Simple Summary
Osteoporosis is a disease that is characterized by bone mass loss and microarchitecture deterioration, due to alterations in the bone remodeling mechanism, which must maintain a balance between bone resorption and formation. In recent years, it has been described that microRNAs (miRNAs) play an essential role in bone activation, differentiation, and homeostasis, since they can act as regulators of genes that participate in different signaling pathways. Therefore, altered expression of distinct miRNAs can affect the pathology of bone diseases such as osteoporosis. This review analyzes the current knowledge on the role of miRNAs in the various signaling pathways that maintain bone homeostasis in humans.
Abstract
Bone remodeling, crucial for maintaining the balance between bone resorption and formation, relies on the coordinated activity of osteoclasts and osteoblasts. During osteoclastogenesis, hematopoietic stem cells (HSCs) differentiate into the osteoclast lineage through the signaling pathways OPG/RANK/RANKL. On the other hand, during osteoblastogenesis, mesenchymal stem cells (MSCs) differentiate into the osteoblast lineage through activation of the signaling pathways TGF-β/BMP/Wnt. Recent studies have shown that bone remodeling is regulated by post-transcriptional mechanisms including microRNAs (miRNAs). miRNAs are small, single-stranded, noncoding RNAs approximately 22 nucleotides in length. miRNAs can regulate virtually all cellular processes through binding to miRNA-response elements (MRE) at the 3’ untranslated region (3′UTR) of the target mRNA. miRNAs are involved in controlling gene expression during osteogenic differentiation through the regulation of key signaling cascades during bone formation and resorption. Alterations of miRNA expression could favor the development of bone disorders, including osteoporosis. This review provides a general description of the miRNAs involved in bone remodeling and their significance in osteoporosis development.
1. Introduction
Osteoporosis (OP) is a metabolic disease characterized by low bone mineral density (BMD), leading to increased susceptibility to bone fragility fractures [1]. It is prevalent among adults over 50, and postmenopausal women are particularly affected [2]. OP stems from dysregulated bone remodeling, where osteoclasts remove old or damaged bone, replaced by osteoblasts with new bone [3]. In recent years, OP management strategies have improved considerably. However, the complexity of the multiple molecular mechanisms responsible for the development of OP, and the lack of specific therapeutic targets, have hindered advances in preventing OP [4]. MicroRNAs (miRNAs) have garnered attention for their involvement in various biological processes, including cell differentiation, migration, invasion, and apoptosis, with their dysregulation linked to metabolic diseases, including osteoporosis [5]. miRNAs are a class of small, noncoding RNAs that regulate gene expression post-transcriptionally through mRNA degradation or translation inhibition [6]. Recent studies have shown that miRNAs play an essential role in bone remodeling since they are involved in osteoclast and osteoblast differentiation. Therefore, they have been proposed as potential biomarkers and possible therapeutic targets for treating [7]. This review aims to synthesize the current evidence on miRNAs participating in bone remodeling and their potential as biomarkers for early OP detection.
2. Bone Remodeling
Bone is a dynamic tissue that is continuously turned over throughout life. Normal bone homeostasis depends on a self-renewal mode named bone remodeling, which is mainly maintained by the balance between osteoclastic bone resorption and osteoblastic bone formation [8]. This process comprises five main stages: activation, resorption, reversion, formation, and termination (Figure 1) [9]. It occurs within the basic multicellular unit (BMU), where osteoblasts (bone tissue-forming cells), osteoclasts (bone tissue resorption cells), osteocytes (mechanosensory and chemotaxis cells), and bone lining cells collaborate harmoniously [10]. The functions of these cell types are precisely controlled through their different intracellular events. The coupling systems regulate them during their interaction, so the regulation of any intracellular event or deterioration in their coupling factors can affect development and bone remodeling. The BMU, covered by bone lining cells, orchestrates resorption and formation processes, maintaining bone volume and responding to mechanical damage. The disruption of this balance leads to mineral diseases like osteopetrosis or osteoporosis [11].
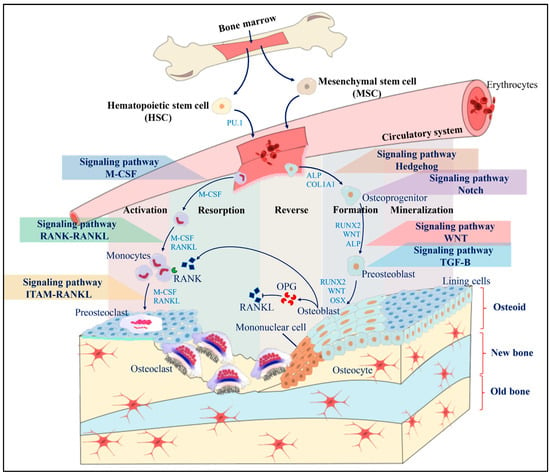
Figure 1.
Scheme of a BMU and the mechanism of bone remodeling. Initially, the resting bone surface is covered by lining cells and pre-osteoblasts. Mononuclear cells secrete OPG, which suppresses osteoclastogenesis until the activation, resorption, reverse, formation and mineralization stages are activated. During the differentiation of osteoclasts and osteoblasts, different signaling pathways are activated that allow obtaining mature bone cells capable of performing specific functions. Osteoclastogenesis can be activated through the M-CSF signaling pathways, the RANK-RANKL pathway, and the ITAM RANKL pathway, while osteoclastogenesis is activated through the Hedgehog signaling pathway, the Notch pathway, the Wnt pathway, and the TGF-B pathway.
The current data demonstrate the genetic and epigenetic factors that can affect bone remodeling [12]. Epigenetics is defined as the mechanisms regulating gene expression resulting in a determined phenotype, without altering the DNA sequence; furthermore, epigenetic mechanisms can be inherited [13,14]. There are currently three recognized epigenetic mechanisms: histone modification, DNA methylation, and noncoding RNAs (ncRNAs). Among (ncRNAs), microRNAs (miRNAs) have attracted special interest for their role in bone development and function [12]. The miRNAs have been found to be involved in multiple biological processes, including cell differentiation and proliferation [15]. Therefore, numerous miRNAs have been found to be involved in the regulation of bone homeostasis, and they play critical roles in bone remodeling.
3. Biogenesis of miRNAs
miRNAs are the most studied noncoding RNAs related to bone metabolism and bone diseases. miRNAs are small ncRNAs ranging from 19 to 25 nucleotides in length that can regulate gene expression post-transcriptionally in eukaryotic organisms. miRNAs genes are transcribed by the RNA pol II/III. The primary transcript is called pri-miRNA. In some cases, several miRNAs from a single locus can be transcribed simultaneously [16,17]; alternatively, miRNAs can be located within an intron or an untranslated region (UTR). There are two main miRNA biogenesis pathways: the canonical and non-canonical [18]. In the canonical pathway, the Drosha/DGCR8 complex and other associated proteins process the pri-miRNA into a miRNA precursor (pre-miRNA), which acquires a hairpin structure of approximately 60–100 nucleotides in length. Pre-miRNA is transported from the nucleus to the cytoplasm through the protein Exportin 5 (XPO5) and Ran-GTP [17].
Conversely, in the non-canonical pathway, the miRNAs lying on introns, called miRtrons, are processed by the spliceosome and are transported directly to the cytoplasm [19]. Once in the cytoplasm, pre-miRNAs are processed by the Dicer complex to generate a mature miRNA duplex. Subsequently, the RNA-induced silencing complex (RISC) is formed by the Argonaute protein (AGO), the PAK activator protein (PACT), the RNA-binding protein (TRBP), and one of the strands from the mature miRNA duplex. The RISC complex targets mRNA through base pairing between the seed region of the miRNA and the miRNA Response Elements (MREs), located at the 3′UTR of the target mRNA. Perfect base complementarity promotes the AGO2 protein to cleave the mRNA, leading to degradation by endonucleases. When base complementarity is imperfect, a hairpin is formed, promoting the translational suppression of the target mRNA. Furthermore, circulating miRNAs are found in various forms in the bloodstream (Figure 2): 90% of extracellular miRNAs are bound to AGO proteins, while the remaining 10% are packaged into exosomes in apoptotic bodies or bound to high-density lipoproteins (HDL) [20].
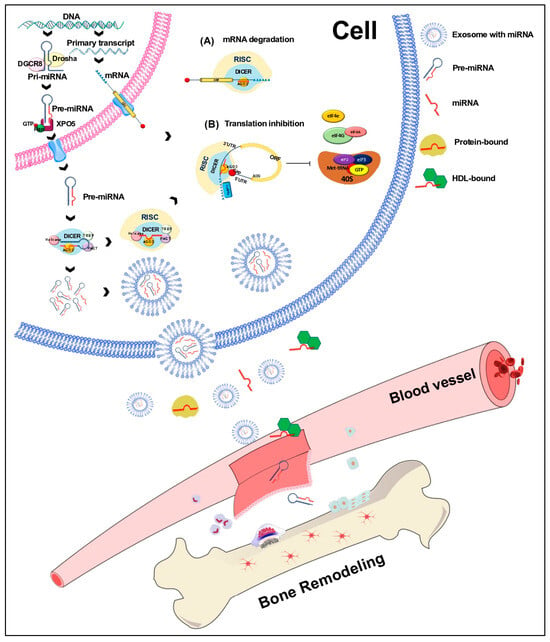
Figure 2.
Biogenesis of miRNAs. miRNAs are transcribed by RNA polymerase II or III into primary miRNAs (pri-miRNAs) that are processed by Drosha/DGCR8 into miRNA precursors (pre-miRNAs). The strand of a mature miRNA is shown in red; the pre-miRNA is transported from the nucleus to the cytoplasm by exportin 5 (XPO5), where it is processed by Dicer/TRBP into a duplex miRNA. A helicase unwinds this miRNA, and the mature strand (red) is incorporated into the RNA-induced silencing complex (RISC). Depending on the complementarity of the miRNA with the seed region of a target mRNA, the RISC complex mediates the downregulation of gene expression either by mRNA degradation (A) or by translational repression (B). Circulating miRNAs are produced within the donor cell and are bound to proteins. They are exported directly or packaged into exosomes and released into circulation. miRNAs are also bound to HDL (high-density lipoproteins); however, the binding mechanisms are still under investigation.
4. Osteoclastogenesis
Bone remodeling is intricately regulated by a delicate balance between bone resorption and formation [21]. The first phase, termed “activation”, commences in the BMU, where mechanical damage stimulates bone resorption through the release of cytokines responsible for recruiting osteoclast precursors to the bone surface to induce bone decalcification. The macrophage colony-stimulating factor (M-CSF) and the receptor activator nuclear kappa B ligand (RANKL) induce the differentiation of hematopoietic progenitor cells into osteoclast. Under physiological conditions, these cytokines are released by osteoblasts and stromal cells. They are also required for the survival, proliferation, and expression of receptor activator nuclear kappa B (RANK) in osteoclast precursors [22]. RANKL can initiate signaling cascades for osteoclast differentiation, such as those dependent on NF-KB, c-Fos, cell nuclear factor transcription factor t (NFATc1), and microphthalmia-induced transcription factor (MITF) [23]. The differentiated osteoclasts will start with the “resorption” phase, forming a wavy edge that allows them to adhere to the bone surface. Between the osteoclasts and the bone surface, an isolated microenvironment is formed where proton pumps release ions for acidifying the medium, dissolving the mineralized component of the bone matrix and forming a cavity known as Howship’s lagoon. Afterwards, the exposed organic matrix is degraded by cathepsin k, initiating the “reverse” phase [24]. In the reverse phase, mononuclear cells prepare the newly resorbed bone surface and recruit osteoblast precursors, which mature into osteoblasts and constitutively express type I collagen. Osteoblasts trapped within the mineralized matrix continue their differentiation to become osteocytes, while osteoblasts on the surface become lining cells [3].
4.1. Signaling Pathways Involved in Osteoclast Differentiation
The connection between the miRNAs and the involved pathways in osteoclastogenesis is shown in Figure 3 and Table 1. In recent years, several studies have unraveled key signaling mechanisms involved in osteoclast differentiation, including the signaling pathways, such as the M-CSF and RANK ligand (RANKL) pathways, which are critical for bone resorption.
