Simple Summary
Photoperiod sensitivity is important for rice breeders to enable crop adaptation to changing climatic conditions. This study aims to pinpoint candidate insertion/deletion (INDEL) markers linked to photoperiod sensitivity in indica rice cultivars. The INDEL marker can be utilized to explore the gene function in rice flowering, potentially facilitating gene editing using CRISPR/Cas9 technology. Applying this INDEL marker in breeding programs can enhance the genetic gain and economic efficiency of a targeted breeding strategy.
Abstract
The current study aims to identify candidate insertion/deletion (INDEL) markers associated with photoperiod sensitivity (PS) in rice landraces from the Vietnamese Mekong Delta. The whole-genome sequencing of 20 accessions was conducted to analyze INDEL variations between two photoperiod-sensitivity groups. A total of 2240 INDELs were identified between the two photoperiod-sensitivity groups. The selection criteria included INDELs with insertions or deletions of at least 20 base pairs within the improved rice group. Six INDELs were discovered on chromosomes 01 (5 INDELs) and 6 (1 INDEL), and two genes were identified: LOC_Os01g23780 and LOC_Os01g36500. The gene LOC_Os01g23780, which may be involved in rice flowering, was identified in a 20 bp deletion on chromosome 01 from the improved rice accession group. A marker was devised for this gene, indicating a polymorphism rate of 20%. Remarkably, 20% of the materials comprised improved rice accessions. This INDEL marker could explain 100% of the observed distinctions. Further analysis of the mapping population demonstrated that an INDEL marker associated with the MADS-box gene on chromosome 01 was linked to photoperiod sensitivity. The F1 population displayed two bands across all hybrid individuals. The marker demonstrates efficacy in distinguishing improved rice accessions within the indica accessions. This study underscores the potential applicability of the INDEL marker in breeding strategies.
1. Introduction
Rice (Oryza sativa) production is important in the global food supply, particularly in Asian countries. Rice is grown in diverse climatic regions with wide variation in their photoperiods [1,2]. It is inherently a short-day plant, displaying pronounced photoperiod sensitivity, where short days encourage flowering while long days inhibit it [3]. The heading date (flowering time) is an important trait for the regional and seasonal adaptation of rice and influences grain yield [4]. Thus, photoperiod-sensitive control in rice is essential for the production of rice adaptive to different climatic conditions [5].
The primary distinction between landrace and improved rice varieties lies in their photoperiod sensitivity [5]. Rice landraces flower under short-day conditions [5], while improved rice can flower throughout the year, given sufficient time for the preproduction stage. Previous studies on heading dates focused on genotypic photoperiod-sensitive rice cultivars [6]. The heading date is regulated by multiple quantitative trait loci (QTLs) [7] with several well-known QTLs identified to date, including se-1 [8,9], Hd1 [10], Hd6 [11], Hd3a [12], Ehd1 [13], Ghd7 [14], DTH8 [15], Ghd8 [16], dth3 [17], Hd17 [18], Ehd4 [19], Hd16 [20], RFT1 [21], DTH2 [22], and DTH7 [23]. Furthermore, the intricate regulation of strong photoperiod sensitivity involves collaborative and competitive interactions among Hd1, Ghd7, and DTH8 in rice heading [3]. Consequently, the heading date exhibits a strong correlation with photoperiod sensitivity. While previous studies focused on identifying QTLs using marker-assisted selection and backcrossing, recent years have seen the adoption of genome-wide association studies (GWASs) as a method for identifying candidate genes associated with phenotypic traits, including the heading date [24].
The application of single-nucleotide polymorphism (SNP) genotyping methods based on next-generation sequencing and high-throughput genotyping has gained popularity in various crops [25,26,27,28,29]. GWAS, facilitated by SNP analysis, enables the study of complex quantitative traits such as plant stress tolerance, growth, and development.
Insertion/deletion (INDEL) markers are valuable markers in rice breeding [30], aiding in distinguishing the phenotypic traits of rice varieties [31,32,33,34,35]. However, existing studies have primarily utilized available INDEL markers from the Nipponbare genomic reference database. The distinction between photoperiod-sensitive and non-sensitive rice based on whole-genome sequencing and INDEL markers has not been a primary focus of previous studies. In this study, we employed the whole-genome sequencing of 20 indica rice accessions (including 16 landraces and four improved rice accessions) collected from the Vietnamese Mekong Delta provinces to analyze the relationship between two distinct groups based on INDEL information.
