Identification, Elucidation and Deployment of a Cytoplasmic Male Sterility System for Hybrid Potato
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Crossing Conditions
2.3. Phenotypic Analysis of Male Fertility
2.4. SeqSNP Genotyping, Linkage Analysis and QTL Mapping of Population BC2(P)-1
2.5. KASP Marker Development and Genotyping
2.6. Field Experiment
3. Results
3.1. Identification of the Antherless Phenotype and Development of a Mapping Population
3.2. Segregation of the Antherless Phenotype in Population BC2(P)-1
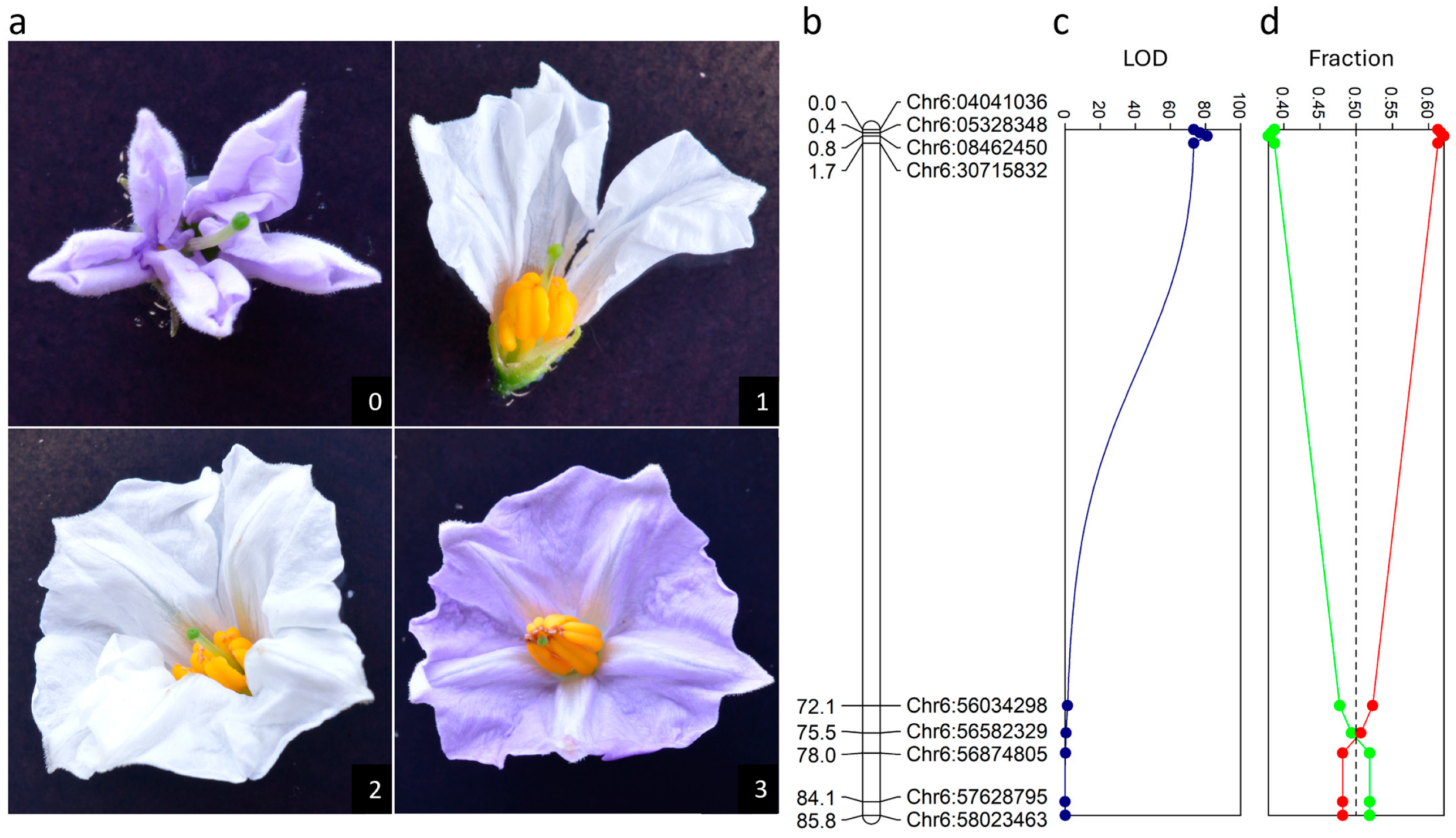
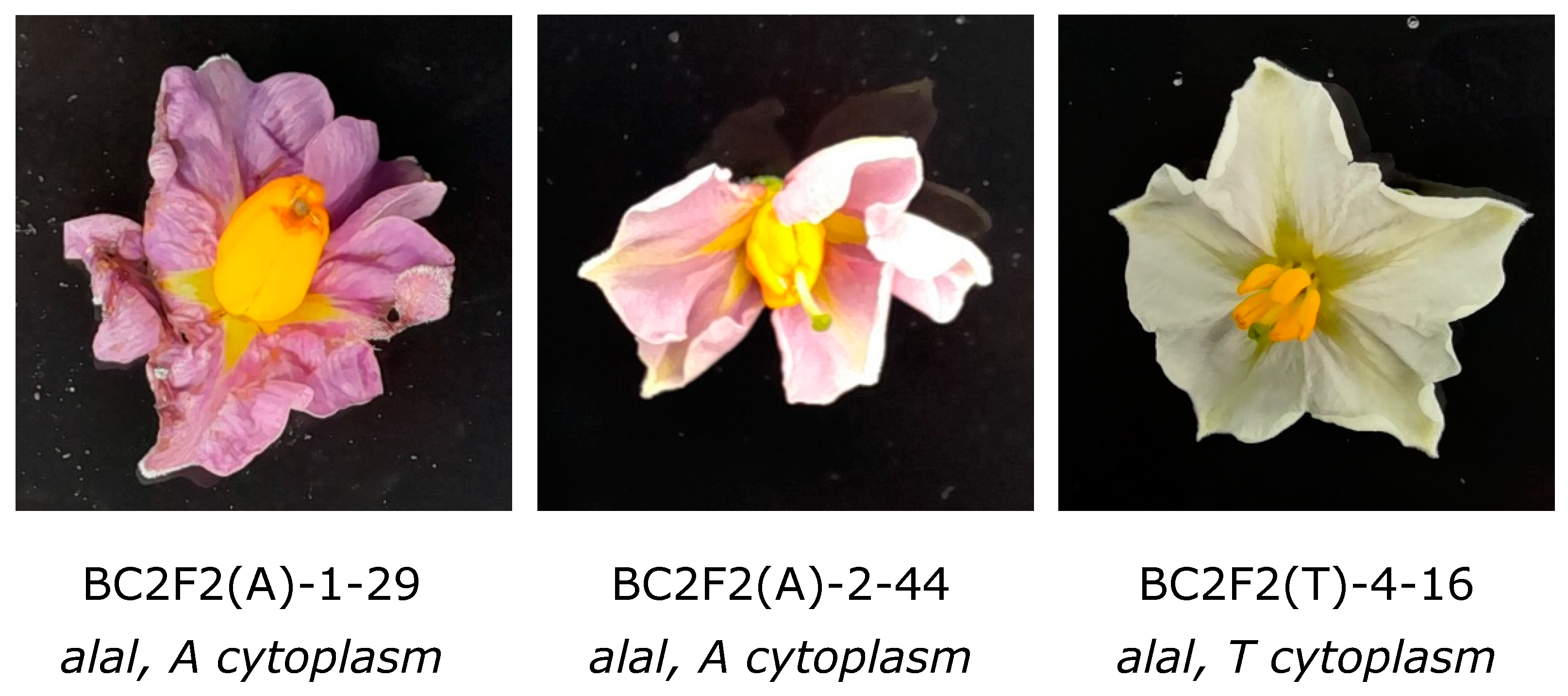
3.3. Expression of the Antherless Phenotype in A and T Cytoplasm Types
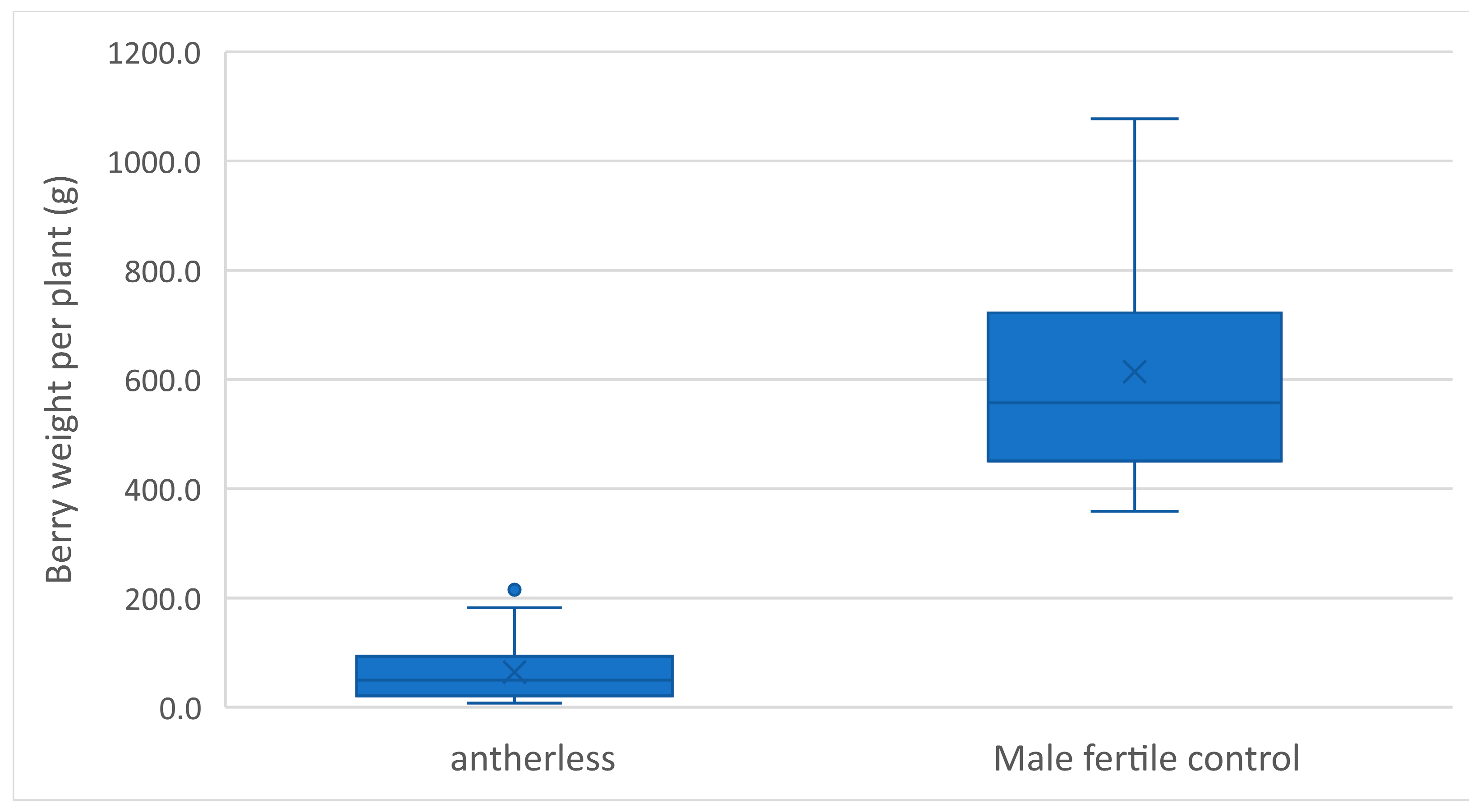
3.4. Application and Deployment of the Antherless Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindhout, P.; de Vries, M.; ter Maat, M.; Ying, S.; Viquez-Zamora, M.; van Heusden, S. Hybrid Potato Breeding for Improved Varieties. In Achieving Sustainable Cultivation of Potatoes; Burleigh Dodds Science Publishing: London, UK, 2016; Volume 1, pp. 1–24. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, Z.; Tang, D.; Zhu, Y.; Wang, P.; Li, D.; Zhu, G.; Xiong, X.; Shang, Y.; Li, C.; et al. Genome Design of Hybrid Potato. Cell 2021, 184, 3873–3883.e12. [Google Scholar] [CrossRef] [PubMed]
- Jansky, S.H.; Charkowski, A.O.; Douches, D.S.; Gusmini, G.; Richael, C.; Bethke, P.C.; Spooner, D.M.; Novy, R.G.; De Jong, H.; De Jong, W.S.; et al. Reinventing Potato as a Diploid Inbred Line-Based Crop. Crop Sci. 2016, 56, 1412–1422. [Google Scholar] [CrossRef]
- Alsahlany, M.; Enciso-Rodriguez, F.; Lopez-Cruz, M.; Coombs, J.; Douches, D.S. Developing Self-Compatible Diploid Potato Germplasm through Recurrent Selection. Euphytica 2021, 217, 47. [Google Scholar] [CrossRef]
- Hosaka, K.; Sanetomo, R. Creation of a Highly Homozygous Diploid Potato Using the S Locus Inhibitor (Sli) Gene. Euphytica 2020, 216, 169. [Google Scholar] [CrossRef]
- Lindhout, P.; Meijer, D.; Schotte, T.; Hutten, R.C.B.; Visser, R.G.F.; van Eck, H.J. Towards F1 Hybrid Seed Potato Breeding. Potato Res. 2011, 54, 301–312. [Google Scholar] [CrossRef]
- Douches, D.S.; Maas, D.; Jastrzebski, K.; Chase, R.W. Assessment of Potato Breeding Progress in the USA over the Last Century. Crop Sci. 1996, 36, 1544–1552. [Google Scholar] [CrossRef]
- Jansky, S. Breeding, Genetics, and Cultivar Development. In Advances in Potato Chemistry and Technology; Singh, J., Kaur, L., Eds.; Academic Press: New York, NY, USA, 2009; pp. 26–27. [Google Scholar]
- Su, Y.; Viquez-Zamora, M.; den Uil, D.; Sinnige, J.; Kruyt, H.; Vossen, J.; Lindhout, P.; van Heusden, S. Introgression of Genes for Resistance against Phytophthora Infestans in Diploid Potato. Am. J. Potato Res. 2020, 97, 33–42. [Google Scholar] [CrossRef]
- Hosaka, K.; Hanneman, R.E. Genetics of Self-Compatibility in a Self-Incompatible Wild Diploid Potato Species Solanum Chacoense. 1. Detection of an S Locus Inhibitor (Sli) Gene. Euphytica 1998, 99, 191–197. [Google Scholar] [CrossRef]
- Clot, C.R.; Polzer, C.; Prodhomme, C.; Schuit, C.; Engelen, C.J.M.; Hutten, R.C.B.; van Eck, H.J. The Origin and Widespread Occurrence of Sli-Based Self-Compatibility in Potato. Theor. Appl. Genet. 2020, 133, 2713–2728. [Google Scholar] [CrossRef]