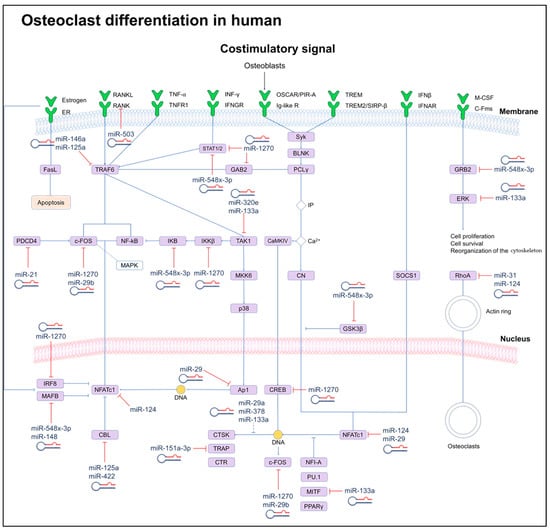
Figure 3.
Scheme of signaling networks (OPG/RANK/RANKL) involved in osteoclast differentiation and their regulation induced by miRNAs. miRNAs can directly (solid lines) or indirectly (dotted lines) inhibit key genes for osteoclast differentiation. Dashed lines indicate cellular processes.

Table 1.
Summary of miRNAs in humans and their targets, expressions, and effects on osteoclast differentiation.
4.1.1. M-CSF Signaling Pathway
Differentiation into osteoclasts begins when M-CSF binds to the c-Fms receptor of a hematopoietic precursor. Signal transduction leads to the activation of the extracellular signal-regulated kinase (ERK) through Grb2 and the phosphoinositide 3-kinase (PI-3K/Akt) [40]. Hematopoietic stem cell differentiation into osteoclasts is induced by the activation of transcription factors such as binding protein 1 (PU.1) and MITF. In hematopoietic stem cells, PU.1 stimulates the expression of CSF1R, the M-CSF receptor.
Another transcription factor that plays an essential role in osteoclastogenesis is the activating protein 1 (AP-1), a member of the Fra, Fos, Jun, and activating transcription factor (ATF) family. On the other hand, the transcription factor MITF is involved in the late stage of osteoclastogenesis through interaction at a mitogen-activated protein kinase (MAPK) consensus site, where M-CSF induces MITF phosphorylation [41]. MITF phosphorylation induces expression of the anti-apoptotic gene of B cell lymphoma 2 (BCL2) and promotes macrophage survival. In addition, MITF and PU.1 can significantly increase RANK promoter activity. On the contrary, MITF levels are regulated by RANKL [42]. In this sense, both PU.1 and MITF have an essential role in the differentiation and survival of osteoclasts by the induction of specific genes [43].
4.1.2. RANKL–RANK Signaling Pathway
Activation of RANKL–RANK pathway induces the expression of genes driving the fusion of cells derived from the monocyte/macrophage lineage, for example, dendritic cell-specific transmembrane protein (CD-STAMP), as well as genes involved in the resorption process of mineral tissue [44], including cathepsin K (CTSK), chloride channel 7 (CIC-7), matrix metalloproteinase 9 (MMP9), and calcitonin receptor (CTR) [45]. The binding of RANKL to RANK induces the recruitment of TRAF-6, which activates the PI3K family of transcription factors, NF-kB, and the MAPK pathways, including ERK, c-Jun N-terminal kinase (JNK), and p38. NF-kB induces the expression of cytokines such as interleukin 6 (IL6), interleukin 1 (IL-1), TNF, and GM-CSF; the p38 protein kinase is activated through phosphorylation of the MAPK kinase (MKK), allowing the downstream activation of MITF. The inhibition of p38 increases ERK phosphorylation while maintaining a balance between ERK and p38 phosphorylation. RANKL induces the expression of the AP-1/c-Fos complex [46]. On the other hand, the expression of NFATc1 depends on the TRAF-6/NF-kB/c-Fos pathways, activated by RANKL and Ca+2 signaling. NFATc1 is a transcription factor that regulates osteoclast-specific gene expression such as tartrate-resistant acid phosphatase (TRAP), CTSK, calcitonin receptor (CGRP), osteoclast-associated receptor (OSCAR), and integrin 3 (ITGB3) [47].
4.1.3. Tyrosine-Based Immunoreceptor (ITAM) Signaling Pathway
Recent studies have demonstrated that ITAM adapter proteins are involved in the formation and function of osteoclasts. They signal to activate Syk kinase and PLCγ2 which initiates Ca2+ oscillations that can result in activation of the key transcription factor, NFATc1, controlling the differentiation of pre-osteoclasts and multinucleation. The activation of M-CSF/RANKL-induced signaling is not sufficient to complete the osteoclast differentiation process, so it is necessary to activate costimulatory signals dependent on the immunoreceptor tyrosine-based activation motif (ITAM); these signals are activated by multiple immunoreceptors involved in osteoclastogenesis, among them are the Fc receptor subunit (FcR) and DNAX activating protein 12 (DAP12), which are part of ITAM. In osteoclast precursor cells, FcR and DAP12 are associated with multiple immunoreceptors that activate the calcium signaling pathway through phospholipase C (PLC) [48]. These receptors include OSCAR, myeloid cell-activating receptor 2 (TREM-2), signal regulatory protein (SIRP1), and paired Ig-like receptor A (PIR-A), which work together with RANKL and ITAM to costimulate RANK [49]. However, although ITAM adapter signaling is critical for normal bone remodeling, estrogen deficiency induces an ITAM adapter-independent bypass mechanism enhancing osteoclastogenesis and activation in specific bony microenvironments [50].
5. The Role of miRNAs in Osteoclastogenesis
In the following sections, we highlight several critical miRNAs playing an essential role in osteoclastogenesis. The have been proposed as potential markers for the early detection of bone-related diseases.
5.1. miR-30
This miRNA targets genes associated with stimulation of osteoclastogenesis (IL-8, IL-11), the inhibition of osteoblastogenesis (DKK-1), tumor cell osteomimetics (RUNX2, CDH11), and invasiveness (CTGF, ITGA5, ITGB3) [51]. Abnormal levels of secret antagonists of Wnt signaling can displace bone remodeling in both directions. DKK1 is considered an inhibitor of Wnt/β-catenin signaling, which is linked to new bone formation by functioning as a positive regulator of osteoblasts. Meanwhile, RUNX2 and Wnt/β-catenin signaling plays a crucial role in MSCs migration and osteoblast differentiation [52,53]. Therefore, the miR-30-induced downregulation of DKK1 and RUNX2 could affect the Wnt/β-catenin signaling pathway and, consequently, the differentiation mechanisms of both the osteoclastic and osteoblastic pathways. On the other hand, the CTGF, ITGA5, and ITGB3 genes are regulators of the PI3K signaling pathway [54]. It has been reported that this signaling pathway is involved in processes of the formation, differentiation, and function of osteoclasts [55].
5.2. miR-320e
This miRNA has been observed in extracellular vesicles (EVs) derived from patients with ossification of the posterior longitudinal ligament (OPLL). miR-320e promotes the osteoblastic differentiation of mesenchymal stem cells (MSC) and inhibits the osteoclastic differentiation of monocytes. miR-30e targets TGF-β-activated kinase 1 (TAK1), which is an essential activator of osteoclastogenesis [56]. TAK1 is a crucial regulator of innate and proliferative immune signaling pathways such as TNF, IL-1R, and TLR. Its activation phosphorylates the IkB kinase (IKK) complex, p38, JNK, and ERK, leading to the activation of nuclear factor (NF)-κB and the MAPK signaling pathways. Therefore, the regulation of TAK1, induced by miR-320e, could be a critical factor in the regulation of osteoclastogenesis via NF-κB [57].
5.3. miR-1270
This miRNA was described as a positive regulator of osteoclastogenesis since it inhibits the expression of interferon regulatory factor 8 (IRF8), which has been described as an anti-osteoclastogenic gene. The positive regulation of this miRNA promotes monocyte/osteoclast differentiation in postmenopausal women, favoring the development of osteoporosis [58]; therefore, this miRNA could be a potential biomarker candidate for the early detection of osteoporosis [59]. It has been reported that the RANK–RANKL signaling pathway activates the NF-κB and c-Fos signaling, which in turn activates the master transcription factor NFATc1, which promotes osteoclastogenesis. However, it has been reported that the activation of factors such as IRF8, MAFB, and IDS can suppress osteoclast differentiation [60]. IRF8 is regulated by miR-1270 in circulating monocytes from postmenopausal women, so this miRNA could stimulate osteoclast differentiation through the RANK–RANKL signaling pathway.
During the process of differentiation and maturation of osteoclasts, other changes in the expression of miRNAs have been described in humans. However, these miRNAs’ molecular mechanisms and functional roles are still unclear. Table 1 describes the role of the most relevant miRNAs [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39] in osteoclastogenesis that have been described in recent years.
6. Osteoblastogenesis
Bone formation begins with MSCs, derived from the mesoderm in the early stages of embryonic development. The differentiation of MSC to osteoblasts is crucial for maintaining bone remodeling. Osteoblasts secrete organic bone matrix, promote bone matrix mineralization, and maintain bone homeostasis. Osteoblast dysfunction can lead to the destruction of bone microarchitecture and defects in bone formation, leading to the development of metabolic diseases such as osteoarthritis and osteoporosis (Figure 4).
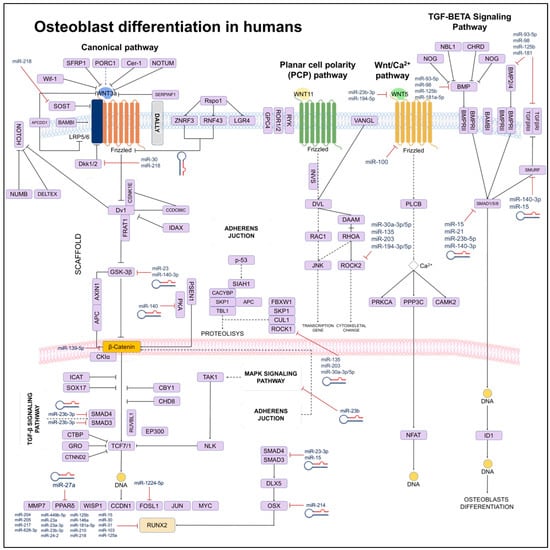
Figure 4.
Scheme of signaling networks (TGF-β/BMP/Wnt) involved in osteoblast differentiation. It is show that miRNAs can directly (solid lines) or indirectly (dotted lines) to inhibit genes regulating differentiation processes, apoptosis, or the phenotype of osteoblasts.
6.1. Signaling Pathways Involved in Osteoblast Differentiation
Osteogenesis is regulated by different signaling pathways, such as, M-CSF, and RANKL, involving several transcription factors, like RUNX2, the nuclear factor of acti-vated T cells (NFAT), and Osterix (Osx). RUNX2 is the determinant transcription; its inactivation can inhibit or delay osteoblast formation. RUNX2 is a critical factor in osteoblast-mediated bone formation and requires precise regulation by mechanisms such as Osx [61]. Additional pathways are also involved in the osteoblast differentiation process and the regulation of the ossification process, such as the Hedgehog pathway (Hh), the Wnt pathway, the BMP pathway, and the Notch pathway [62].