2. Methods
2.1. Plant Materials
A total of twenty rice accessions from a pool of 99 rice accessions were selected following the criteria outlined by Tam et al. [36]. The selected accessions comprised sixteen landraces and four improved rice accessions originating from the Vietnamese Mekong Delta provinces (Table 1). The origin locations of the studied rice landraces are characterised by salinity-affected and rainfed agro-ecology. In contrast, the origins of the improved rice accessions are considered freshwater and irrigated agro-ecology.

Table 1.
Rice accession codes and names, origin provinces, and photoperiod sensitivity.
2.2. Genome Sequencing and Data Analysis
2.2.1. Genomic DNA Isolation
Genomic DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany), adhering to the manufacturer’s protocol. Sample preparation of leaf specimens and DNA quality assessment procedures were followed [36]. Sterilized seeds of each accession were placed in Petri dishes for germination. Three days after germination, Petri dishes were transferred into a growth chamber at 27 °C. In a period of 7 to 10 days after germination, leaves were harvested for DNA extraction. The quantity of DNA was measured using a Thermo Scientific NanoDrop 2000 spectrophotometer (Fisher Scientific, Hampton, NH, USA) with a volume of 1 µL. DNA quality of samples was checked by 1% (w/v) agarose gel electrophoresis.
2.2.2. Whole-Genome Sequencing
The whole genome of the selected varieties was sequenced using Illumina’s next-generation sequencing approach. Library preparation and sequencing were performed at each step of the procedure. The workflow was as follows: (1) checking of DNA; (2) construction of libraries; (3) checking of libraries; (4) sequencing.
We performed two main methods of quality control for DNA samples: (1) agarose gel electrophoresis to test DNA degradation and potential contamination, and (2) Qubit 2.0 to quantify DNA concentration precisely.
Library construction and quality control: A total amount of 1.0 μg DNA per sample was used as input material for DNA sample preparation. Sequencing libraries were generated using NEBNextR DNA Library Prep Kit (Novogene corporation INC (823 Anchorage Place, Chula Vista, CA, USA)) following manufacturer’s recommendations and indices were added to each sample. Genomic DNA was randomly fragmented to a size of 350 bp by shearing; then, DNA fragments were end-polished, A-tailed, and ligated with an NEBNext adapter for Illmina sequencing, and further PCR-enriched by P5 and indexed P7 oligos. PCR products were purified (AMPure XP system), and resulting libraries were analyzed for size distribution by an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) and quantified using real-time PCR.
Sequencing: Qualified libraries were fed into Illumina sequencers after pooling according to their effective concentration and expected data volume.
2.2.3. Data Processing
Processing of INDELs for the twenty varieties followed the protocol outlined by Tam et al. [36]. Sequenced raw reads underwent filtering and sorting based on original sample names. Sequence trimming was executed to a length of 350 bp using a Trimmomatic with the following specified parameters: LEADING:19, TRAILING:19, SLIDINGWINDOW:30:20, AVGQUAL:20, and MINLEN:51. High-quality reads were mapped to the Nipponbare IRGSP1.0 reference genome using Bowtie2 [37], available in Galaxy (www.https://usegalaxy.org, accessed on 24 November 2023). Further filtering was performed using Picard in Galaxy, and alignments were adjusted around INDELs using the INDELRealigner tool in Genome Analysis Toolkit v2.8 [38]. INDEL calling utilized the Unified Genotyper tool in GATK v2.8 [39], and the initial INDEL dataset was filtered based on the following parameters: missing call rate (MCR), 100%; minor allele frequency (MAF), 0.05; and heterozygosity rate (HR), 0.0. Subsequent dataset filtering included functions to separate accessions into two groups (landrace and improved), leading to the identification of 2240 INDELs for further analysis.
2.3. Identifying INDELs between Photoperiod-Sensitive and Non-Sensitive Rice Groups
The INDELs, derived from dataset filtering (2240 INDELs) between the two groups, with a minimum length of 20 bp, were specifically chosen to emphasize the distinct variations between landrace and improved rice. In a previous study, Adedze et al. selected INDEL sizes starting from 30 bp as agarose gel PCR-based markers [40]. The selected INDELs (candidates) were employed for gene filtering using the Rice SNP-Seek Database (https://snp-seek.irri.org/, accessed on 7 December 2023). Candidate INDEL positions were utilized to search the Rice SNP-Seek Database using the Search/Genotype function.