- Birhman, R.K.; Hosaka, K. Production of Inbred Progenies of Diploid Potatoes Using an S -Locus Inhibitor (Sli) Gene, and Their Characterization. Genome 2000, 502, 495–502. [Google Scholar] [CrossRef]
- Eggers, E.J.; van der Burgt, A.; van Heusden, S.A.W.; de Vries, M.E.; Visser, R.G.F.; Bachem, C.W.B.; Lindhout, P. Neofunctionalisation of the Sli Gene Leads to Self-Compatibility and Facilitates Precision Breeding in Potato. Nat. Commun. 2021, 12, 4141. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhang, C.; Zhang, B.; Tang, F.; Li, F.; Liao, Q.; Tang, D.; Peng, Z.; Jia, Y.; Gao, M.; et al. A NonS-Locus F-Box Gene Breaks Self-Incompatibility in Diploid Potatoes. Nat. Commun. 2021, 12, 4142. [Google Scholar] [CrossRef] [PubMed]
- Phumichai, C.; Mori, M.; Kobayashi, A.; Kamijima, O.; Hosaka, K. Toward the Development of Highly Homozygous Diploid Potato Lines Using the Self-Compatibility Controlling Sli Gene. Genome 2005, 48, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, N.R.; Billings, G.; Coombs, J.; Buell, C.R.; Enciso-Rodríguez, F.; Douches, D.S. Self-Fertility and Resistance to the Colorado Potato Beetle (Leptinotarsa Decemlineata) in a Diploid Solanum Chacoense Recombinant Inbred Line Population. Crop Sci. 2021, 61, 3392–3414. [Google Scholar] [CrossRef]
- Jansky, S.H.; Chung, Y.S.; Kittipadukal, P. M6: A Diploid Potato Inbred Line for Use in Breeding and Genetics Research. J. Plant Regist. 2014, 8, 195–199. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, P.; Tang, D.; Yang, Z.; Lu, F.; Qi, J.; Tawari, N.R.; Shang, Y.; Li, C.; Huang, S. The Genetic Basis of Inbreeding Depression in Potato. Nat. Genet. 2019, 51, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Veerman, A.; Van Loon, C.D. Prevention of Berry Formation in Potato Plants (Solanum tuberosum L.) by Single Foliar Applications of Herbicides or Growth Regulators. Potato Res. 1993, 36, 135–142. [Google Scholar] [CrossRef]
- Bartholdi, W.L. Influence of Flowering and Fruiting Upon Vegetative Growth and Tuber Yield in the Potato; Minn. Tech. Bull 150; University of Minnesota: Minneapolis, MN, USA, 1942. [Google Scholar]
- Ter Steeg, E.M.S.; Struik, P.C.; Visser, R.G.F.; Lindhout, P. Crucial Factors for the Feasibility of Commercial Hybrid Breeding in Food Crops. Nat. Plants 2022, 8, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Gramaje, L.V.; Caguiat, J.D.; Enriquez, J.O.S.; dela Cruz, Q.D.; Millas, R.A.; Carampatana, J.E.; Tabanao, D.A.A. Heterosis and Combining Ability Analysis in CMS Hybrid Rice. Euphytica 2020, 216, 1–22. [Google Scholar] [CrossRef]
- Martín, A.C.; Atienza, S.G.; Ramírez, M.C.; Barro, F.; Martín, A. Male Fertility Restoration of Wheat in Hordeum Chilense Cytoplasm Is Associated with 6HchS Chromosome Addition. Aust. J. Agric. Res. 2008, 59, 206–213. [Google Scholar] [CrossRef]
- Martín, A.C.; Castillo, A.; Atienza, S.G.; Rodríguez-Suárez, C. A Cytoplasmic Male Sterility (CMS) System in Durum Wheat. Mol. Breed. 2018, 38, 90. [Google Scholar] [CrossRef]