6.1.1. Wnt Signaling Pathway
The Wnt family comprises a set of highly conserved genes that regulate cell behavior, expression, adhesion, and polarity. The Wnt-induced signaling was thought to be mediated by β-catenin. However, current evidence supports that the Wnt pathway activates β-catenin-independent mechanisms by what is known as the “non-canonical pathway” to regulate vertebrate development [63]. Just as the canonical pathway is essential in vertebrate development and bone diseases, the non-canonical Wnt pathway is involved in bone formation. The non-canonical pathway is divided into two main sub-pathways: the Wnt–planar-cell-polarity pathway (Wnt–PCP pathway) and the Wnt–calcium pathway (Wnt–Ca2+ pathway). The Wnt5a protein regulates limb morphogenesis, chondrogenesis, and osteoblastogenesis through the receptor tyrosine kinase (Ror) proteins in the Wnt–PCP pathway. Furthermore, this pathway also regulates osteoclastogenesis. The Wnt5a-Ror2 signaling activates the expression of JNK, which is responsible for recruiting c-Jun from promoting the expression of RANK on the surface of osteoclast precursor cells. On the other hand, in the Wnt–Ca2+ pathway, the Wnt5a ligand binds to the Frizzled receptor, triggering an increase of inositol 1,4,5-trisphosphate (IP3), 1,2-diacylglycerol (DAC), and Ca2+ with PLC. This interaction triggers the activation of the NF-kB signaling pathway and the expression of transcription factors such as NFAT, which regulates osteoclastogenesis [64]. Different studies have reported that the Wnt–Lrp5 interaction can induce mTORC2-AKT signaling activity and triggers glycolytic enzymes in bone cells to promote bone formation. These findings indicate that Wnt signaling can regulate bone homeostasis [65].
6.1.2. Ligands and Agonists of the Wnt Pathway in Bone
Wnt ligands are cysteine-rich proteins that contain an N-terminal signal peptide for secretion and have effects at various stages of bone development, including chondro-genesis, osteoclastogenesis and osteoblastogenesis. Mutations of the Wnt1 gene have been found in children with osteogenesis imperfecta, which is a disorder characterized by increased bone fragility and other connective tissue manifestations [66]. Wnt3a regulates the fate of the dorsal mesoderm and is required during the early stages of limb formation and craniofacial development [67]. In a mouse model of R26floxneo Wnt4 with a Col2a1-Cre mutation, the conditional expression of Wnt4 results in dwarfism and an increased number of hypertrophic chondrocytes. In addition, it was also observed that Wnt5a and Wnt5b activate the proliferation and differentiation of chondrocytes [68]. In different studies with mouse models, it was observed that Wnt3a +/− and Wnt5a +/− presented a low bone mass phenotype [69]. Wnt6, Wnt10a, and Wnt10b have also been reported to stimulate osteo-blastogenesis and inhibit adipogenesis [70].
Mutations in the ligands Wnt7 and Wnt11 activate the differentiation of chondrocytes and osteoblasts, while deficiency of Wnt16 decreases BMD and increases fracture risk [71]. The current data report that parathyroid hormone (PTH) influences Wnt signaling at different stages of bone development. PTH decreases the expression of Wnt inhibitors such as Sost, leading to an increase of Wnt signaling. In a study by Tian Y. et al. 2011, in a model of MC3T. E1 osteoblasts treated with PTH, the characteristic markers of osteoblast differentiation were blocked by decreasing B-catenin expression [72]. On the other hand, transgenic mice expressing a constitutively active PTH receptor in osteocytes showed an increase in the number of osteoblasts and bone mass and the downregulation of Sost [73]. In postmenopausal women, PTH can reduce circulating sclerostin levels. The effects of PTH on the canonical Wnt signaling pathway can upregulate FZD-1 or LRP6 receptor complex proteins and decrease the Dkk-1 antagonist [74]. Although PTH treatment reduces Dkk-1 expression, Dkk-1 upregulation does not inhibit the anabolic response to PTH in vivo [75]. Evidence suggests that in both in vivo and in vitro models, there is a direct crosstalk between PTH1R and the Wnt signaling pathway. PTH binding to the PTHR receptor induces the association of the PTH-PTH1R complex with the extracellular domain of the Lrp6/Wnt receptor in the absence of the Wnt ligand, resulting in the phosphorylation of Rlp6, [76]. These findings suggest that PTH induces osteoblast differentiation by activating the canonical pathway of Wnt.
6.1.3. Notch Signaling Pathway
The Notch signaling pathway requires cell-to-cell interaction. It is initiated when Jagged ligands 1/2 (JAG) and delta-like ligands 1/3/4 (DLL) interact with Notch receptors (1-4 in mammals). Ligand binding leads to the proteolytic cleavage of the Notch receptor by the action of the γ-secretase complex, releasing the Notch intracellular domain (NICD). The NICD translocates to the nucleus and interacts with the DNA-binding protein RBPjk/CBF1, displacing corepressors and allowing the assembly of an activator complex that includes transcriptional coactivators similar to the Mastermind protein type 1 (MAML1), which is involved in cell differentiation. Notch target genes include the Hairy transcription receptor (HES-1) and HES associated with the YRPW-like motif (HEY) [77]. The Notch signaling pathway interacts with the Wnt signaling pathway and bone morphogenic protein (BMP) to regulate skeletal development and homeostasis. In addition, in vitro experiments have showed that NICD overexpression opposes the differentiation of osteoblasts induced by exogenous Wnt. This effect is regulated by HES-1 [78]. Other in vitro studies investigating the relationship between Notch signaling and BMP report opposite results, showing that NICD overexpression blocks the differentiation of osteoblast precursors in the presence of BMP2 [79]. In summary, the signaling induced by Notch enhances osteoblast precursor differentiation [80], whereas Notch inhibition impairs BMP2-induced osteoblast differentiation [81].
6.1.4. Notch Signaling Pathway in Osteoblastogenesis
Studies in mouse models suggest an essential role of the Notch signaling pathway in skeletal development. Knockdown of Notch-1 and Notch-2 in skeletal mesenchymal tissue led to the accumulation of bone tissue within the medullary canal and a severe reduction in trabecular bone mass [82]. These observations suggest that Notch signaling is responsible for suppressing the differentiation of mesenchymal progenitor cells into osteoblasts. Later, it was reported that Notch-2 has a predominant role in suppressing osteoblastogenesis, mainly mediated by Notch-1 and RBPjk13. It has also been observed that downstream of RBPjk, the HEY family of transcriptional suppressors is responsible for the inhibition of RUNX2 activity and the suppression of NFATc1 [83]. In murine models, it has been observed that the Hey transcription factor plays an important role in osteoblastogenesis. Mice expressing Hey present an osteopenic phenotype, while those expressing RBPjk do not show this phenotype. It seems that the suppressive role of Notch-RBPjk signaling in osteoblastogenesis is limited to the early stages of osteoblast differentiation [84]. Studies in transgenic mice overexpressing NICD have revealed different effects depending on the differentiation stage of the osteoblasts. The overexpression of NICD in osteoblastic cells, under the control of the constitutive COL1A1 promoter, generates a phenotype with a high bone mass, but with an excess of immature bone tissue and fibrotic cells in the bone marrow [85]. The expression of NICD stimulates cell proliferation of the osteoblastic lineage in the early stages but prevents the differentiation of mature osteoblasts [86].
6.1.5. TGF-β Signaling Pathway in Osteoblastogenesis
Signaling induced by transforming growth factor-beta (TGF-β) and BMP plays a fundamental role in embryonic skeletal development and bone homeostasis. TGF-β and BMPs act as a tetrameric receptor complex. They are responsible for transducing signals in both the Smad-dependent canonical signaling pathway and the Smad-independent non-canonical signaling pathway. Activated mitogen p38/p38 MAPK is activated to regulate the differentiation of mesenchymal stem cells during skeletal development, bone formation, and bone homeostasis [87]. The transforming growth factors bind to a tetrameric receptor complex comprising two TGF-β receptors of the type I (TβRI/ALK5) and two type II receptor kinases (TβRII) [88]. Transphosphorylases TβRII are responsible for phosphorylating the Smad proteins, activated by Smad 2/3 receptors that subsequently interact with Smad and Smad 4, that are translocated to the nucleus, where they recruit cofactors for gene regulation, such as Creb-binding protein (CBP) and p300 [89].
On the other hand, it has been reported that TGF-β activates a group of R-Smad receptors (Smad1/5/8) through binding to ALK. In addition, it has also been observed that as an alternative pathway, not dependent on Smad, TGF-β activates the protein kinase 1 (TAK1) and the TAK1 binding protein (TAB1), allowing the activation of the p-38/MAPK signaling cascade [89]. TGF-β isoforms are expressed in the perichondrium, periosteum, and epiphyseal growth plate. Mouse models with an attenuated Tgfb2 expression showed severe skeletal abnormalities in both endochondral and intramembranous bone. Meanwhile, Tgfb1- and Tgfb2-deficient mice had no defects. Therefore, it can be assumed that TGF-β1 and TGF-β3 are not essential for the development of the embryonic skeleton. On the contrary, TGF-β2 is necessary for the maintenance of postnatal bone mass by coupling bone resorption and formation. TGF-β proteins are synthesized as precursor molecules containing the latency-associated protein (LAP), which remains non-covalently bound to TGF-β. The cleavage of LAP, induced by bone resorption, allows the release of active TGF-β1, which induces the enrichment of osteoprogenitor cells in the Howship’s cavities [90]. These observations support that the BMP and TGF-β signaling pathways play an essential role in skeletal development and bone homeostasis by interacting with other signaling pathways previously discussed, such as Wnt, Hedgehog, and Notch.
7. The Role of miRNAs in Osteoblastogenesis
The osteoblasts cells are involved in bone formation and, thus, maintain homeostasis and metabolism. These cells also play an important role in the development of osteoporosis. Some miRNAs that play an essential role in osteoblastogenesis and bone-related diseases are described below.
7.1. miR-23b-3p/miR-885, miR-140-3p and miR-885
In a study carried out by Ramírez et al., the miRNAs miR-23b-3p, miR-140-3p, and miR-885 showed differential expression in the serum of postmenopausal women with low BMD or with osteoporotic fracture, compared to a group with normal BMD. The bioinformatic analysis revealed that these miRNAs regulate genes involved in the Wnt, MAPK, and TGF-β signaling pathways. The authors suggested that these miRNAs could be associated with variation in BMD and highlighted their potential as biomarkers for the early detection of osteoporosis [91].
7.2. miR-29-3p, miR-324-3p, and miR-550a-3p
These miRNAs showed a significant correlation with histomorphometric parameters of bone formation and microarchitectural parameters. In addition, these miRNAs were downregulated in patients treated with antiresorptive therapy. The authors proposed these three miRNAs as potential biomarkers for the diagnosis of osteoporosis [92]. Although the potential targets of miR-29b-3p are not mentioned, this miRNA is involved in osteoclast differentiation and bone resorption, mainly at the phase of differentiation from monocyte precursor [93]. miR-324-3p regulates genes such as BSP, RUNX2, OCN, and ALP, which are key in osteoblast differentiation and participate in the Wnt signaling pathway. On the other hand, miR-324-3p can also promote osteoblastogenesis through the regulation of SMAD7 [94]. The miRNA miR-550a-3p was identified in the serum of postmenopausal women and has been suggested as a potential biomarker for the early diagnosis of osteoporosis. This miRNA targets osteocalcin (OCN), which is a crucial participant in the Wnt signaling pathway [95].
7.3. miR-30a-3p/5p, miR-194-3p/5p, miR-27b-3p/5p and miR-34a-3p/5p
In a work published by Zhou et al., miRNAs and differentially expressed genes (DEGs) were identified in the serum of a group of postmenopausal women with both high and low BMD. The authors identify 34 miRNAs, of which hsa-miR-30a-3p/5p, hsa-miR-194-3p/5p, hsa-miR-27b-3p/5p, and hsa-miR-34a-3p/5p were downregulated in the samples from women with osteoporosis compared to a control group. The authors propose that the expression of these miRNAs could be related to the suppression of osteoclast survival and the promotion of osteoblast activity. Therefore, the authors suggested that these miRNAs could play a key role in regulating bone formation since the overexpression of miR-30a suppress TGFB1, which in turn regulates RUNX2, a transcription factor specific to osteoblasts and bone formation [96].
7.4. miR-194
A previous study analyzing blood samples from postmenopausal women with osteopenia and osteoporosis revealed the overexpression of miR-194-5p, hsa-miR-454-3p, hsa-miR-151a-3p, and hsa-miR-590-5p and downregulation of hsa-miR-574-3p, hsa-miR-3907, hsa-miR-4767, and hsa-miR-1972 [97]. From these miRNAs, hsa-miR-194 and hsa-miR-590-5p have been related to bone metabolism, mainly in osteogenesis. They promote osteoblast differentiation through the regulation of the SMAD family genes and RUNX2, which participate in the STAT1 signaling pathway and the Wnt pathway [98,99].