2.4. Primer Design for Testing Improved Rice Varieties
Primer design, confirming the differences between landrace and improved groups, was utilized on the INDEL located on the gene in the Rice SNP-Seek Database. The primer, named Photoperiod-sensitivity (PS), was designed using Primer3Plus (http://primer3plus.ut.ee/cgi-bin/primer3plus/primer3plus.cgi, accessed on 7 December 2023). The information began with the specified primers (5′-GAGGGAGCTCTCCATCCTCT-3′ for the left primer and 5′GCTTCAACTCGAGGCACTCT-3′ for the right primer), targeting the region on chromosome 01. This amplification product was 203 from the standard sequence referenced for the Nipponbare variety.
2.5. Validation of INDEL Candidate from the Rice SNP-Seek Database
The validation of potential INDEL candidates based on the Rice SNP-Seek Database (3K) is a pivotal step. This database is well known for SNP, INDEL, gene, and other genomic data from 3024 whole-genome sequencing accessions [41]. Mansueto et al. categorized these accessions into twelve distinct sub-populations based on SNP and INDEL profiles, including admix, aro, aus, Ind1a, ind1b, ind2, ind3, indx, japx, subtrop, temp, and trop [41]. In order to ascertain the credibility of INDEL candidates in our study, an investigation was conducted within the rice genome utilizing the 3K database. This involved the genomic positions of the INDEL candidates across the 3K dataset. The diversity of INDEL markers was assessed, focusing on the occurrence of Nipponbare reference genome sequences or deletions within each sub-population.
2.6. Data Analysis
The filtering process utilized TASSEL 5.2.50 (Trait Analysis by Association Evolution and Linkage) [42]. Candidate genes were scrutinized using the QTL and gene database in the Rice SNP-Seek Database [41]. The variations among the twelve rice groups in the Rice SNP-Seek Database, and the ratio of deletion accessions to total accessions was calculated for each group.
3. Results
3.1. INDEL Information on Selected Landraces and Improved Accessions
A total of 9690 INDELs were identified through the whole-genome sequencing of 20 accessions. The highest count of INDELs was observed on Chr. 01 (2970 INDELs), followed by Chr. 06 (1300 INDELs), while the lowest count was on Chr. 09, with 241 INDELs (Table 2). Applying filtering criteria (MCR: 100%, MAF: 0.0, max allele frequency: 0.0), the total INDELs in the landrace group (2240) were fewer than those in the improved group (5740) (Table 2). After combining and further filtering with MCR (100%), MAF (0.2), max allele frequency (1.0), and HR (0.0), 2240 INDELs were identified, with a noteworthy location on Chr. 01 (1623 out of 2240 INDELs). No differences were found between the two groups in Chr. 09 and Chr. 10. Chr. 02, Chr. 05, Chr. 11, and Chr. 12 exhibited only a few INDELs (Table 2).

Table 2.
Number of INDELs identified in rice accessions based on 20 whole-genome sequences.
This study focused on insertions or deletions between the two groups, with a minimum length of 20 bp. This analysis revealed 22 INDEL markers, of which 14 were on Chr. 01, 1 was on Chr. 03, 2 were on Chr. 04, 2 were on Chr. 06, and 3 were on Chr. 07 (Table 3). Additionally, only one INDEL marker (S01_14359423) involved an insertion; the rest were deletions. This implies that 21 INDEL markers differed from the Nipponbare genome (Japonica rice cultivar). Almost all genome types of the improved group differed from the genome reference, except for three INDEL markers (S01_14359423, S06_24284080, and S07_1972486) (Table 3).

Table 3.
Different characteristics of INDELs with minimum lengths of 20 bp between two accession groups.
3.2. Gene Related to Photoperiod Sensitivity
To identify candidate genes that are associated with photoperiod sensitivity, the locations of the 22 INDELs were examined on the website https://snp-seek.irri.org/ (accessed 10 June 2023). Ten INDELs located in 10 genes were found in landrace and improved rice accessions. Each gene had one INDEL. Seven genes were located on chromosome 01, two on chromosome 04, and one on chromosome 07 (Table 4). The functions of these genes included MADS-box family genes, acyl-ACP thioesterase, DUF26 kinases, carboxyl-terminal peptidase, pentatricopeptide, CBS domain-containing protein, homeobox-associated leucine zipper, WD domain, G-beta repeat domain-containing protein, and CAMK_KIN1/SNF1/Nim1_like_AMPKh.1—CAMK, the latter of which includes calcium/calmodulin-dependent protein kinases. One gene (LOC_Os01g23780), encoding a putative regulator of rice flowering [43], was identified from 13373620 to 13374665 on chromosome 01. In this gene’s position, the improved rice accession group exhibited a 20 bp deletion compared to the landrace accession group.

