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery. Biology, 13(8), 563. https://doi.org/10.3390/biology13080563