7.5. miR-1224-5p
This miRNA was identified as downregulated in plasma derived from patients with osteoporotic fractures, suggesting that it plays a role in osteoclastogenesis. Experimental data showed that expression of miR-1224-5p was positively correlated with the progression of fracture healing. The authors observed that miR-1224-5p slowed down osteoclast differentiation induced by RANKL. Additionally, the expression of miR-1224-5p promoted osteoblast differentiation through the Rap1 signaling pathway, by regulating the ADCY2 gene. Furthermore, the in vivo overexpression of miR-1224-5p significantly promoted fracture healing and facilitated the progression of osteoporosis caused by estrogen deficiency or aging. These results suggest that miR-1224-5p is a critical regulator of osteogenesis and may be a potential therapeutic target for osteoporosis and fragility fractures [100]. Table 2 [28,91,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137] describes the function of the most relevant miRNAs involved in osteoblastogenesis that have been described in recent years. Beyond the osteoclastogenesis and osteoblastogenesis pathways, several pathways interact and participate in the bone remodeling process [138].

Table 2.
Summary of miRNAs, study models, and miRNAs’ targets, expressions, and effects on osteoblast differentiation.
8. miRNAs Involved in Osteocyte Differentiation
Osteocytes are the last level of differentiation of osteoblasts and are surrounded by the mineralized bone matrix. They act as sensors of chemical signals and mechanical loads that control bone activity through communication with cell effectors that participate in bone remodeling [139]. Osteocytes secrete different molecules that can modulate the function of osteoclasts and osteoblasts on the bone surface, in addition to the mineralized bone matrix and cells of other tissues and organs. Their differentiation involves changes in their morphology that allow them to develop numerous cytoplasmic projections and changes in miRNA expression profiles. Recent studies have reported that osteocytes can release extracellular vesicles containing miRNAs, which can affect the function of skeletal muscle and adipose tissue [140]. It has been shown that miRNAs induce or inhibit the differentiation of osteoblastic cells derived from MSC [141] while, in cell lines, as well as in primary osteocytes, the role of miRNAs in osteocyte differentiation processes has been demonstrated and summarize in Table 3 [142,143,144,145,146,147,148,149,150]. These miRNAs involved in the differentiation of bone cells are known as osteomiR and participate in the differentiation of terminal osteoblasts through the regulation of osteocytic genes [151]. However, as cells progress in their differentiation pathway, they lose the ability to express miRNAs that regulate the activation or inhibition of differentiation, which is why they have been proposed as characteristic markers of osteocytes known as Snord85. In contrast, others are considered negative markers; among them are miR-101a, miR-10a, and the let-7 family, as well as other members of the miR-30 family that increase their expression during late osteocyte differentiation and suppress genes that are important for osteoblast differentiation, such as Runx2, Smad1/2, and CCN3 [152]. The role of miRNAs in osteocytes is a field that has been little explored, so new research is necessary to determine how miRNAs modulate osteocyte differentiation and participate in bone remodeling.

Table 3.
Summary of miRNAs in humans and their targets, expressions, and effects on osteocyte differentiation.
9. The Role of miRNA in Osteoporosis
Bone metabolism is a multifactorial process that involves different types of cells, such as mesenchymal stem cells, hematopoietic stem cells, osteoclasts, osteoblasts, and osteocytes, in which different biological processes, such as proliferation, angiogenesis, differentiation, migration, and apoptosis, are developed, which are regulated by miRNAs [144]. Alterations in the expression profiles of miRNAs can affect the functions of osteoclasts, osteoblasts, and osteocytes, with a consequent imbalance in bone remodeling, leading to the development of OP. However, miRNAs not only regulate bone remodeling but also participate in fracture repair. The fracture repair process consists of several interdependent stages: hematoma formation, inflammation, osteogenesis, chondrogenesis, endochondral ossification, and remodeling, where miRNAs miR-21, miR-140, miR-214 have been proposed as potential biomarkers for monitoring the overall fracture healing process [153]. On the other hand, it has been reported that the blood serum is an ideal sample for the identification of different miRNAs that can be used as biomarkers for the detection of OP, and miR-365a-3p is upregulated in the blood serum. In patients with OP, the levels gradually decrease as osteoinduction in MSC is prolonged, while the downregulation of miR-365a-3p reduces the expression of osteocalcin (OCN), RUNX2, Osteopontin (OPN), and COL1A1, which reduces the bone formation potential [154]. Other studies have reported that miR-579-3p regulates the expression of sirtuin 1 (Sirt1), which is highly expressed in MSCs and is associated with the maintenance of bone homeostasis, so its regulation, induced by miR-579-3p, inhibits the differentiation of MSC into osteogenic cells [155]. Another miRNA identified as upregulated in the blood serum and MSCs of OP patients is miR-96, which was associated with the decreased osteogenic differentiation of MSCs. At the same time, the inhibition of miR-96 mitigated bone loss caused by aging [156]. These studies have demonstrated the presence of miRNAs, in the serum of patients with OP, which show variable expression and can act as potential biomarkers since they act on MSCs, osteoblasts, osteoclasts, and other bone cells. However, using miRNAs as biomarkers may have some disadvantages, such as the lack of standardization during the selection of circulating miRNAs, their identification in a large population, the lack of exhaustive studies on specific diseases, and the effect of confounding variables such as age, sex, and the lack of standards on accurately identifying the origin of miRNAs [157]. Table 4 [106,158,159,160,161,162,163,164,165,166,167,168] shows the miRNAs identified in humans that are involved in OP.

Table 4.
Summary of miRNAs in humans and their targets, expressions, and effects on osteoporosis.
10. Effect of Drugs or Biomaterials in the Expression of miRNAs
10.1. Bisphosphonates in the Treatment of OP and Changes in the Expression of miRNAs
Current drug treatments for postmenopausal OP (PMO) aim to prevent fractures by inhibiting bone resorption and stimulating bone formation. Oral bisphosphonates (BPs) are a first-line treatment for postmenopausal OP and are well-effective in the prevention of fragility fractures; these drugs decrease bone turnover by inhibiting osteoclast function [169].
Still, few studies have examined the effect of antiosteoporotic treatment on miRNA expression.
Previous studies [170] have investigated the differential expression of miRNAs in previously treatment-naïve women with PMO who were treated with either the potent antiresorptive agent denosumab (Dmab) or the osteoanabolic agent teriparatide (TPTD). The authors reported that while Dmab did not change the relative serum expression of the analyzed miRNAs, TPTD affected the expression of miRNAs that are related to the critical genes regulating osteoblastogenesis, that is, RUNX2 and DKK-1 [170], during the first year of treatment. In a subsequent study, the circulating miRNA expression profile of women with PMO who had received sequential antiosteoporotic treatments was analyzed. The authors observed that circulating miRNAs are differentially affected by treatment with TPTD and Dmab. The TPTD treatment potentially affects the expression of the pro-osteoclastogenic miR-21a-5p and the miRNAs (miR23a-3p, miR-29a-3p, and miR-2861), related to the key osteoblastic genes RUNX2, COL1, and HDAC5, while progressive treatment with Dmab acts in the opposite direction [171]. Recent research by Lia et al. has shed light on the practical implications of miRNA expression in postmenopausal osteoporosis treatment. They found that miR-30a-5p was significantly increased in patients undergoing long-term BP treatment for post-menopausal OP. This miRNA was found to be negatively correlated with bone formation, suggesting that it could serve as a novel mediator of long-term BP treatment that regulates bone formation. The mechanism of action was found to be a direct targeting of RUNX1 to inhibit osteoblastic differentiation in postmenopausal OP patients [172].
Furthermore, the potential of miRNA expression in monitoring treatment response was highlighted in a study on two years of denosumab (DMAB) treatment. The study revealed an upregulation of 7 miRNAs (miR-101-3p, miR-191-5p, miR-26b-5p, miR-32-5p, miR-4508, miR-454-3p, and miR-584-5p) after the treatment period. Four of these miRNAs were primarily expressed in monocytes, indicating a significant impact of DMAB on circulating osteoclast precursor cells. Notably, miR-454-3p, miR-26b-5p, and miR-584-5p emerged as the top biomarker candidates, showing the strongest association with the persistent effect of denosumab on bone in osteoporotic patients. These changes were also associated with BMD gain, further highlighting the potential of miRNA expression in monitoring DMAB treatment response [173]. Additional studies are necessary to confirm circulating miRNAs in serum as reliable indicators of the efficacy of sequential treatment in osteoporosis.
10.2. Effect of Biomaterials in the Expression of miRNAs
In recent years, with the increase in studies on miRNAs, the concepts related to biomaterials for repairing bone defects and restoring bone functions have expanded. Previous research focused on different materials and constructs that induce different miRNAs in the bone formation process, such as inorganic silicate-based PerioGlas, which act to alter osteoblast activity due to the exposure of different cell-binding domains and miRNA translations [132,174,175].
Lithium (Li) is a metal with critical therapeutic properties. This metal can be integrated into the structure of various biomaterials. Li-doped bioceramics (capable of immunomodulation), lithium-doped bioactive-glass nanoparticles, and gelatin/PRGF have been used extensively for bone and tooth regeneration. Combining bioactive glass as a bone-inducing mineral phase, gelatin as a biocompatible polymer, PRGF as a source of growth factor, and lithium-ion as a promoting bone repair agent can be considered as an approach to repair bone lesions, and may be helpful as functional bone tissue engineering materials using miRNA as an osteogenic factor. [176,177]. More recently, it has been reported that platelets release growth factors that can modify the expression profiles of miRNAs, and although platelets cannot transcribe a gene, megakaryocytes direct the translational and post-transcriptional regulation of mRNAs and ncRNAs, including miRNAs in bone metabolism cells, so that platelet activation can induce changes in the expression profiles of miRNAs and influence the function of osteoblasts and osteoclasts [178].
11. Conclusions
Bone remodeling is a balanced process between bone resorption and formation. However, this process is complicated and involves a large number of signaling pathways and modifications to the expression of regulatory molecules such as miRNAs. miRNAs are associated with numerous biological functions at different stages of bone remodeling due to their capacity to control gene expression, and they can regulate osteoclast and osteoblast differentiation, affecting bone formation and resorption. Therefore, alterations in the expression profiles of miRNAs can affect cell function, leading to bone diseases such as OP. The origin of OP is centered on the excessive activity of osteoclasts; therefore, understanding osteoclast differentiation and maturation has allowed to learn the molecular mechanisms leading to the development of OP and to propose new treatment strategies, based on these molecules. Novel knowledge will support the discovery of molecules with clinical utility as biomarkers for early detection and as therapeutic targets for bone disorders, including OP. The identification of precise and reliable biomarkers for early detection of OP is crucial for intervening in the initial stages of the disease, potentially enabling more effective therapeutic interventions and ultimately improving clinical outcomes and patients’ quality of life. Moreover, understanding how specific molecules, such as miRNAs, regulate the cellular processes involved in OP may lead to the identification of more effective therapeutic targets. miRNAs and other regulatory molecules could offer opportunities to develop highly specific therapeutic approaches that address the underlying causes of the disease rather than merely treating the symptoms. These approaches may include therapies aimed at restoring the balance between bone formation and resorption, which could have a significant impact on the prevention and treatment of OP.
As molecules that can regulate the differentiation of osteoblasts and osteoclasts through multiple pathways, miRNAs can be used as therapeutic agents, because of their ability to regulate the translation of several proteins that are involved in a particular gene’s expression by choosing a variety of methods like degradation or deadenylation of the mRNAs [176]. It may be possible to treat osteoporosis by adjusting their content and expression levels in patients. In this sense, the understanding of miRNAs is still in the initial stages, and studies focused on achieving safety, efficacy, and specific administration systems that allow using a miRNA as a modulator are necessary. Therefore, advancements in the understanding of molecules involved in the pathophysiology of OP will not only provide tools for earlier and more accurate detection of the disease but also open new avenues for the development of more effective and personalized therapies. However, further research is needed to validate these findings and translate them into practical clinical applications that benefit patients with bone disorders. In this context, studying the biology of HSCs and MSCs is necessary to understand the complex interactions between miRNAs and other types of noncoding RNA (ncRNA) as well as their target genes.