Table 4.
Comprehensive INDEL information for the 20 selected accessions.
3.3. Differences in Photoperiod-Sensitivity Group in Gene LOC_Os01g23780
The MADS-box family gene LOC_Os01g23780, which may be involved in rice flowering [43], was located in the INDEL region between the landrace (photoperiod sensitive) and improved varieties (photoperiod insensitive). We therefore designed the primer based on Primer3Plus (http://primer3plus.ut.ee/cgi-bin/primer3plus/primer3plus.cgi, accessed on 7 December 2023), and we obtained the photoperiod-sensitive primer (PS primer) with the following information: left primer 5′-GAGGGAGCTCTCCATCCTCT-3′ and right primer 5′GCTTCAACTCGAGGCACTCT-3′). It occurred at positions 13373620 to 13373910 in chromosome 01, and produced two prominent diagnostic fragments of approximately 203 bp and 183 bp for landrace rice and improved rice, respectively. In this position, the improved rice accession group was deleted at 20bp from the normal sequence of Nipponbare references or indica landrace rice.
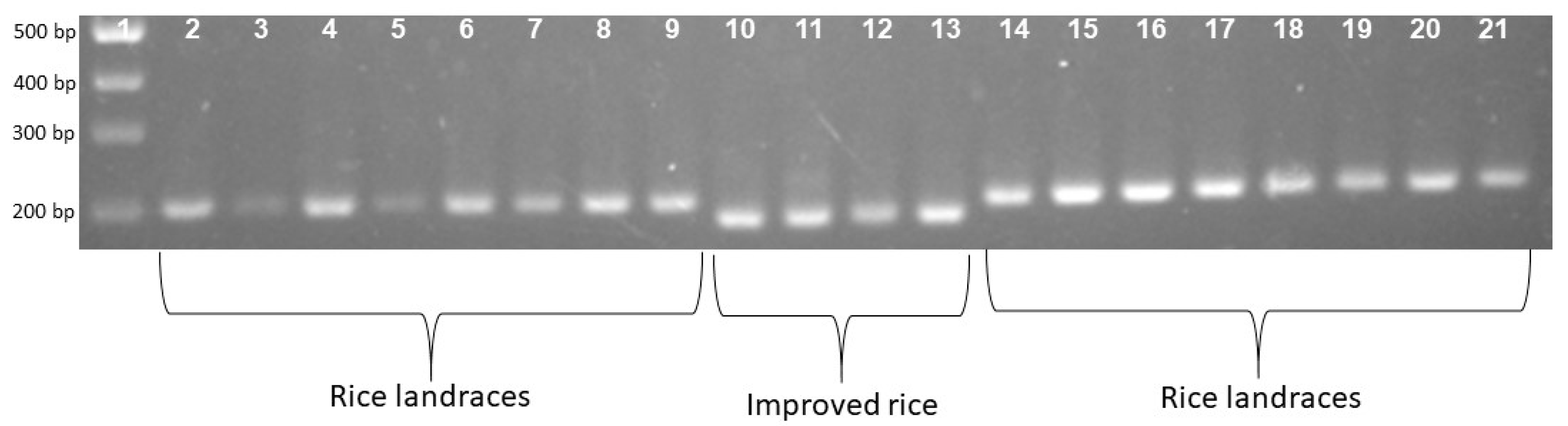
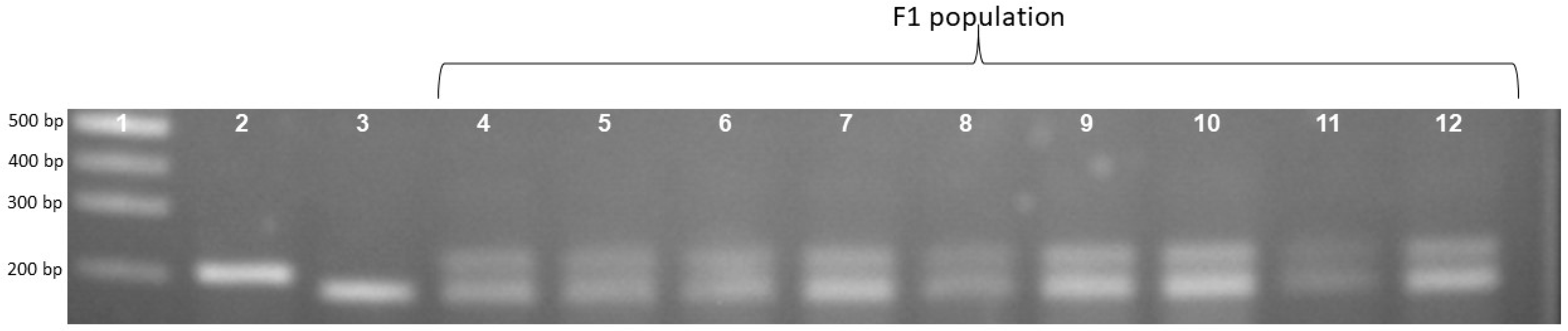
To confirm the phenotype diversity of two distinct photoperiod-sensitive groups, we designed a primer to screen the action of the gene LOC_Os01g23780. The results indicate that the PCR product of the landrace accession group was larger than that of the improved group at about 20bp (Figure 1). Each of the 16 landrace accessions exhibited a PCR product measuring 203 bp. In contrast, the four improved rice accessions displayed a band of 183 bp (Figure 1). In addition, Figure 2 illustrates that the F1 population resulting from the cross between Nang Thom Cho Dao and MTL372 exhibited two bands across all hybrid individuals. Specifically, the Nang Thom Cho Dao accession displayed a band of 203 bp, while MTL372 exhibited a band of 183 bp.
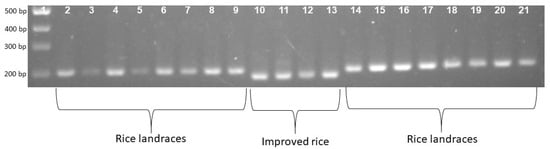
Figure 1.
Gel electrophoresis patterns depicting locus phase amplification across all 20 genotypes. Rice landrace produces a PCR product with a size of 203 bp. Improved rice produces a PCR product with a size of 183 bp. 1: Marker labels; 2–9: rice landraces; 10–13: improved rice; 14–21: rice landraces.
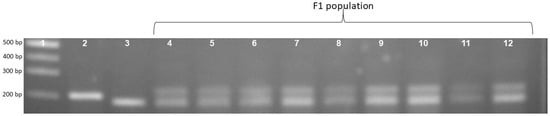
Figure 2.
Gel electrophoresis image illustrating locus phase amplification patterns in the F1 population. Rice landrace produces a PCR product with a size of 203 bp and improved rice produces a PCR product with a size of 183 bp, whereas F1 population has bands with sizes of 203 bp and 183 bp for whole hybrid population. 1: Marker labels; 2: Nang Thom Cho Dao (rice landraces); 3: MTL372 (Improved rice); 4–12: F1.