- Tyagi, V.; Dhillon, S.K.; Kaushik, P.; Kaur, G. Characterization for Drought Tolerance and Physiological Efficiency in Novel Cytoplasmic Male Sterile Sources of Sunflower (Helianthus annuus L.). Agronomy 2018, 8, 232. [Google Scholar] [CrossRef]
- Li, J.; Nadeem, M.; Sun, G.; Wang, X.; Qiu, L. Male Sterility in Soybean: Occurrence, Molecular Basis and Utilization. Plant Breed. 2019, 138, 659–676. [Google Scholar] [CrossRef]
- Bruns, H.A. Southern Corn Leaf Blight: A Story Worth Retelling. Agron. J. 2017, 109, 1218–1224. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Palmer, R.G. Male Sterility in Soybean (Glycine max). I. Phenotypic Expression of the Ms2 Mutant. Am. J. Bot. 1985, 72, 1738–1750. [Google Scholar] [CrossRef]
- Balaji Suresh, P.; Srikanth, B.; Hemanth Kishore, V.; Subhakara Rao, I.; Vemireddy, L.R.; Dharika, N.; Sundaram, R.M.; Ramesha, M.S.; Sambasiva Rao, K.R.S.; Viraktamath, B.C.; et al. Fine Mapping of Rf3 and Rf4 Fertility Restorer Loci of WA-CMS of Rice (Oryza sativa L.) and Validation of the Developed Marker System for Identification of Restorer Lines. Euphytica 2012, 187, 421–435. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, M.; Zhang, B.; Zhang, X.; Guo, L.; Qi, T.; Wang, H.; Zhang, J.; Xing, C. Genome-Wide Comparative Transcriptome Analysis of CMS-D2 and Its Maintainer and Restorer Lines in Upland Cotton. BMC Genom. 2017, 18, 454. [Google Scholar] [CrossRef]
- El-Rady, A.; Ayman, G.; Soliman, G. Identification of Maintainer and Restorer Lines Based on CMS System for Developing Hybrid Wheat in Egypt. Egypt. J. Agric. Res. 2023, 101, 322–330. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D. Molecular Control of Male Fertility for Crop Hybrid Breeding. Trends Plant Sci. 2018, 23, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, K.; Sanetomo, R. Comparative Differentiation in Mitochondrial and Chloroplast DNA among Cultivated Potatoes and Closely Related Wild Species. Genes Genet. Syst. 2009, 84, 371–378. [Google Scholar] [CrossRef]
- Dionne, L.A. Cytoplasmic Sterility in Derivatives of Solanum Demissum. Am. Potato J. 1961, 38, 117–120. [Google Scholar] [CrossRef]
- Ortiz, R.; Iwanaga, M.; Peloquin, S.J. Male Sterility and 2n Pollen in 4x Progenies Derived from 4x × 2x and 4x × 4x Crosses in Potatoes. Potato Res. 1993, 36, 227–236. [Google Scholar] [CrossRef]
- Lössl, A.; Götz, M.; Braun, A.; Wenzel, G. Molecular Markers for Cytoplasm in Potato: Male Sterility and Contribution of Different Plastid-Mitochondrial Configurations to Starch Production. Euphytica 2000, 116, 221–230. [Google Scholar] [CrossRef]
- Hoopes, R.W.; Plaisted, R.L.; Cubillos, A.G. Yield and Fertility of Reciprocal-Cross Tuberosum-Andigena Hybrids. Am. Potato J. 1980, 57, 275–284. [Google Scholar] [CrossRef]
- Staub, J.E.; Grun, P.; Amoah, V. Cytoplasmic Evaluations during Substitution Backcrossing in Solanum. Potato Res. 1982, 25, 299–319. [Google Scholar] [CrossRef]
- Grun, P. Cytoplasmic Sterilities That Separate the Group Tuberosum Cultivated Potato from Its Putative Tetraploid Ancestor. Evolution 1973, 27, 633. [Google Scholar] [CrossRef]
- Santayana, M.; Aponte, M.; Kante, M.; Gastelo, M. Cytoplasmic Male Sterility Incidence in Potato Breeding Populations with Late Blight Resistance and Identification of Breeding Lines with a Potential Fertility Restorer Mechanism. Plants 2022, 11, 3093. [Google Scholar] [CrossRef] [PubMed]