Author Contributions
Conceptualization, R.F.J.-O. and R.V.-C.; investigation, A.I.O.-M., N.P., B.R.-P. and R.F.J.-O.; data curation, A.I.O.-M., N.P. and B.R.-P.; writing—original draft preparation, R.F.J.-O.; writing—review and editing, R.F.J.-O., A.H.-B., and R.V.-C.; supervision, R.V.-C.; funding acquisition, A.H.-B., and R.V.-C. All authors have read and agreed to the published version of the manuscript.
Funding
R.F.J.-O. is supported by a Postdoctoral Fellowship from the Consejo Nacional de Ciencia y Tecnología (Grant Ciencia de Frontera CF 2019-102962). R.V.-C. was supported partly by INMEGEN (132-13/2013/I, 198–11/2015/I and 266–17/2016/I).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Porter, J.L.; Varacallo, M. Osteoporosis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar] [PubMed]
- Rashki Kemmak, A.; Rezapour, A.; Jahangiri, R.; Nikjoo, S.; Farabi, H.; Soleimanpour, S. Economic burden of Osteoporosis in the world: A systematic review. Med. J. Islam. Repub. Iran 2020, 34, 154. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Lin, C.; Stavre, Z.; Greenblatt, M.B.; Shim, J.H. Osteoblast-Osteoclast Communication and Bone Homeostasis. Cells 2020, 9, 2073. [Google Scholar] [CrossRef] [PubMed]
- Tu, K.N.; Lie, J.D.; Wan, C.K.V.; Cameron, M.; Austel, A.G.; Nguyen, J.K.; Van, K.; Hyun, D. Osteoporosis: A Review of Treatment Options. Pharm. Ther. 2018, 43, 92–104. [Google Scholar]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Macvanin, M.; Obradovic, M.; Zafirovic, S.; Stanimirovic, J.; Isenovic, E.R. The role of miRNAs in metabolic diseases. Curr. Med. Chem. 2022, 30, 1922–1944. [Google Scholar] [CrossRef] [PubMed]
- Materozzi, M.; Merlotti, D.; Gennari, L.; Bianciardi, S. The Potential Role of miRNAs as New Biomarkers for Osteoporosis. Int. J. Endocrinol. 2018, 2018, 2342860. [Google Scholar] [CrossRef] [PubMed]
- Krane, S.M. Identifying genes that regulate bone remodeling as potential therapeutic targets. J. Exp. Med. 2005, 201, 841–843. [Google Scholar] [CrossRef]
- Katsimbri, P. The biology of normal bone remodeling. Eur. J. Cancer Care 2017, 26, e12740. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef]
- Raut, N.; Wicks, S.M.; Lawal, T.O.; Mahady, G.B. Epigenetic regulation of bone remodeling by natural compounds. Pharmacol. Res. 2019, 147, 104350. [Google Scholar] [CrossRef]
- Oton-Gonzalez, L.; Mazziotta, C.; Iaquinta, M.R.; Mazzoni, E.; Nocini, R.; Trevisiol, L.; D’Agostino, A.; Tognon, M.; Rotondo, J.C.; Martini, F. Genetics and Epigenetics of Bone Remodeling and Metabolic Bone Diseases. Int. J. Mol. Sci. 2022, 23, 1500. [Google Scholar] [CrossRef]
- Stotz, K.; Griffiths, P. Epigenetics: Ambiguities and implications. Hist. Philos. Life Sci. 2016, 38, 22. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sultana, A.; Abdullah, K.M.; Pothuraju, R.; Nasser, M.W.; Batra, S.K.; Siddiqui, J.A. Epigenetic regulation of bone remodeling and bone metastasis. Semin. Cell Dev. Biol. 2024, 154 (Pt C), 275–285. [Google Scholar] [CrossRef]
- Suzuki, H.I. Roles of MicroRNAs in Disease Biology. JMA J. 2023, 6, 104–113. [Google Scholar] [CrossRef]
- Bravo Vázquez, L.A.; Moreno Becerril, M.Y.; Mora Hernández, E.O.; León Carmona, G.G.; Aguirre Padilla, M.E.; Chakraborty, S.; Bandyopadhyay, A.; Paul, S. The Emerging Role of MicroRNAs in Bone Diseases and Their Therapeutic Potential. Molecules 2021, 27, 211. [Google Scholar] [CrossRef]
- Alva-Partida, I.; Espinosa-Zavala, L.I.; Jiménez-Ortega, R.F. Biogenesis of miRNAs and their role as biomarkers in detection of diabetic nephropathy. ALAD 2022, 12, 15–25. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef] [PubMed]
- Sreedharam, S.; Puthamohan, V.M.; Valiya Parambil, S. MicroRNAs in cancer as biomarkers and therapeutic keys. ExRNA 2020, 2, 9. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 38. [Google Scholar] [CrossRef]
- Wang, L.; You, X.; Zhang, L.; Zhang, C.; Zou, W. Mechanical regulation of bone remodeling. Bone Res. 2022, 10, 16. [Google Scholar] [CrossRef]
- Ikeda, K.; Takeshita, S. The role of osteoclast differentiation and function in skeletal homeostasis. J. Biochem. 2016, 159, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Chen, X.; Yu, X. MicroRNAs in Osteoclastogenesis and Function: Potential Therapeutic Targets for Osteoporosis. Int. J. Mol. Sci. 2016, 17, 349. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast-osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, F.; Murakami, Y.; Saito, T.; Miyasaka, N.; Kohsaka, H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res. Ther. 2013, 15, R102. [Google Scholar] [CrossRef] [PubMed]
- Minamizaki, T.; Nakao, Y.; Irie, Y.; Ahmed, F.; Itoh, S.; Sarmin, N.; Yoshioka, H.; Nobukiyo, A.; Fujimoto, C.; Niida, S.; et al. The matrix vesicle cargo miR-125b accumulates in the bone matrix, inhibiting bone resorption in mice. Commun. Biol. 2020, 3, 30. [Google Scholar] [CrossRef]
- Guo, L.J.; Liao, L.; Yang, L.; Li, Y.; Jiang, T.J. MiR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp. Cell Res. 2014, 321, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Kelch, S.; Balmayor, E.R.; Seeliger, C.; Vester, H.; Kirschke, J.S.; van Griensven, M. miRNAs in bone tissue correlate to bone mineral density, and circulating miRNAs are gender independent in osteoporotic patients. Sci. Rep. 2017, 7, 15861. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Pitari, M.R.; Amodio, N.; Di Martino, M.T.; Conforti, F.; Leone, E.; Botta, C.; Paolino, F.M.; Del Giudice, T.; Iuliano, E.; et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: Implications for treating multiple mye-loma-related bone disease. J. Cell. Physiol. 2013, 228, 1506–1515. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Moore, B.T.; Peng, X.H.; Fang, X.; Lappe, J.M.; Recker, R.R.; Xiao, P. MiR-133a in human circulating monocytes: A potential biomarker associated with postmenopausal Osteoporosis. PLoS ONE 2012, 7, e34641. [Google Scholar] [CrossRef]
- Cheng, P.; Chen, C.; He, H.B.; Hu, R.; Zhou, H.D.; Xie, H.; Zhu, W.; Dai, R.-C.; Wu, X.-P.; Liao, E.-Y.; et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J. Bone Miner. Res. 2013, 28, 1180–1190. [Google Scholar] [CrossRef]
- He, Y.; Chen, D.; Guo, Q.; Shi, P.; You, C.; Feng, Y. MicroRNA-151a-3p Functions in the Regulation of Osteoclast Differentiation: Significance to Postmenopausal Osteoporosis. Clin. Interv. Aging 2021, 16, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Deng, Z.; Chen, Y.; Giannopoulou, E.; Xu, R.; Gong, S.; Greenblatt, M.B.; Mangala, L.S.; Lopez-Berestein, G.; Kirsch, D.G.; et al. Bone protection by inhibition of microRNA-182. Nat. Commun. 2018, 9, 4108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, W.; Zhang, P.; Ling, S.; Li, Y.; Zhao, D.; Peng, J.; Wang, A.; Li, Q.; Song, J.; et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol. 2015, 12, 343–353. [Google Scholar] [CrossRef] [PubMed]
- De-Ugarte, L.; Balcells, S.; Nogues, X.; Grinberg, D.; Diez-Perez, A.; Garcia-Giralt, N. Pro-osteoporotic miR-320a impairs osteoblast function and induces oxidative stress. PLoS ONE 2018, 13, e0208131. [Google Scholar] [CrossRef] [PubMed]
- Ke, K.; Sul, O.J.; Rajasekaran, M.; Choi, H.S. MicroRNA-183 increases osteoclastogenesis by repressing heme oxygenase-1. Bone 2015, 81, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cheng, P.; Xie, H.; Zhou, H.D.; Wu, X.P.; Liao, E.-Y.; Luo, X.-H. MiR-503 regulates osteoclastogenesis via targeting RANK. J. Bone Miner. Res. 2014, 29, 338–347. [Google Scholar] [CrossRef]
- Ramírez-Salazar, E.G.; Almeraya, E.V.; López-Perez, T.V.; Patiño, N.; Salmeron, J.; Velázquez-Cruz, R. MicroRNA-548-3p overexpression inhibits proliferation, migration and invasion in osteoblast-like cells by targeting STAT1 and MAFB. J. Biochem. 2020, 168, 203–211. [Google Scholar] [CrossRef]
- Hodge, J.M.; Collier, F.M.; Pavlos, N.J.; Kirkland, M.A.; Nicholson, G.C. M-CSF potently augments RANKL-induced resorption activation in mature human osteoclasts. PLoS ONE 2011, 6, e21462. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.H.; Park, P.S.U.; Park-Min, K.H. The M-CSF receptor in osteoclasts and beyond. Exp. Mol. Med. 2020, 52, 1239–1254. [Google Scholar] [CrossRef]
- Oppezzo, A.; Rosselli, F. The underestimated role of the microphthalmia-associated transcription factor (MiTF) in normal and pathological haematopoiesis. Cell Biosci. 2021, 11, 18. [Google Scholar] [CrossRef]
- Park, B.; Yu, S.N.; Kim, S.H.; Lee, J.; Choi, S.J.; Chang, J.H.; Yang, E.J.; Kim, K.-Y.; Ahn, S.-C. Inhibitory Effect of Biotransformed-Fucoidan on the Differentiation of Osteoclasts Induced by Receptor for Activation of Nuclear Factor-κB Ligand. J. Microbiol. Biotechnol. 2022, 32, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.Y.; Li, M.; Lin, Y.L. Mitf regulates osteoclastogenesis by modulating NFATc1 activity. Exp. Cell Res. 2014, 328, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Ritchlin, C.T. DC-STAMP: A Key Regulator in Osteoclast Differentiation. J. Cell. Physiol. 2016, 231, 2402–2407. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X.; Gu, J.H.; Zhang, Y.R.; Tong, X.S.; Zhao, H.Y.; Yuan, Y.; Liu, X.-Z.; Bian, J.-C.; Liu, Z.-P. Osteoprotegerin influences the bone resorption activity of osteoclasts. Int. J. Mol. Med. 2013, 31, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Chen, M.; Song, R.; Zhao, H.; Bian, J.; Gu, J.; Liu, Z. Overexpression of c-Fos reverses osteoprotegerin-mediated suppression of osteoclastogenesis by increasing the Beclin1-induced autophagy. J. Cell. Mol. Med. 2021, 25, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, N. Regulation of NFATc1 in Osteoclast Differentiation. J. Bone Metab. 2014, 21, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Inui, M.; Inoue, K.; Kim, S.; Suematsu, A.; Kobayashi, E.; Iwata, T.; Ohnishi, H.; Matozaki, T.; Kodama, T.; et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature 2004, 428, 758–763. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, N.K.; Lee, S.Y. Current Understanding of RANK Signaling in Osteoclast Differentiation and Maturation. Mol. Cells 2017, 40, 706–713. [Google Scholar] [CrossRef]