Phenotypic analyses of heading dates and agronomic traits were carried out on 20 accessions in Hong Dan district, Bac Lieu province (latitude: 9°29′20.6″ N; longitude: 105°30′24.1″ E) from September 2020 to March 2021. Among these accessions, the heading dates of 16 landraces ranged from 24 December 2020, to 31 January 2021 (Supplementary Table S1). In contrast, four improved accessions were not affected by short-day conditions, displaying heading patterns determined by their genetic traits (Supplementary Table S1). The improved-accession group displayed shorter plant heights in comparison to the landrace group (Supplementary Table S1).
The INDEL marker S01_13373751, located in the gene LOC_Os01g23780, was employed to investigate the genetic diversity within the rice genome via the Rice SNP-Seek database (3K). It revealed that five sub-populations (aro, japx, subtrop, temp, and trop) exhibited identical genotypes to those of the Nipponbare and landrace groups (Table 5). Therefore, this INDEL marker distinguishes landrace and improved groups and separates the genotype of the five subgroups above from the improved indica rice group (Table 5).

Table 5.
Number of accessions displaying deletion at position 13373751 on chromosome 01 from the 3K database.
4. Discussion
Culturing improved rice varieties with photoperiod insensitivity and a short growth duration has been prominent in Vietnam and other Asian countries, driven by agricultural intensification for enhanced food security and export [44]. Photoperiod sensitivity is important to rice breeders [45]. In the Vietnamese Mekong Delta, since the 1980s, improved rice varieties have gradually replaced rice landraces with photoperiod sensitivity and long growth durations [46]. The key distinction between landrace and improved rice lies in their responsiveness to day length, impacting crucial aspects such as flowering, grain yield, and overall rice productivity. To investigate photoperiod sensitivity, numerous earlier studies have concentrated on the heading date [3,6,45] because rice typically utilizes photoperiodism to regulate the timing of flowering. The progression of rice development leading up to floral initiation is characterized by two consecutive phases: the basic vegetative growth phase, which is photoperiod-insensitive, and the photoperiod-sensitive phase [47].