- Sanetomo, R.; Akai, K.; Nashiki, A. Discovery of a Novel Mitochondrial DNA Molecule Associated with Tetrad Pollen Sterility in Potato. BMC Plant Biol. 2022, 22, 302. [Google Scholar] [CrossRef]
- Sanetomo, R.; Nashiki, A. Identification of the Tetrad-Sterility-Causing Solanum Stoloniferum Schltdl. & Bouché Cytoplasm in Interspecific Hybrids with S. tuberosum L. Genet Resour. Crop Evol. 2021, 68, 3383–3397. [Google Scholar] [CrossRef]
- Endelman, J.B.; Jansky, S.H. Genetic Mapping with an Inbred Line-Derived F2 Population in Potato. Theor. Appl. Genet. 2016, 129, 935–943. [Google Scholar] [CrossRef]
- Spooner, D.M.; Ghislain, M.; Simon, R.; Jansky, S.H.; Gavrilenko, T. Systematics, Diversity, Genetics, and Evolution of Wild and Cultivated Potatoes. Bot. Rev. 2014, 80, 283–383. [Google Scholar] [CrossRef]
- Lin, X.; Torres Ascurra, Y.C.; Fillianti, H.; Dethier, L.; de Rond, L.; Domazakis, E.; Aguilera-Galvez, C.; Kiros, A.Y.; Jacobsen, E.; Visser, R.G.F.; et al. Recognition of Pep-13/25 MAMPs of Phytophthora Localizes to an RLK Locus in Solanum microdontum. Front. Plant Sci. 2023, 13, 1037030. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, J.; Fartmann, B.; Winkler, S.; Arvidsson, S.; Osterkamp, S.; Zimmermann, W.; Heath, J.D. SeqSNP, a Massively Parallel Marker Screening Approach–High-Speed and High-Throughput Genomic Selection for Plants and Livestock. In Proceedings of the Plant and Animal Genome XXVI Conference, San Diego, CA, USA, 13–17 January 2018. [Google Scholar]
- Adams, J.; de Vries, M.; van Eeuwijk, F. Efficient Genomic Prediction of Yield and Dry Matter in Hybrid Potato. Plants 2023, 12, 2617. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap®4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wagening, The Netherlands, 2006. [Google Scholar]
- van Ooijen, J.W. Accuracy of Mapping Quantitative Trait Loci in Autogamous Species. Theor. Appl. Genet. 1992, 84, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Kacheyo, O.C.; de Vries, M.E.; van Dijk, L.C.M.; Schneider, H.M.; Struik, P.C. Agronomic Consequences of Growing Field-Transplanted Hybrid Potato Seedlings. Crop Sci 2024, 64, 1093–1111. [Google Scholar] [CrossRef]
- van Lieshout, N.; van der Burgt, A.; de Vries, M.E.; ter Maat, M.; Eickholt, D.; Esselink, D.; van Kaauwen, M.P.W.; Kodde, L.P.; Visser, R.G.F.; Lindhout, P.; et al. Solyntus, the New Highly Contiguous Reference Genome for Potato (Solanum tuberosum). G3 Genes Genomes Genet. 2020, 10, 3489–3495. [Google Scholar] [CrossRef]
- Pham, G.M.; Hamilton, J.P.; Wood, J.C.; Burke, J.T.; Zhao, H.; Vaillancourt, B.; Ou, S.; Jiang, J.; Robin Buell, C. Construction of a Chromosome-Scale Long-Read Reference Genome Assembly for Potato. Gigascience 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Hosaka, K.; Sanetomo, R. Development of a Rapid Identification Method for Potato Cytoplasm and Its Use for Evaluating Japanese Collections. Theor. Appl. Genet. 2012, 125, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.N.; Manrique-Carpintero, N.C.; Coombs, J.J.; Zarka, D.G.; Boone, A.E.; Kirk, W.W.; Hackett, C.A.; Bryan, G.J.; Douches, D.S. Genetic Linkage Mapping of Economically Important Traits in Cultivated Tetraploid Potato (Solanum tuberosum L.). G3 Genes Genomes Genet. 2015, 5, 2357–2364. [Google Scholar] [CrossRef]