- Galson, D.L.; Roodman, G.D. Origin of osteoclasts. In Osteoimmunology. Interactions of the Immune and Skeletal Systems; Lorenzo, J., Choi, Y., Horowitz, M., Takayanagi, H., Eds.; Academic Press: Cambridge, MA, USA, 2011; pp. 7–41. [Google Scholar] [CrossRef]
- Croset, M.; Pantano, F.; Kan, C.W.S.; Bonnelye, E.; Descotes, F.; Alix-Panabières, C.; Lecellier, C.-H.; Bachelier, R.; Allioli, N.; Hong, S.-S.; et al. miRNA-30 Family Members Inhibit Breast Cancer Invasion, Osteomimicry, and Bone Destruction by Directly Targeting Multiple Bone Metastasis-Associated Genes. Cancer Res. 2018, 78, 5259–5273. [Google Scholar] [CrossRef]
- Fujita, K.; Janz, S. Attenuation of WNT signaling by DKK-1 and -2 regulates BMP2-induced osteoblast differentiation and expression of OPG, RANKL and M-CSF. Mol. Cancer 2007, 6, 71. [Google Scholar] [CrossRef]
- Chen, H.; Hu, Y.; Xu, X.; Dai, Y.; Qian, H.; Yang, X.; Liu, J.; He, Q.; Zhang, H. DKK1 activates the PI3K/AKT pathway via CKAP4 to balance the inhibitory effect on Wnt/β-catenin signaling and regulates Wnt3a-induced MSC migration. Stem Cells 2024, 42, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Clézardin, P.; Coleman, R.; Puppo, M.; Ottewell, P.; Bonnelye, E.; Paycha, F.; Confavreux, C.B.; Holen, I. Bone metastasis: Mechanisms, therapies, and biomarkers. Physiol. Rev. 2021, 101, 797–855. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Liu, H.; Peng, F.; Liu, Z.; Ding, K.; Song, J.; Li, L.; Chen, J.; Shao, Q.; Yan, S.; et al. Complement C3a activates osteoclasts by regulating the PI3K/PDK1/SGK3 pathway in patients with multiple myeloma. Cancer Biol. Med. 2021, 18, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhang, Z.; Liu, N.; Li, L.; Zhong, H.; Wang, R.; Shi, Q.; Zhang, Z.; Wei, L.; Hu, B.; et al. Small extracellular vesicle-mediated miR-320e transmission promotes osteogenesis in OPLL by targeting TAK1. Nat. Commun. 2022, 13, 2467. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, C.; Nie, L.; Gu, M.; Wu, A.; Han, X.; Wang, X.; Shao, J.; Xia, Z. Transforming growth factor (TGF)-β-activated kinase 1 (TAK1) activation requires phosphorylation of serine 412 by protein kinase A catalytic subunit α (PKACα) and X-linked protein kinase (PRKX). J. Biol. Chem. 2014, 289, 24226–24237. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ortega, R.F.; Ramírez-Salazar, E.G.; Parra-Torres, A.Y.; Muñoz-Montero, S.A.; Rangel-Escareňo, C.; Salido-Guadarrama, I.; Rodriguez-Dorantes, M.; Quiterio, M.; Salmerón, J.; Velázquez-Cruz, R. Identification of microRNAs in human circulating monocytes of postmenopausal osteoporotic Mexican-Mestizo women: A pilot study. Exp. Ther. Med. 2017, 14, 5464–5472. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Salazar, E.G.; Almeraya, E.V.; López-Perez, T.V.; Jiménez-Salas, Z.; Patiño, N.; Velázquez-Cruz, R. MicroRNA-1270 Inhibits Cell Proliferation, Migration, and Invasion via Targeting IRF8 in Osteoblast-like Cell Lines. Curr. Issues Mol. Biol. 2022, 44, 1182–1190. [Google Scholar] [CrossRef] [PubMed]
- Wehrhan, F.; Gross, C.; Creutzburg, K.; Amann, K.; Ries, J.; Kesting, M.; Geppert, C.I.; Weber, M. Osteoclastic expression of higher-level regulators NFATc1 and BCL6 in medication-related osteonecrosis of the jaw secondary to bisphosphonate therapy: A comparison with osteoradionecrosis and osteomyelitis. J. Transl. Med. 2019, 17, 69. [Google Scholar] [CrossRef]
- Koga, T.; Matsui, Y.; Asagiri, M.; Kodama, T.; de Crombrugghe, B.; Nakashima, K.; Takayanagi, H. NFAT, and Osterix cooperatively regulate bone formation. Nat. Med. 2005, 11, 880–885. [Google Scholar] [CrossRef]
- Hojo, H.; Ohba, S.; Chung, U.I. Signaling pathways regulating the specification and differentiation of the osteoblast lineage. Regen. Ther. 2015, 1, 57–62. [Google Scholar] [CrossRef]
- Maupin, K.A.; Droscha, C.J.; Williams, B.O. A Comprehensive Overview of Skeletal Phenotypes Associated with Alterations in Wnt/β-catenin Signaling in Humans and Mice. Bone Res. 2013, 1, 27–71. [Google Scholar] [CrossRef] [PubMed]
- De, A. Wnt/Ca2+ signaling pathway: A brief overview. Acta Biochim. Biophys. Sin. 2011, 43, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.P.; Paulson, C.; Shao, J.Z.; Zhang, X.; Wu, M.; Chen, W. Wnt and the Wnt signaling pathway in bone development and disease. Front. Biosci. 2014, 19, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Rauch, F.; Glorieux, F.H. Osteogenesis imperfecta. Lancet 2004, 363, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Niemann, S.; Zhao, C.; Pascu, F.; Stahl, U.; Aulepp, U.; Niswander, L.; Weber, J.L.; Müller, U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am. J. Hum. Genet. 2004, 74, 558–563. [Google Scholar] [CrossRef]
- Yang, Y.; Topol, L.; Lee, H.; Wu, J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development 2003, 130, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Haÿ, E.; Buczkowski, T.; Marty, C.; Da Nascimento, S.; Sonnet, P.; Marie, P.J. Peptide-based mediated disruption of N-cadherin-LRP5/6 interaction promotes Wnt signaling and bone formation. J. Bone Miner. Res. 2012, 27, 1852–1863. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.N.; Ouyang, H.; Ma, Y.L.; Zeng, Q.; Gerin, I.; Sousa, K.M.; Lane, T.F.; Krishnan, V.; Hankenson, K.D.; A MacDougald, O. Wnt10b increases postnatal bone formation by enhancing osteoblast differentiation. J. Bone Miner. Res. 2007, 22, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.S.; Oyserman, S.M.; Hankenson, K.D. Wnt11 promotes osteoblast maturation and mineralization through R-spondin 2. J. Biol. Chem. 2009, 284, 14117–14125. [Google Scholar] [CrossRef]
- Tian, Y.; Xu, Y.; Fu, Q.; He, M. Parathyroid hormone regulates osteoblast differentiation in a Wnt/β-catenin-dependent manner. Mol. Cell Biochem. 2011, 355, 211–216. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Plotkin, L.I.; Galli, C.; Goellner, J.J.; Gortazar, A.R.; Allen, M.R.; Robling, A.G.; Bouxsein, M.; Schipani, E.; Turner, C.H.; et al. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS ONE 2008, 3, e2942. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.H.; Halladay, D.L.; Miles, R.R.; Gilbert, L.M.; Frolik, C.A.; Galvin, R.J.; Martin, T.; Gillespie, M.; Onyia, J. Effects of parathyroid hormone on Wnt signaling pathway in bone. J. Cell. Biochem. 2005, 95, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, M.; Yang, D.; Bouxsein, M.L.; Saito, H.; Galvin, R.S.; Kuhstoss, S.A.; Thomas, C.C.; Schipani, E.; Baron, R.; et al. Suppression of Wnt signaling by Dkk1 attenuates PTH-mediated stromal cell response and new bone formation. Cell Metab. 2010, 11, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Sneddon, W.B.; Yang, Y.; Wheeler, D.; Blair, H.C. Parathyroid hormone receptors directly interact with disheveled to regulate beta-catenin signaling and osteoclastogenesis. J. Biol. Chem. 2010, 285, 14756–14763. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.; Long, F. Notch signaling and bone remodeling. Curr. Osteoporos. Rep. 2013, 11, 126–129. [Google Scholar] [CrossRef]
- Deregowski, V.; Gazzerro, E.; Priest, L.; Rydziel, S.; Canalis, E. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J. Biol. Chem. 2006, 281, 6203–6210. [Google Scholar] [CrossRef] [PubMed]
- Sciaudone, M.; Gazzerro, E.; Priest, L.; Delany, A.M.; Canalis, E. Notch 1 impairs osteoblastic cell differentiation. Endocrinology 2003, 144, 5631–5639. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, K.; Yasuda, M.; Watanabe, N.; Morimura, N.; Kuroda, K.; Miyatani, S.; Hozumi, N. Stimulation of osteoblastic cell differentiation by Notch. J. Bone Miner. Res. 2002, 17, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Nobta, M.; Tsukazaki, T.; Shibata, Y.; Xin, C.; Moriishi, T.; Sakano, S.; Shindo, H.; Yamaguchi, A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J. Biol. Chem. 2005, 280, 15842–15848. [Google Scholar] [CrossRef]
- Hilton, M.J.; Tu, X.; Wu, X.; Bai, S.; Zhao, H.; Kobayashi, T.; Kronenberg, H.M.; Teitelbaum, S.L.; Ross, F.P.; Kopan, R.; et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008, 14, 306–314. [Google Scholar] [CrossRef]
- Tu, X.; Chen, J.; Lim, J.; Karner, C.M.; Lee, S.Y.; Heisig, J.; Wiese, C.; Surendran, K.; Kopan, R.; Gessler, M.; et al. Physiological notch signaling maintains bone homeostasis via RBPjk and Hey upstream of NFATc1. PLoS Genet. 2012, 8, e1002577. [Google Scholar] [CrossRef]
- Salie, R.; Kneissel, M.; Vukevic, M.; Zamurovic, N.; Kramer, I.; Evans, G.; Gerwin, N.; Mueller, M.; Kinzel, B.; Susa, M. Ubiquitous overexpression of the Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone 2010, 46, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Chen, S.; Yang, T.; Dawson, B.; Munivez, E.; Bertin, T.; Lee, B. Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J. Bone Miner. Res. 2010, 25, 2175–2183. [Google Scholar] [CrossRef]
- Zanotti, S.; Smerdel-Ramoya, A.; Stadmeyer, L.; Durant, D.; Radtke, F.; Canalis, E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 2008, 149, 3890–3899. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chen, G.; Li, Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019, 12, eaav5183. [Google Scholar] [CrossRef]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-β Family Signaling in Mesenchymal Differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10, a022202. [Google Scholar] [CrossRef]
- Crane, J.L.; Cao, X. Bone marrow mesenchymal stem cells and TGF-β signaling in bone remodeling. J. Clin. Investig. 2014, 124, 466–472. [Google Scholar] [CrossRef]
- Ramírez-Salazar, E.G.; Carrillo-Patiño, S.; Hidalgo-Bravo, A.; Rivera-Paredez, B.; Quiterio, M.; Ramírez-Palacios, P.; Patiño, N.; Valdés-Flores, M.; Salmerón, J.; Velázquez-Cruz, R. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene 2018, 679, 19–27. [Google Scholar] [CrossRef]
- Feichtinger, X.; Muschitz, C.; Heimel, P.; Baierl, A.; Fahrleitner-Pammer, A.; Redl, H.; Resch, H.; Geiger, E.; Skalicky, S.; Dormann, R.; et al. Bone-related Circulating MicroRNAs miR-29b-3p, miR-550a-3p, and miR-324-3p and their Association to Bone Microstructure and Histomorphometry. Sci. Rep. 2018, 8, 4867. [Google Scholar] [CrossRef]
- Weivoda, M.M.; Lee, S.K.; Monroe, D.G. miRNAs in osteoclast biology. Bone 2021, 143, 115757. [Google Scholar] [CrossRef]