INDEL polymorphisms are the second most prevalent type of genetic variation, following SNPs, in humans and plants [48]. In addition, INDEL markers easily distinguish the genotypes among individuals based on their size. Therefore, developing INDEL markers is becoming popular for crop genetic studies [48].
In our study, identifying 2240 INDELs from the genome sequencing of 20 accessions, representing two distinct groups of landrace and improved rice, revealed significant differences. Chr. 01 exhibited the highest discrepancy, with 1263 INDELs, followed by chr. 04 (220 INDELs), 06 (171 INDELs), and 03 (123 INDELs), collectively accounting for 79.3% of all INDELs (1777 INDELs). The information on these INDELs can be found in the 3K database, where the highest number of INDELs is in chr. 01 [41]. Fourteen INDELs in Chr. 01 exhibited insertions or deletions of at least 20 bp, suggesting their potential utility for distinguishing between the two groups. An INDEL at the position of 13373751 bp on chr. 01, involving a 20 bp deletion in the improved rice group compared to the landrace group, was identified. This INDEL resides in the codon of the gene LOC_Os01g23780, classified as OsMADS95—MADS-box, a gene known for its involvement in floral organ development, flower development, and overall plant growth and development [43]. LOC_Os01g23780 plays a crucial role during the reproduction phase of rice [43].
The INDEL candidate marker for the gene LOC_Os01g23780 successfully distinguished landrace and improved rice groups. Utilizing this INDEL marker allows breeders to assess the outputs of crosses between landrace and improved rice parents. Successful crossings can be identified by the presence of two bands in the F1 individual using the PS marker. Additionally, applying this marker can enhance the genetic gain and economic efficiency of a targeted breeding strategy [49]. Furthermore, this INDEL marker exhibited broader applicability, enabling the classification of five subgroups (aro, japx, subtrop, temp, and trop) within the indica improved rice category (Table 5). This finding underscores the potential significance of the identified INDEL marker in both distinguishing and categorizing rice accessions, providing valuable insights for further breeding programs and genetic studies.
5. Conclusions
This study was undertaken to elucidate the genetic distinctions, specifically the number of INDELs, between landrace and improved rice groups. A significant discovery is that a substantial proportion of the identified INDELs, totaling almost 2240, were concentrated on chromosome 01. In addition, the specific INDEL position S01_13373751 bp within the gene LOC_Os01g23780 emerged as a focal point of significance.
The outcomes of the current study lend strong support to the proposition that the INDEL marker S01_13373751 holds practical utility. This marker demonstrates efficacy in distinguishing improved rice accessions within the indica accessions. Further, it serves as a discriminative tool between improved rice accessions originating from indica and Japonica rice accessions. This study underscores the potential applicability of the INDEL marker in breeding strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology13050358/s1, Table S1: Variation in heading date, days to flowering, and plant height.
Author Contributions
N.T.T. designed and carried out the research, analyzed the data, and wrote and improved the manuscript. D.K.N. discussed the initial ideas and preliminary results of the research and improved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded in part by the Can Tho University Improvement Project VN14-P6, supported by a Japanese ODA loan.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequence data of the 20-accession MDI set generated in this study have been deposited in the NCBI database (accession number PRJNA787403).
Acknowledgments
We gratefully acknowledge Kishima and Koide (Research Faculty of Agriculture, Hokkaido University) for their valuable suggestions concerning this study.
Conflicts of Interest
The authors declare that there are no conflicts of interest associated with this publication. The manuscript has been read and approved for publication by all authors.