- da Silva Pereira, G.; Mollinari, M.; Schumann, M.J.; Clough, M.E.; Zeng, Z.B.; Yencho, G.C. The Recombination Landscape and Multiple QTL Mapping in a Solanum Tuberosum Cv. ‘Atlantic’-Derived F1 Population. Heredity 2021, 126, 817–830. [Google Scholar] [CrossRef]
- Bourke, P.M.; Voorrips, R.E.; Kranenburg, T.; Jansen, J.; Visser, R.G.F.; Maliepaard, C. Integrating Haplotype-Specific Linkage Maps in Tetraploid Species Using SNP Markers. Theor. Appl. Genet. 2016, 129, 2211–2226. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Bolser, D.; de Boer, J.; Sønderkær, M.; Amoros, W.; Carboni, M.F.; D’Ambrosio, J.M.; de la Cruz, G.; Di Genova, A.; Douches, D.S.; et al. Construction of Reference Chromosome-Scale Pseudomolecules for Potato: Integrating the Potato Genome with Genetic and Physical Maps. G3 Genes Genomes Genet. 2013, 3, 2031–2047. [Google Scholar] [CrossRef]
- Harkess, A.; Zhou, J.; Xu, C.; Bowers, J.E.; Van Der Hulst, R.; Ayyampalayam, S.; Mercati, F.; Riccardi, P.; McKain, M.R.; Kakrana, A.; et al. The Asparagus Genome Sheds Light on the Origin and Evolution of a Young Y Chromosome. Nat. Commun. 2017, 8, 1279. [Google Scholar] [CrossRef] [PubMed]
- Harkess, A.; Huang, K.; van der Hulst, R.; Tissen, B.; Caplan, J.L.; Koppula, A.; Batish, M.; Meyers, B.C.; Leebens-Mack, J. Sex Determination by Two Y-Linked Genes in Garden Asparagus. Plant Cell 2020, 32, 1790–1796. [Google Scholar] [CrossRef] [PubMed]
- Underwood, C.J.; Vijverberg, K.; Rigola, D.; Okamoto, S.; Oplaat, C.; Camp, R.H.M.O.d.; Radoeva, T.; Schauer, S.E.; Fierens, J.; Jansen, K.; et al. A PARTHENOGENESIS Allele from Apomictic Dandelion Can Induce Egg Cell Division without Fertilization in Lettuce. Nat. Genet. 2022, 54, 84–93. [Google Scholar] [CrossRef]
- Dahan, J.; Mireau, H. The Rf and Rf-like PPR in Higher Plants, a Fast-Evolving Subclass of PPR Genes. RNA Biol. 2013, 10, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Gaborieau, L.; Brown, G.G.; Mireau, H. The Propensity of Pentatricopeptide Repeat Genes to Evolve into Restorers of Cytoplasmic Male Sterility. Front. Plant Sci. 2016, 7, 225677. [Google Scholar] [CrossRef]
- Jo, Y.D.; Ha, Y.; Lee, J.H.; Park, M.; Bergsma, A.C.; Choi, H.I.; Goritschnig, S.; Kloosterman, B.; van Dijk, P.J.; Choi, D.; et al. Fine Mapping of Restorer-of-Fertility in Pepper (Capsicum annuum L.) Identified a Candidate Gene Encoding a Pentatricopeptide Repeat (PPR)-Containing Protein. Theor. Appl. Genet. 2016, 129, 2003–2017. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; et al. Cytoplasmic Male Sterility of Rice with Boro II Cytoplasm Is Caused by a Cytotoxic Peptide and Is Restored by Two Related PPR Motif Genes via Distinct Modes of MRNA Silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef]
- Brown, G.G.; Formanová, N.; Jin, H.; Wargachuk, R.; Dendy, C.; Patil, P.; Laforest, M.; Zhang, J.; Cheung, W.Y.; Landry, B.S. The Radish Rfo Restorer Gene of Ogura Cytoplasmic Male Sterility Encodes a Protein with Multiple Pentatricopeptide Repeats. Plant J. 2003, 35, 262–272. [Google Scholar] [CrossRef]