- Xu, W.; Xia, R.; Tian, F.; Liu, L.; Li, M.; Fang, S. microRNA-324-3p Promotes Osteoblasts Differentiation via Suppressing SMAD7. Hard Tissue Biol. 2022, 31, 263–268. [Google Scholar] [CrossRef]
- Zarecki, P.; Hackl, M.; Grillari, J.; Debono, M.; Eastell, R. Serum microRNAs as novel biomarkers for osteoporotic vertebral fractures. Bone 2020, 130, 115105. [Google Scholar] [CrossRef]
- Zhou, H.; Jiang, J.; Chen, X.; Zhang, Z. Differentially expressed genes and miRNAs in female osteoporosis patients. Medicine 2022, 101, e29856. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, Z.; Feng, S. Systematic analysis of miRNAs in patients with postmenopausal Osteoporosis. Gynecol. Endocrinol. 2020, 36, 997–1001. [Google Scholar] [CrossRef]
- Li, J.; He, X.; Wei, W.; Zhou, X. MicroRNA-194 promotes osteoblast differentiation via downregulating STAT1. Biochem. Biophys. Res. Commun. 2015, 460, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, C.; Lu, J.; Zou, L.; Hu, M.; Yang, Z.; Xu, Y. MicroRNA-590-5p antagonizes the inhibitory effect of high glucose on osteoblast differentiation by suppressing Smad7 in MC3T3-E1 cells. J. Int. Med. Res. 2019, 47, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xie, X.; Xue, H.; Wang, T.; Panayi, A.C.; Lin, Z.; Xiong, Y.; Cao, F.; Yan, C.; Chen, L.; et al. MiR-1224-5p modulates osteogenesis by coordinating osteoblast/osteoclast differentiation via the Rap1 signaling target ADCY2. Exp. Mol. Med. 2022, 54, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, H.; Wang, S.; Li, T.; Fan, J.; Liang, X.; Li, J.; Han, Q.; Zhu, L.; Fan, L.; et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014, 23, 1452–1463. [Google Scholar] [CrossRef]
- Fan, J.B.; Liu, W.; Zhu, X.H.; Cui, S.Y.; Cui, Z.M.; Zhao, J.N. microRNA-7 inhibition protects human osteoblasts from dexamethasone via activation of epidermal growth factor receptor signaling. Mol. Cell. Biochem. 2019, 460, 113–121. [Google Scholar] [CrossRef]
- Vimalraj, S.; Partridge, N.C.; Selvamurugan, N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J. Cell. Physiol. 2014, 229, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, Y.; Zheng, Y.; Chen, B. The miRNA-15b/USP7/KDM6B axis engages in the initiation of Osteoporosis by modulating osteoblast differentiation and autophagy. J. Cell. Mol. Med. 2021, 25, 2069–2081. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Zhu, S.; Wang, L.; Yang, L.; He, C. MiR-30 Family: A Novel Avenue for Treating Bone and Joint Diseases? Int. J. Med. Sci. 2023, 20, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wu, S.; Zhou, H.; Bi, X.; Wang, Y.; Hu, Y.; Gu, P.; Fan, X. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 2013, 22, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Q.S.; Ding, W.B.; Zhang, L.L.; Wang, H.C.; Zhu, Y.J.; He, W.; Chai, Y.N.; Liu, Y.W. Increased microRNA-93-5p inhibits osteogenic differentiation by targeting bone morphogenetic protein-2. PLoS ONE 2017, 12, e0182678. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.P.; Zhang, J.; Zhu, C.H.; Lin, L.; Wang, J.; Zhang, H.J.; Li, J.; Yu, X.G.; Zhao, Z.S.; Dong, W.; et al. MicroRNA-98 regulates osteogenic differentiation of human bone mesenchymal stromal cells by targeting BMP2. J. Cell. Mol. Med. 2017, 21, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Dole, N.S.; Yoon, J.; Monteiro, D.A.; Yang, J.; Mazur, C.M.; Kaya, S.; Belair, C.D.; Alliston, T. Mechanosensitive miR-100 coordinates TGFβ and Wnt signaling in osteocytes during fluid shear stress. FASEB J. 2021, 35, e21883. [Google Scholar] [CrossRef]
- Zuo, B.; Zhu, J.; Li, J.; Wang, C.; Zhao, X.; Cai, G.; Li, Z.; Peng, J.; Wang, P.; Shen, C.; et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Miner. Res. 2015, 30, 330–345. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Liu, Q.; Li, C.; He, P. miR-125a-5p Regulates Osteogenic Differentiation of Human Adipose-Derived Mesenchymal Stem Cells under Oxidative Stress. BioMed Res. Int. 2021, 2021, 6684709. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, L.; Yan, C.; Wang, F.; Zhang, Y. Overexpression of miR125b Promotes Osteoporosis Through miR-125b-TRAF6 Pathway in Postmenopausal Ovariectomized Rats. Diabetes Metab. Syndr. Obes. 2021, 14, 671–682. [Google Scholar] [CrossRef]
- Eskildsen, T.; Taipaleenmäki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, K.; Hu, Z.; Zhou, H.; Zhang, L.; Wang, H.; Li, G.; Zhang, S.; Cao, X.; Shi, F. MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell Death Dis. 2018, 9, 1107. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Park, S.K.; Lee, H.Y.; Kim, S.W.; Lee, J.S.; Choi, E.K.; You, D.; Kim, C.S.; Suh, N. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014, 588, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Huszar, J.M.; Payne, C.J. MIR146A inhibits JMJD3 expression and osteogenic differentiation in human mesenchymal stem cells. FEBS Lett. 2014, 588, 1850–1856. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, H.; Ding, D.; Wang, S.; Dai, X.; Zhu, Y. The interaction of miR-181a-5p and sirtuin 1 regulated human bone marrow mesenchymal stem cells differentiation and apoptosis. Bioengineered 2021, 12, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Dou, P.; Cai, W.; Mao, M.; Wu, R. MiR-181a-5p promotes osteogenesis by targeting BMP3. Aging 2023, 15, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Mi, B.; Yan, C.; Xue, H.; Chen, L.; Panayi, A.C.; Hu, L.; Hu, Y.; Cao, F.; Sun, Y.; Zhou, W.; et al. Inhibition of Circulating miR-194-5p Reverses Osteoporosis through Wnt5a/β-Catenin-Dependent Induction of Osteogenic Differentiation. Mol. Ther. Nucleic Acids 2020, 21, 814–823. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Zhan, J.; Yan, Z.; Chen, D.; Xue, X.; Pan, X. Bone marrow mesenchymal stem cell-derived exosomal miR-206 promotes osteoblast proliferation and differentiation in osteoarthritis by reducing Elf3. J. Cell. Mol. Med. 2021, 25, 7734–7745. [Google Scholar] [CrossRef]
- Asgharzadeh, A.; Alizadeh, S.; Keramati, M.R.; Soleimani, M.; Atashi, A.; Edalati, M.; Khatib, Z.K.; Rafiee, M.; Barzegar, M.; Razavi, H. Upregulation of miR-210 promotes differentiation of mesenchymal stem cells (MSCs) into osteoblasts. Bosn. J. Basic Med. Sci. 2018, 18, 328–335. [Google Scholar] [CrossRef]
- Gao, Y.; Patil, S.; Qian, A. The Role of MicroRNAs in Bone Metabolism and Disease. Int. J. Mol. Sci. 2020, 21, 6081. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hassan, M.Q.; Gordon, J.A.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.; Zheng, Z.; Zeng, X.; Liu, D.; Wang, C.; Ting, K. MicroRNA-223 Suppresses Osteoblast Differentiation by Inhibiting DHRS3. Cell. Physiol. Biochem. 2018, 47, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Nie, H.; Liu, P.; Wang, Z.; Yao, B.; Yang, L. Down-regulation of miR-339 promotes differentiation of BMSCs and alleviates Osteoporosis by targeting DLX5. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wei, X.; Sun, Q.; Zhao, X.Q.; Zheng, C.Y.; Bai, C.X.; Du, J.; Zhang, Z.; Zhu, L.G.; Jia, Y.S. MicroRNA-449b-5p promotes the progression of Osteoporosis by inhibiting osteogenic differentiation of BMSCs via targeting Satb2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6394–6403. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; You, Y.; Zhang, L.; Hou, D.; Gu, G.; Chen, Y.; Jiao, G. M2 macrophage-derived exosomal miR-486-5p influences the differentiation potential of bone marrow mesenchymal stem cells and Osteoporosis. Aging 2023, 15, 9499–9520. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wu, P.; Fu, W.; Xiong, Y.; Zhang, L.; Gao, Y.; Deng, G.; Zong, S.; Zeng, G. The Role and Mechanism of miRNA-1224 in the Polygonatum sibiricum Polysaccharide Regulation of Bone Marrow-Derived Macrophages to Osteoclast Differentiation. Rejuvenation Res. 2019, 22, 420–430. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Q.; Maeda, Y.; Taipaleenmaki, H.; Zhang, W.; Jafferji, M.; Gordon, J.A.; Li, Z.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J. Biol. Chem. 2012, 287, 42084–42092. [Google Scholar] [CrossRef]
- Fukuda, T.; Ochi, H.; Sunamura, S.; Haiden, A.; Bando, W.; Inose, H.; Okawa, A.; Asou, Y.; Takeda, S. MicroRNA-145 regulates osteoblastic differentiation by targeting the transcription factor Cbfb. FEBS Lett. 2015, 589, 3302–3308. [Google Scholar] [CrossRef]
- Taipaleenmäki, H.; Browne, G.; Akech, J.; Zustin, J.; van Wijnen, A.J.; Stein, J.L.; Hesse, E.; Stein, G.S.; Lian, J.B. Targeting of Runx2 by miR-135 and miR-203 Impairs Progression of Breast Cancer and Metastatic Bone Disease. Cancer Res. 2015, 75, 1433–1444. [Google Scholar] [CrossRef]
- Sharma, A.R.; Lee, Y.H.; Lee, S.S. Recent advancements of miRNAs in the treatment of bone diseases and their delivery potential. Curr. Res. Pharmacol. Drug Discov. 2022, 4, 100150. [Google Scholar] [CrossRef]
- Daamouch, S.; Emini, L.; Rauner, M.; Hofbauer, L.C. MicroRNA and Diabetic Bone Disease. Curr. Osteoporos. Rep. 2022, 20, 194–201. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Huang, H.T.; Cheng, T.L.; Lu, Y.M.; Lin, S.Y.; Ho, C.J.; Lee, T.C.; Hsu, C.H.; Huang, P.J.; Huang, H.H. Application of microRNA in Human Osteoporosis and Fragility Fracture: A Systemic Review of Literatures. Int. J. Mol. Sci. 2021, 22, 5232. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, R.L.; Croce, C.M.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. USA 2011, 108, 9863–9868. [Google Scholar] [CrossRef]
- Chen, H.; Ji, X.; She, F.; Gao, Y.; Tang, P. miR-628-3p regulates osteoblast differentiation by targeting RUNX2: Possible role in atrophic nonunion. Int. J. Mol. Med. 2017, 39, 279–286. [Google Scholar] [CrossRef]
- Mizoguchi, F.; Izu, Y.; Hayata, T.; Hemmi, H.; Nakashima, K.; Nakamura, T.; Kato, S.; Miyasaka, N.; Ezura, Y.; Noda, M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 2010, 109, 866–875. [Google Scholar] [CrossRef]
- Kerschan-Schindl, K.; Hackl, M.; Boschitsch, E.; Föger-Samwald, U.; Nägele, O.; Skalicky, S.; Weigl, M.; Grillari, J.; Pietschmann, P. Diagnostic Performance of a Panel of miRNAs (OsteomiR) for Osteoporosis in a Cohort of Postmenopausal Women. Calcif. Tissue Int. 2021, 108, 725–737. [Google Scholar] [CrossRef]