Abbreviations
| INDEL | Insertion/deletion |
| QTL | Quantitative trait locus |
| GWAS | Genome-wide association study |
| SNP | Single-nucleotide polymorphism |
| PHISE | Photoperiod insensitivity |
| PCR | Polymorphism chain reaction |
| MCR | Missing call rate |
| MAF | Minor allele frequency |
| HR | Heterozygosity rate |
| MDI | Mekong Delta Development Research Institute |
References
- Izawa, T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007, 58, 3091–3097. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.; Kumar, M.; Molla, K.A.; Chakraborty, K. Abiotic stresses in rice production: Impacts and management. Oryza-Int. J. Rice 2021, 58, 103–125. [Google Scholar] [CrossRef]
- Zong, W.; Ren, D.; Huang, M.; Sun, K.; Feng, J.; Zhao, J.; Xiao, D.; Xie, W.; Liu, S.; Zhang, H. Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading. New Phytol. 2021, 229, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A. Flowering Time and Photoperiod Sensitivity in Rice: Key Players and Their Interactions Identified; Oxford University Press: Oxford, UK, 2022. [Google Scholar]
- Padukkage, D.; Senanayake, G.; Geekiyanage, S. Photoperiod sensitivity of very early maturing Sri Lankan rice for flowering time and plant architecture. Open Agric. 2017, 2, 580–588. [Google Scholar] [CrossRef]
- Chandraratna, M.F. Genetics of photoperiod sensitivity in rice. J. Genet. 1955, 53, 215–223. [Google Scholar] [CrossRef]
- Sun, C.; Chen, D.; Fang, J.; Wang, P.; Deng, X.; Chu, C. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 2014, 5, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Ichitani, K.; Okumoto, Y.; Tanisaka, T. Photoperiod sensitivity gene of Se-1 locus found in photoperiod insensitive rice cultivars of the northern limit region of rice cultivation. Jpn. J. Breed. 1997, 47, 145–152. [Google Scholar] [CrossRef][Green Version]
- Izawa, T.; Oikawa, T.; Sugiyama, N.; Tanisaka, T.; Yano, M.; Shimamoto, K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002, 16, 2006–2020. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef]
- Takahashi, Y.; Shomura, A.; Sasaki, T.; Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 2001, 98, 7922–7927. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.-H.; Wang, P.; Chen, H.-X.; Zhou, H.-J.; Li, Q.-P.; Wang, C.-R.; Ding, Z.-H.; Zhang, Y.-S.; Yu, S.-B.; Xing, Y.-Z. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Liu, X.; Zhao, Z.; Jiang, L.; Gao, H.; Zhang, Y.; Zheng, M.; Chen, L.; Liu, S.; Zhai, H. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep. 2011, 30, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Ogiso-Tanaka, E.; Hori, K.; Ebana, K.; Ando, T.; Yano, M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 2012, 53, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zheng, X.-M.; Fei, G.; Chen, J.; Jin, M.; Ren, Y.; Wu, W.; Zhou, K.; Sheng, P.; Zhou, F. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013, 9, e1003281. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. H d16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013, 76, 36–46. [Google Scholar] [CrossRef]
- Ogiso-Tanaka, E.; Matsubara, K.; Yamamoto, S.-i.; Nonoue, Y.; Wu, J.; Fujisawa, H.; Ishikubo, H.; Tanaka, T.; Ando, T.; Matsumoto, T. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS ONE 2013, 8, e75959. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, X.-M.; Lu, G.; Zhong, Z.; Gao, H.; Chen, L.; Wu, C.; Wang, H.-J.; Wang, Q.; Zhou, K. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 2775–2780. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, M.; Zheng, X.-M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Inoue, H.; Okumoto, Y.; Tanisaka, T. A novel gene ef1-h conferring an extremely long basic vegetative growth period in rice. Crop. Sci. 2002, 42, 348–354. [Google Scholar] [CrossRef]
- Yang, H.; Tao, Y.; Zheng, Z.; Li, C.; Sweetingham, M.W.; Howieson, J.G. Application of next-generation sequencing for rapid marker development in molecular plant breeding: A case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genom. 2012, 13, 318. [Google Scholar] [CrossRef]