- Koizuka, N.; Imai, R.; Fujimoto, H.; Hayakawa, T.; Kimura, Y.; Kohno-Murase, J.; Sakai, T.; Kawasaki, S.; Imamura, J. Genetic Characterization of a Pentatricopeptide Repeat Protein Gene, Orf687, That Restores Fertility in the Cytoplasmic Male-Sterile Kosena Radish. Plant J. 2003, 34, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Madugula, P.; Uttam, A.G.; Tonapi, V.A.; Ragimasalawada, M. Fine Mapping of Rf2, a Major Locus Controlling Pollen Fertility Restoration in Sorghum A1 Cytoplasm, Encodes a PPR Gene and Its Validation through Expression Analysis. Plant Breed. 2018, 137, 148–161. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, L.; Li, J.; Hu, G.; Wu, Q.; Jiang, H.; Huang, Z. The Restorer Gene for Soybean M-Type Cytoplasmic Male Sterility, Rf-m, Is Located in a PPR Gene-Rich Region on Chromosome 16. Plant Breed. 2016, 135, 342–348. [Google Scholar] [CrossRef]
- Gao, B.; Ren, G.; Wen, T.; Li, H.; Zhang, X.; Lin, Z. A Super PPR Cluster for Restoring Fertility Revealed by Genetic Mapping, Homocap-Seq and de Novo Assembly in Cotton. Theor. Appl. Genet. 2022, 135, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, X.; Guo, L.; Qi, T.; Wang, H.; Tang, H.; Qiao, X.; Shahzad, K.; Xing, C.; Wu, J. Genome-Wide Analysis of Rf-PPR-like (RFL) Genes and a New InDel Marker Development for Rf1 Gene in Cytoplasmic Male Sterile CMS-D2 Upland Cotton. J. Cotton Res. 2018, 1, 1–11. [Google Scholar] [CrossRef]
- Wang, C.; Lezhneva, L.; Arnal, N.; Quadrado, M.; Mireau, H. The Radish Ogura Fertility Restorer Impedes Translation Elongation along Its Cognate CMS-Causing MRNA. Proc. Natl. Acad. Sci. USA 2021, 118, e2105274118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, Z.; Wang, X.; Li, K.; An, H.; Liu, J.; Yang, G.; Fu, T.; Yi, B.; Hong, D. A Mitochondria-Targeted PPR Protein Restores Pol Cytoplasmic Male Sterility by Reducing Orf224 Transcript Levels in Oilseed Rape. Mol. Plant 2016, 9, 1082–1084. [Google Scholar] [CrossRef]
- Tang, H.; Luo, D.; Zhou, D.; Zhang, Q.; Tian, D.; Zheng, X.; Chen, L.; Liu, Y.-G. The Rice Restorer Rf4 for Wild-Abortive Cytoplasmic Male Sterility Encodes a Mitochondrial-Localized PPR Protein That Functions in Reduction of WA352 Transcripts. Mol. Plant 2014, 9, 1497–1500. [Google Scholar] [CrossRef]
- Anisimova, I.N.; Alpatieva, N.V.; Karabitsina, Y.I.; Gavrilenko, T.A. Nucleotide Sequence Polymorphism in the RFL-PPR Genes of Potato. J. Genet. 2019, 98, 87. [Google Scholar] [CrossRef]
- Phumichai, C.; Hosaka, K. Cryptic Improvement for Fertility by Continuous Selfing of Diploid Potatoes Using Sli Gene. Euphytica 2006, 149, 251–258. [Google Scholar] [CrossRef]
- Peterson, B.A.; Holt, S.H.; Laimbeer, F.P.E.; Doulis, A.G.; Coombs, J.; Douches, D.S.; Hardigan, M.A.; Buell, C.R.; Veilleux, R.E. Self-Fertility in a Cultivated Diploid Potato Population Examined with the Infinium 8303 Potato Single-Nucleotide Polymorphism Array. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- De Jong, H.; Rowe, P.R. Inbreeding in Cultivated Diploid Potatoes. Potato Res. 1971, 14, 74–83. [Google Scholar] [CrossRef]