- Palumbo, C.; Ferretti, M. The Osteocyte: From “Prisoner” to “Orchestrator”. J. Funct. Morphol. Kinesiol. 2021, 6, 28. [Google Scholar] [CrossRef]
- Schaffler, M.B.; Cheung, W.Y.; Majeska, R.; Kennedy, O. Osteocytes: Master orchestrators of bone. Calcif. Tissue Int. 2014, 94, 5–24. [Google Scholar] [CrossRef]
- Peng, S.; Gao, D.; Gao, C.; Wei, P.; Niu, M.; Shuai, C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review). Mol. Med. Rep. 2016, 14, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, H.; Leung, R.K.K.; Choy, K.W.; Lam, T.P.; Ng, B.K.W.; Qiu, Y.; Feng, J.Q.; Cheng, J.C.Y.; Lee, W.Y.W. Aberrant miR-145-5p/β-catenin signal impairs osteocyte function in adolescent idiopathic scoliosis. FASEB J. 2018, 32, 6537–6549. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Tang, C.Y.; Man, X.F.; Tang, H.N.; Tang, J.; Wang, F.; Zhou, C.L.; Tan, S.W.; Feng, Y.Z.; Zhou, H.D. Insulin receptor substrate-1 time-dependently regulates bone formation by controlling collagen Iα2 expression via miR-342. FASEB J. 2016, 30, 4214–4226. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, T.; Watanabe, K.; Hara, E.S.; Ono, M.; Kuboki, T.; Calderwood, S.K. OstemiR: A novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS ONE 2013, 8, e58796. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.M.; Deosthale, P.J.; Pacheco-Costa, R.; Essex, A.L.; Atkinson, E.G.; Aref, M.W.; Dilley, J.E.; Bellido, T.; Ivan, M.; Allen, M.R.; et al. Osteocytic miR21 deficiency improves bone strength independent of sex despite having sex divergent effects on osteocyte viability and bone turnover. FEBS J. 2020, 287, 941–963. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.C.; Bae, Y.; Dawson, B.C.; Chen, Y.; Bertin, T.; Munivez, E.; Campeau, P.M.; Tao, J.; Chen, R.; Lee, B.H. MicroRNA miR-23a cluster promotes osteocyte differentiation by regulating TGF-β signalling in osteoblasts. Nat. Commun. 2017, 8, 15000. [Google Scholar] [CrossRef]
- Qin, Y.; Peng, Y.; Zhao, W.; Pan, J.; Ksiezak-Reding, H.; Cardozo, C.; Wu, Y.; Divieti Pajevic, P.; Bonewald, L.F.; Bauman, W.A.; et al. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. J. Biol. Chem. 2017, 292, 11021–11033. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Hao, L.; Tian, Y.; Liu, Y.; Gu, Y.; Wu, J. miR-199a-3p is involved in estrogen-mediated autophagy through the IGF-1/mTOR pathway in osteocyte-like MLO-Y4 cells. J. Cell. Physiol. 2018, 233, 2292–2303. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nakashima, T.; Yoshimura, N.; Okamoto, K.; Tanaka, S.; Takayanagi, H. Autoregulation of Osteocyte Sema3A Orchestrates Estrogen Action and Counteracts Bone Aging. Cell Metab. 2019, 29, 627–637.e5. [Google Scholar] [CrossRef]
- Lv, P.Y.; Gao, P.F.; Tian, G.J.; Yang, Y.Y.; Mo, F.F.; Wang, Z.H.; Sun, L.; Kuang, M.J.; Wang, Y.L. Osteocyte-derived exosomes induced by mechanical strain promote human periodontal ligament stem cell proliferation and osteogenic differentiation via the miR-181b-5p/PTEN/AKT signaling pathway. Stem Cell Res. Ther. 2020, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.I.; Wallace, J.M. MicroRNAs and osteocytes. Bone 2021, 150, 115994. [Google Scholar] [CrossRef]
- Trojniak, J.; Sendera, A.; Banas-Zabczyk Kopanska, M. The MicroRNAs in the Pathophysiology of Osteoporosis. Int. J. Mol. Sci. 2024, 25, 6240. [Google Scholar] [CrossRef]
- Donati, S.; Ciuffi, S.; Palmini, G.; Brandi, M.L. Circulating miRNAs: A New Opportunity in Bone Fragility. Biomolecules 2020, 10, 927. [Google Scholar] [CrossRef]
- Cheng, F.; Yang, M.M.; Yang, R.H. MiRNA-365a-3p promotes the progression of osteoporosis by inhibiting osteogenic differentiation via targeting RUNX2. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7766–7774. [Google Scholar] [CrossRef]
- Luo, B.; Yang, J.F.; Wang, Y.H.; Qu, G.B.; Hao, P.D.; Zeng, Z.J.; Yuan, J.; Yang, R.; Yuan, Y. MicroRNA-579-3p promotes the progression of osteoporosis by inhibiting osteogenic differentiation of mesenchymal stem cells through regulating Sirt1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6791–6799. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Q.; Wu, X.P.; He, H.B.; Fu, L. MiR-96 regulates bone metabolism by targeting osterix. Clin. Exp. Pharmacol. Physiol. 2018, 45, 602–613. [Google Scholar] [CrossRef]
- Zhao, S.L.; Wen, Z.X.; Mo, X.Y.; Zhang, X.Y.; Li, H.N.; Cheung, W.H.; Fu, D.; Zhang, S.H.; Wan, Y.; Chen, B.L. Bone-Metabolism-Related Serum microRNAs to Diagnose Osteoporosis in Middle-Aged and Elderly Women. Diagnostics 2022, 12, 2872. [Google Scholar] [CrossRef]
- Guo, Y.; Li, L.; Gao, J.; Chen, X.; Sang, Q. miR-214 suppresses the osteogenic differentiation of bone marrow-derived mesenchymal stem cells and these effects are mediated through the inhibition of the JNK and p38 pathways. Int. J. Mol. Med. 2017, 39, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Abak, A.; Tavakkoli Avval, S.; Rahmani, S.; Shoorei, H.; Taheri, M.; Samadian, M. Contribution of miRNAs and lncRNAs in osteogenesis and related disorders. Biomed. Pharmacother. 2021, 142, 111942. [Google Scholar] [CrossRef]
- Bedene, A.; Mencej Bedrač, S.; Ješe, L.; Marc, J.; Vrtačnik, P.; Preželj, J.; Kocjan, T.; Kranjc, T.; Ostanek, B. MiR-148a the epigenetic regulator of bone homeostasis is increased in plasma of osteoporotic postmenopausal women. Wien. Klin. Wochenschr. 2016, 128 (Suppl. S7), 519–526. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Pan, L.; Chen, L.; Gu, W.; Chen, J. MiR-27a is Essential for the Shift from Osteogenic Differentiation to Adipogenic Differentiation of Mesenchymal Stem Cells in Postmenopausal Osteoporosis. Cell. Physiol. Biochem. 2016, 39, 253–265. [Google Scholar] [CrossRef]
- Hasanzad, M.; Hassani Doabsari, M.; Rahbaran, M.; Banihashemi, P.; Fazeli, F.; Ganji, M.; Manavi Nameghi, S.; Sarhangi, N.; Nikfar, S.; Aghaei Meybodi, H.R. A systematic review of miRNAs as biomarkers in osteoporosis disease. J. Diabetes Meta.b Disord. 2021, 20, 1391–1406. [Google Scholar] [CrossRef]
- Mandourah, A.Y.; Ranganath, L.; Barraclough, R.; Vinjamuri, S.; Hof, R.V.; Hamill, S.; Czanner, G.; Dera, A.A.; Wang, D.; Barraclough, D.L. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci. Rep. 2018, 8, 8421. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, H.; Wang, L.; Jiang, Y.; Xu, Y. Reduced miR-144-3p expression in serum and bone mediates osteoporosis pathogenesis by targeting RANK. Biochem. Cell Biol. 2018, 96, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.L.; Wang, Y.; Sun, Q.D.; Du, X.F. MiR-203 is involved in osteoporosis by regulating DKK1 and inhibiting osteogenic differentiation of MSCs. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 5098–5105. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yu, S.; Jin, R.; Xiao, Y.; Pan, M.; Pei, F.; Zhu, X.; Huang, H.; Zhang, Z.; Chen, S.; et al. Circulating miR-338 Cluster activities on osteoblast differentiation: Potential Diagnostic and Therapeutic Targets for Postmenopausal Osteoporosis. Theranostics 2019, 9, 3780–3797. [Google Scholar] [CrossRef] [PubMed]
- Vail, D.J.; Somoza, R.A.; Caplan, A.I. MicroRNA Regulation of Bone Marrow Mesenchymal Stem Cell Chondrogenesis: Toward Articular Cartilage. Tissue Eng. Part A 2022, 28, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, W.; Ji, F.; Wu, D. MicroRNA-410 participa en el proceso patológico de la osteoporosis postmenopáusica mediante la regulación de la proteína morfogenética ósea-2. Exp. Ther. Med. 2019, 18, 3659–3666. [Google Scholar] [CrossRef] [PubMed]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Anastasilakis, A.D.; Makras, P.; Pikilidou, M.; Tournis, S.; Makris, K.; Bisbinas, I.; Tsave, O.; Yovos, J.G.; Yavropoulou, M.P. Changes of Circulating MicroRNAs in Response to Treatment with Teriparatide or Denosumab in Postmenopausal Osteoporosis. J. Clin. Endocrinol. Metab. 2018, 103, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Yavropoulou, M.P.; Anastasilakis, A.D.; Makras, P.; Papatheodorou, A.; Rauner, M.; Hofbauer, L.C.; Tsourdi, E. Serum Profile of microRNAs Linked to Bone Metabolism During Sequential Treatment for Postmenopausal Osteoporosis. J. Clin. Endocrinol. Metab. 2020, 105, E2885–E2894. [Google Scholar] [CrossRef]
- Li, X.; Xu, R.; Ye, J.X.; Yuan, F.L. Suppression of bone remodeling associated with long-term bisphosphonate treatment is mediated by microRNA-30a-5p. Bioengineered 2022, 13, 9741–9753. [Google Scholar] [CrossRef]
- Messner, Z.; Carro Vázquez, D.; Haschka, J.; Grillari, J.; Resch, H.; Muschitz, C.; Pietschmann, P.; Zwerina, J.; Hackl, M.; Kocijan, R. Circulating miRNAs Respond to Denosumab Treatment after 2 Years in Postmenopausal Women with Osteoporosis-the MiDeTe study. J. Clin. Endocrinol. Metab. 2023, 108, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Jing, D.; Hao, J.; Shen, Y.; Tang, G.; Li, M.-L.; Huang, S.-H.; Zhao, Z.-H. The role of microRNAs in bone remodeling. Int. J. Oral. Sci. 2015, 7, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, A.; Pezzetti, F.; Brunelli, G.; Zollino, I.; Scapoli, L.; Martinelli, M.; Arlotti, M.; Carinci, F. Differences in osteoblast miRNA induced by cell binding domain of collagen and silicate-based synthetic bone. J. Biomed. Sci. 2007, 14, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Ding, J.; Dai, M.; Liu, L.; Fang, Q.; Wang, D.W.; Wu, L.; Wang, Y. Insights Into Platelet-Derived MicroRNAs in Cardiovascular and Oncologic Diseases: Potential Predictor and Therapeutic Target. Front. Cardiovasc. Med. 2022, 9, 879351. [Google Scholar] [CrossRef]
- Farmani, A.R.; Nekoofar, M.H.; Ebrahimi-Barough, S.; Azami, M.; Najafipour, S.; Moradpanah, S.; Ai, J. Preparation and In Vitro Osteogenic Evaluation of Biomimetic Hybrid Nanocomposite Scaffolds Based on Gelatin/Plasma Rich in Growth Factors (PRGF) and Lithium-Doped 45s5 Bioactive Glass Nanoparticles. J. Polym. Environ. 2023, 31, 870–885. [Google Scholar] [CrossRef]
- Farmani, A.R.; Salmeh, M.A.; Golkar, Z.; Moeinzadeh, A.; Ghiasi, F.F.; Amirabad, S.Z.; Shoormeij, M.H.; Mahdavinezhad, F.; Momeni, S.; Moradbeygi, F.; et al. Li-Doped Bioactive Ceramics: Promising Biomaterials for Tissue Engineering and Regenerative Medicine. J. Funct. Biomater. 2022, 13, 162. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).