- Shavrukov, Y.; Suchecki, R.; Eliby, S.; Abugalieva, A.; Kenebayev, S.; Langridge, P. Application of next-generation sequencing technology to study genetic diversity and identify unique SNP markers in bread wheat from Kazakhstan. BMC Plant Biol. 2014, 14, 258. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Satya, P. Next generation sequencing technologies for next generation plant breeding. Front. Plant Sci. 2014, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.M.; Arif, M.; Aslam, K.; Shabir, G.; Thomson, M.J. Genetic diversity analysis of Pakistan rice (Oryza sativa) germplasm using multiplexed single nucleotide polymorphism markers. Genet. Resour. Crop. Evol. 2016, 63, 1113–1126. [Google Scholar] [CrossRef]
- Chen, K.; Luan, M.; Xiong, H.; Chen, P.; Chen, J.; Gao, G.; Huang, K.; Zhu, A.; Yu, C. Genome-wide association study discovered favorable single nucleotide polymorphisms and candidate genes associated with ramet number in ramie (Boehmeria nivea L.). BMC Plant Biol. 2018, 18, 345. [Google Scholar] [CrossRef]
- Wu, D.-H.; Wu, H.-P.; Wang, C.-S.; Tseng, H.-Y.; Hwu, K.-K. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Steele, K.A.; Ogden, R.; McEwing, R.; Briggs, H.; Gorham, J. InDel markers distinguish Basmatis from other fragrant rice varieties. Field Crops Res. 2008, 105, 81–87. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhang, S.; Wang, Y.; Ford-Lloyd, B.V.; Tu, M.; Jin, X.; Wu, Y.; Yan, H.; Yang, X.; Liu, P. Differentiation and distribution of indica and japonica rice varieties along the altitude gradients in Yunnan Province of China as revealed by InDel molecular markers. Genet. Resour. Crop. Evol. 2010, 57, 891–902. [Google Scholar] [CrossRef]
- Ji, Q.; Lu, J.; Chao, Q.; Zhang, Y.; Zhang, M.; Gu, M.; Xu, M. Two sequence alterations, a 136 bp InDel and an A/C polymorphic site, in the S5 locus are associated with spikelet fertility of indica-japonica hybrid in rice. J. Genet. Genom. 2010, 37, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Yoshida, H.; Ashikawa, I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Rao, I.S.; Srikanth, B.; Kishore, V.H.; Suresh, P.B.; Chaitanya, U.; Vemireddy, L.; Voleti, S.; Subbarao, L.; Rani, N.S.; Sundaram, R. Indel polymorphism in sugar translocation and transport genes associated with grain filling of rice (Oryza sativa L.). Mol. Breed. 2011, 28, 683–691. [Google Scholar]
- Tam, N.T.; Dwiyanti, M.S.; Koide, Y.; Nagano, A.J.; Ky, H.; Tin, H.Q.; Hien, N.L.; Dung, L.V.; Kishima, Y. Profiling SNP and Nucleotide Diversity to Characterize Mekong Delta Rice Landraces in Southeast Asian Populations. Plant Genome 2019, 12, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, U357–U359. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491. [Google Scholar] [CrossRef]
- Adedze, Y.M.N.; Lu, X.; Xia, Y.; Sun, Q.; Nchongboh, C.G.; Alam, M.A.; Liu, M.; Yang, X.; Zhang, W.; Deng, Z.; et al. Agarose-resolvable InDel markers based on whole genome re-sequencing in cucumber. Sci. Rep. 2021, 11, 3872. [Google Scholar] [CrossRef] [PubMed]
- Mansueto, L.; Fuentes, R.R.; Borja, F.N.; Detras, J.; Abriol-Santos, J.M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A.; et al. Rice SNP-seek database update: New SNPs, indels, and queries. Nucleic Acids Res. 2016, 45, D1075–D1081. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Li, C.-C. Te-Tzu Chang International Rice Research Institute. In Rice: Volume I. Production; Spring Science+Business Media, LLC: Berlin/Heidelberg, Germany, 2013; p. 23. [Google Scholar]
- Poonyarit, M.; Mackill, D.; Vergara, B. Genetics of photoperiod sensitivity and critical daylength in rice. Crop. Sci. 1989, 29, 647–652. [Google Scholar] [CrossRef]
- Le Coq, J.-F.; Trébuil, G.; Dufumier, M. History of rice production in the Mekong Delta. In Smallholders and Stockbreeders; Brill: Leyton, The Netherlands, 2004; pp. 163–185. [Google Scholar]
- Yoshitake, Y.; Yokoo, T.; Saito, H.; Tsukiyama, T.; Quan, X.; Zikihara, K.; Katsura, H.; Tokutomi, S.; Aboshi, T.; Mori, N. The effects of phytochrome-mediated light signals on the developmental acquisition of photoperiod sensitivity in rice. Sci. Rep. 2015, 5, 7709. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wei, H.; Cheng, W.; Yang, S.; Zhao, Y.; Li, X.; Luo, D.; Zhang, H.; Feng, X. Development of INDEL Markers for Genetic Mapping Based on Whole Genome Resequencing in Soybean. G3 Genes|Genomes|Genet. 2015, 5, 2793–2799. [Google Scholar] [CrossRef]
- Kuchel, H.; Fox, R.; Reinheimer, J.; Mosionek, L.; Willey, N.; Bariana, H.; Jefferies, S. The successful application of a marker-assisted wheat breeding strategy. Mol. Breed. 2007, 20, 295–308. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).