- Hutten, R.C.B.; Soppe, W.J.J.; Hermsen, J.G.T.; Jacobsen, E. Evaluation of Dihaploid Populations from Potato Varieties and Breeding Lines. Potato Res. 1995, 38, 77–86. [Google Scholar] [CrossRef]
| Population | Antherless Genotype | χ2 | p-Value | ||
|---|---|---|---|---|---|
| AlAl | Alal | alal | |||
| BC2F2(A)-1 | 33 | 38 | 6 | 18.95 | 0.00008 |
| BC2F2(A)-2 | 23 | 34 | 7 | 8.25 | 0.01616 |
| BC2F2(T)-6 | 16 | 21 | 8 | 3.04 | 0.21823 |
| BC2F2(T)-1 | 12 | 23 | 10 | 0.2 | 0.90484 |
| BC2F2(T)-2 | 10 | 19 | 9 | 0.05 | 0.97404 |
| BC2F2(T)-3 | 11 | 22 | 11 | 0 | 1 |
| BC2F2(T)-4 | 8 | 31 | 20 | 5.03 | 0.08071 |
| BC2F2(T)-5 | 1 | 45 | 39 | 34.27 | 0.00001 |
| Genotype | Cytoplasm Type | # Observed Flowers | Anther Phenotype (0–3) | Pollen Shed (0–3) | # Self-Berries | # Spontaneous Berries | # Seeds | Antherless Genotype | F3 Population |
|---|---|---|---|---|---|---|---|---|---|
| BC2F2(A)-1-29 | A | 20 | 3 | 3 | 5 | 2 | 23 | alal | BC2F3(A)-1 |
| BC2F2(A)-2-44 | A | 11 | 2 | 2 | 3 | 0 | 305 | alal | BC2F3(A)-2 |
| BC2F2(T)-2-07 | T | 23 | 3 | 1 | 2 | 1 | 106 | Alal | BC2F3(T)-1 |
| BC2F2(T)-2-20 | T | 25 | 3 | 3 | 5 | 2 | 220 | Alal | BC2F3(T)-2 |
| BC2F2(T)-3-06 | T | 6 | 3 | 1 | 3 | 0 | 97 | Alal | BC2F3(T)-3 |
| BC2F2(T)-4-16 | T | 30 | 3 | 3 | 10 | 16 | 140 | alal | BC2F3(T)-4 |
| BC2F2(T)-4-17 | T | 14 | 2 | 3 | 2 | 0 | 6 | Alal | |
| BC2F2(T)-6-02 | T | 27 | 2 | 1 | 2 | 1 | 9 | Alal | BC2F3(T)-5 |
| Genotype | Cytoplasm Type | Antherless Genotype | # Flowers | Anther Phenotype | Pollen Shed (0–3) | # Selfings | # Berries | # Seeds | Pollen Viability |
|---|---|---|---|---|---|---|---|---|---|
| BC2F2(A)-1-29-C1 | A | alal | 11 | 3 | 1-2 | 6 | 4 | 1296 | 0.9 |
| BC2F2(A)-1-29-C2 | A | alal | 7 | 3 | 1 | 4 | 2 | 135 | 0.9 |
| BC2F2(A)-1-29-C3 | A | alal | 17 | 3 | 1 | 4 | 1 | 22 | 0.9 |
| BC2F2(A)-1-29-C4 | A | alal | 9 | 3 | 1-2 | 3 | 3 | 4 | 0.9 |
| BC2F2(A)-1-29-C5 | A | alal | 11 | 3 | 1-2 | 1 | 1 | 49 | 0.9 |
| BC2F2(A)-2-09-C1 | A | alal | 17 | 2-3 | 1 | 6 | 26 | 12 | 0.8 |
| BC2F2(A)-2-09-C2 | A | alal | 0 | N.D | N.D | 0 | N.D | N.D | N.D |
| BC2F2(A)-2-09-C3 | A | alal | 3 | 3 | 1 | 1 | 13 | 1 | 0.8 |
| BC2F2(A)-2-09-C4 | A | alal | 7 | 2-3 | 1-2 | 2 | 25 | 0 | 0.8 |
| BC2F2(A)-2-09-C5 | A | alal | 11 | 2-3 | 1 | 1 | 12 | 3 | 0.8 |
| BC2F2(A)-2-39-C1 | A | alal | 4 | 2 | 0 | 0 | N.D | N.D | N.D. |
| BC2F2(A)-2-39-C2 | A | alal | 3 | 2 | 0 | 0 | N.D | N.D | N.D. |
| BC2F2(A)-2-39-C3 | A | alal | 0 | N.D | N.D | 0 | N.D | N.D | N.D. |
| BC2F2(A)-2-39-C4 | A | alal | 1 | 2 | 0 | 0 | N.D | N.D | N.D. |
| BC2F2(A)-2-39-C5 | A | alal | 0 | N.D | N.D | 0 | N.D | N.D | N.D. |
| BC2F2(A)-2-44-C1 | A | alal | 32 | 2-3 | 3 | 17 | 19 | 740 | 0.9 |
| BC2F2(A)-2-44-C2 | A | alal | 32 | 2-3 | 3 | 24 | 28 | 2457 | 0.9 |
| BC2F2(A)-2-44-C3 | A | alal | 11 | 2-3 | 3 | 10 | 23 | 2320 | 0.9 |
| BC2F2(A)-2-44-C4 | A | alal | 15 | 2-3 | 3 | 12 | 19 | 719 | 0.9 |
| BC2F2(A)-2-44-C5 | A | alal | 24 | 2-3 | 3 | 13 | 34 | 1246 | 0.9 |
| BC2F2(T)-3-07-C1 | T | alal | 17 | 3 | 1 | 4 | 2 | 18 | N.D. |
| BC2F2(T)-3-07-C2 | T | alal | 20 | 3 | 1 | 6 | 3 | 40 | N.D. |
| BC2F2(T)-3-07-C3 | T | alal | 24 | 3 | 0-1 | 3 | 2 | 3 | N.D. |
| BC2F2(T)-3-07-C4 | T | alal | 33 | 3 | 0-1 | 8 | 4 | 82 | N.D. |
| BC2F2(T)-3-07-C5 | T | alal | 38 | 3 | 0-1 | 8 | 7 | 75 | N.D. |
| BC2F2(T)-4-16-C1 | T | alal | 52 | 3 | 3 | 16 | 21 | 550 | 0.9 |
| BC2F2(T)-4-16-C2 | T | alal | 47 | 3 | 3 | 17 | 21 | 906 | 0.9 |
| BC2F2(T)-4-16-C3 | T | alal | 40 | 3 | 3 | 10 | 7 | 177 | 0.9 |
| BC2F2(T)-4-16-C4 | T | alal | 44 | 3 | 3 | 14 | 10 | 199 | 0.9 |
| BC2F2(T)-4-16-C5 | T | alal | 43 | 3 | 3 | 20 | 37 | 843 | 0.9 |
| Genotype | Parent | Cytoplasm Type | Antherless Genotype | # Flowers | Anther Phenotype (0–3) | Pollen Shed (0–3) | # Selfings | # Berries | # Seeds | Pollen Viability |
|---|---|---|---|---|---|---|---|---|---|---|
| BC2F3(A)-1-11 | BC2F2(A)-1-29 | A | alal | 21 | 3 | 1 | 5 | 5 | 15 | N.D. |
| BC2F3(A)-1-12 | BC2F2(A)-1-29 | A | alal | 12 | 3 | 3 | 9 | 3 | 48 | N.D. |
| BC2F3(A)-2-04 | BC2F2(A)-2-44 | A | alal | 9 | 2 | 2 | 7 | 14 | 98 | N.D. |
| BC2F3(A)-2-05 | BC2F2(A)-2-44 | A | alal | 2 | 2 | 3 | 2 | 1 | 125 | N.D. |
| BC2F3(A)-2-14 | BC2F2(A)-2-44 | A | alal | 5 | 2 | 3 | 5 | 3 | 242 | 85% |
| BC2F3(A)-2-17 | BC2F2(A)-2-44 | A | alal | 7 | 2 | 3 | 7 | 10 | 278 | 90% |
| BC2F3(T)-2-06 | BC2F2(T)-2-20 | T | AlAl | 5 | 3 | 3 | 5 | 4 | 83 | 60% |
| BC2F3(T)-2-09 | BC2F2(T)-2-20 | T | Alal | 9 | 3 | 2-3 | 8 | 5 | 30 | 70% |
| BC2F3(T)-2-24 | BC2F2(T)-2-20 | T | Alal | 13 | 3 | 2-3 | 18 | 11 | 336 | 70% |
| BC2F3(T)-3-27 | BC2F2(T)-3-06 | T | Alal | 22 | 2 | 3 | 18 | 12 | 1514 | 90% |
| BC2F3(T)-3-29 | BC2F2(T)-3-06 | T | Alal | 45 | 3 | 1-2 | 22 | 21 | 707 | 90% |
| BC2F3(T)-4-01 | BC2F2(T)-4-16 | T | alal | 35 | 2-3 | 2-3 | 24 | 10 | 55 | 85% |
| BC2F3(T)-4-07 | BC2F2(T)-4-16 | T | alal | 32 | 3 | 2-3 | 19 | 1 | 10 | 60% |
| BC2F3(T)-4-13 | BC2F2(T)-4-16 | T | alal | 15 | 3 | 3 | 24 | 21 | 122 | 70% |
| BC2F3(T)-4-25 | BC2F2(T)-4-16 | T | alal | 17 | 3 | 1-2 | 7 | 10 | 59 | 60% |
| BC2F3(T)-4-31 | BC2F2(T)-4-16 | T | alal | 49 | 3 | 2 | 23 | 11 | 69 | N.D. |
| BC2F3(T)-5-02 | BC2F2(T)-4-16 | T | alal | 22 | 3 | 3 | 19 | 13 | 297 | 90% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eggers, E.-J.; Su, Y.; van der Poel, E.; Flipsen, M.; de Vries, M.E.; Bachem, C.W.B.; Visser, R.G.F.; Lindhout, P. Identification, Elucidation and Deployment of a Cytoplasmic Male Sterility System for Hybrid Potato. Biology 2024, 13, 447. https://doi.org/10.3390/biology13060447
Eggers E-J, Su Y, van der Poel E, Flipsen M, de Vries ME, Bachem CWB, Visser RGF, Lindhout P. Identification, Elucidation and Deployment of a Cytoplasmic Male Sterility System for Hybrid Potato. Biology. 2024; 13(6):447. https://doi.org/10.3390/biology13060447
Chicago/Turabian StyleEggers, Ernst-Jan, Ying Su, Esmee van der Poel, Martijn Flipsen, Michiel E. de Vries, Christian W. B. Bachem, Richard G. F. Visser, and Pim Lindhout. 2024. "Identification, Elucidation and Deployment of a Cytoplasmic Male Sterility System for Hybrid Potato" Biology 13, no. 6: 447. https://doi.org/10.3390/biology13060447
APA StyleEggers, E.-J., Su, Y., van der Poel, E., Flipsen, M., de Vries, M. E., Bachem, C. W. B., Visser, R. G. F., & Lindhout, P. (2024). Identification, Elucidation and Deployment of a Cytoplasmic Male Sterility System for Hybrid Potato. Biology, 13(6), 447. https://doi.org/10.3390/biology13060447







