Genome-Wide Association Study of Early Vigour-Related Traits for a Rice (Oryza sativa L.) japonica Diversity Set Grown in Aerobic Conditions
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experiment Location
2.2. Plant Material and Experimental Design
2.3. Cultural Details of Glasshouse Experiments
2.4. Field Experiments Management
2.5. Measurements of Traits
2.5.1. Glasshouse Traits Measurements
2.5.2. Field Traits Measurements
2.6. Phenotypic Data Analysis
- vblup—the average standard error of differences between BLUPs squared.
- var_g—the genotypic variance.
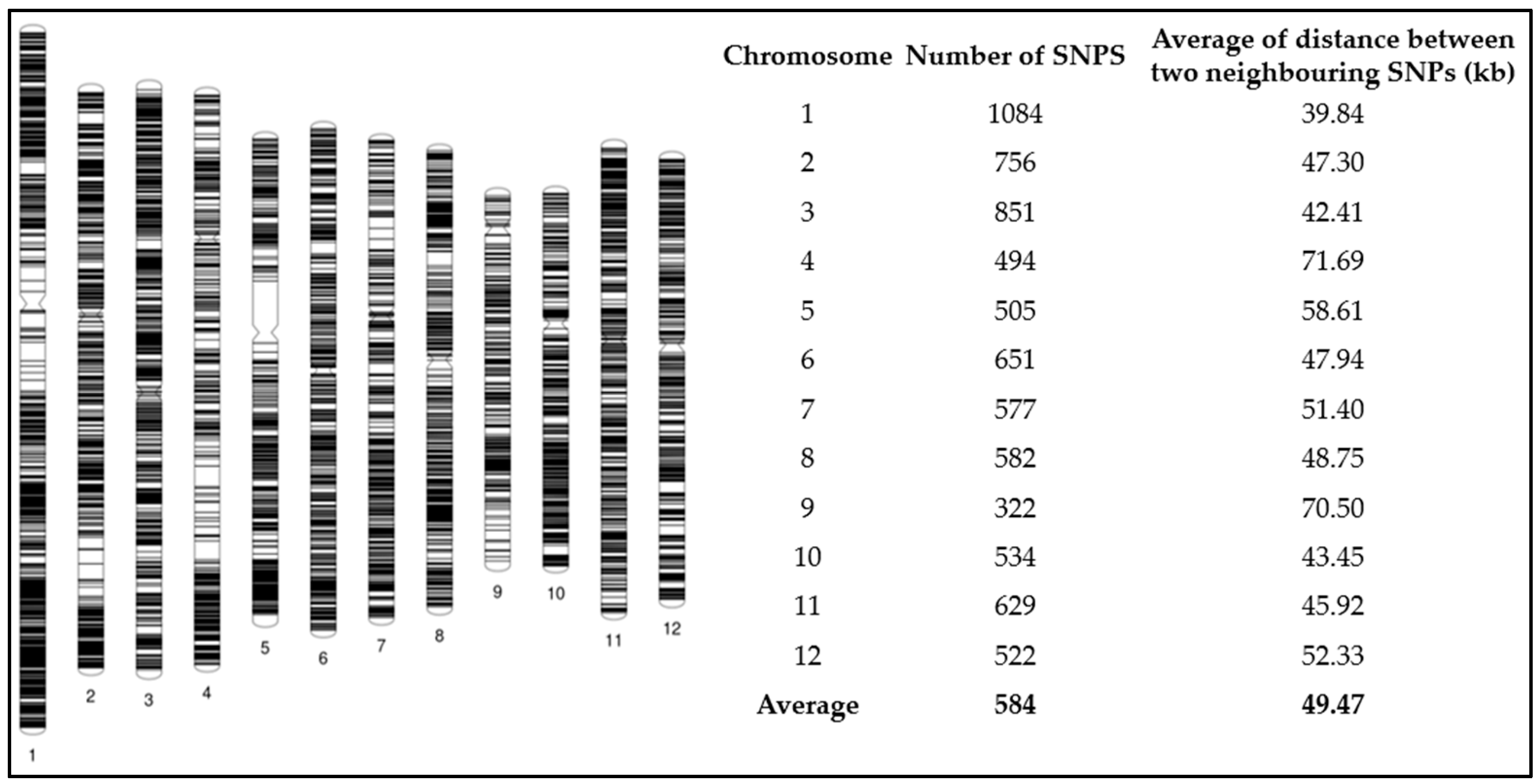
2.7. SNP Genotyping
2.8. Genome-Wide Association Study
2.9. QTL Nomenclature and Identification of Candidate Genes
3. Results
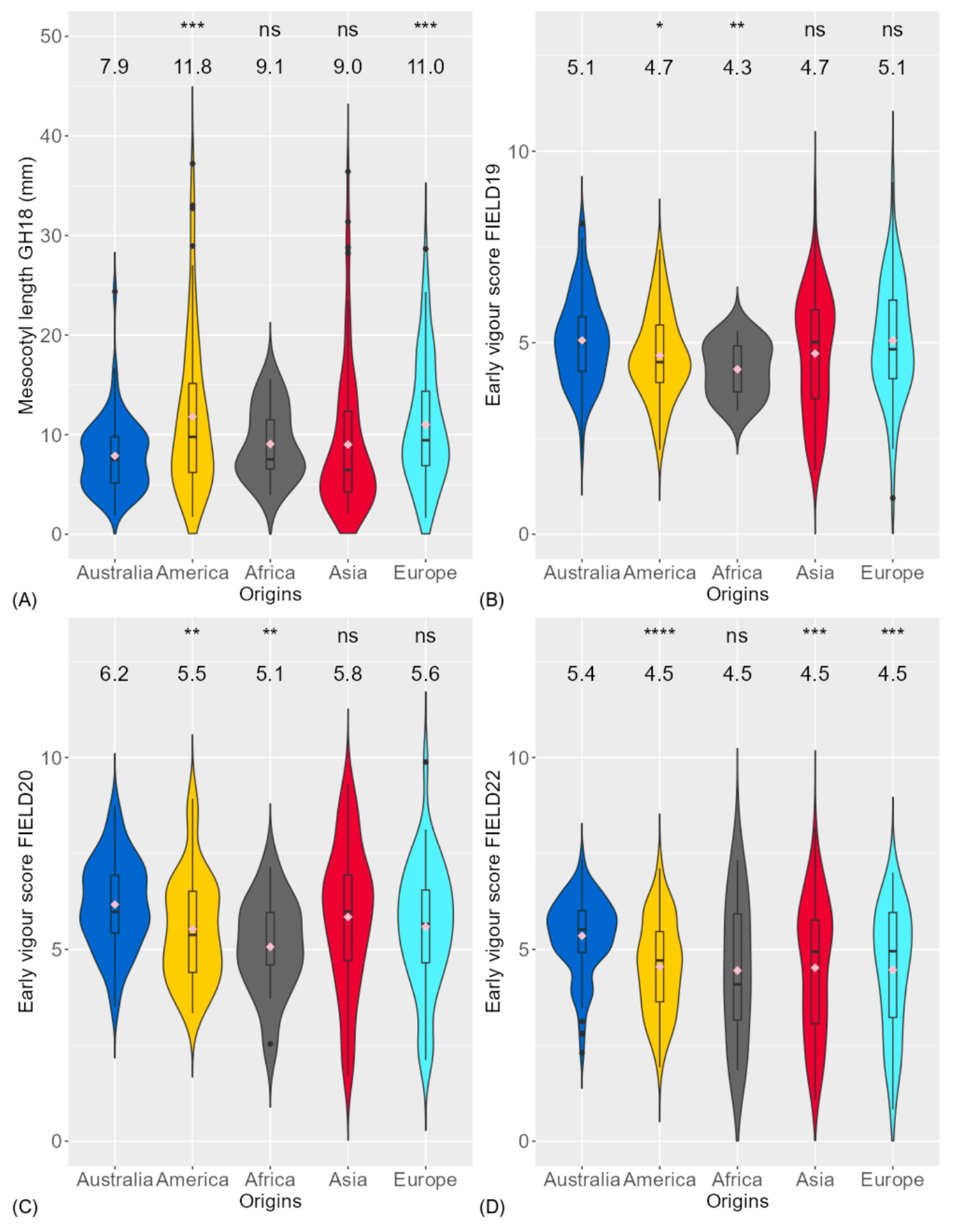
3.1. Phenotypic Variation of Traits
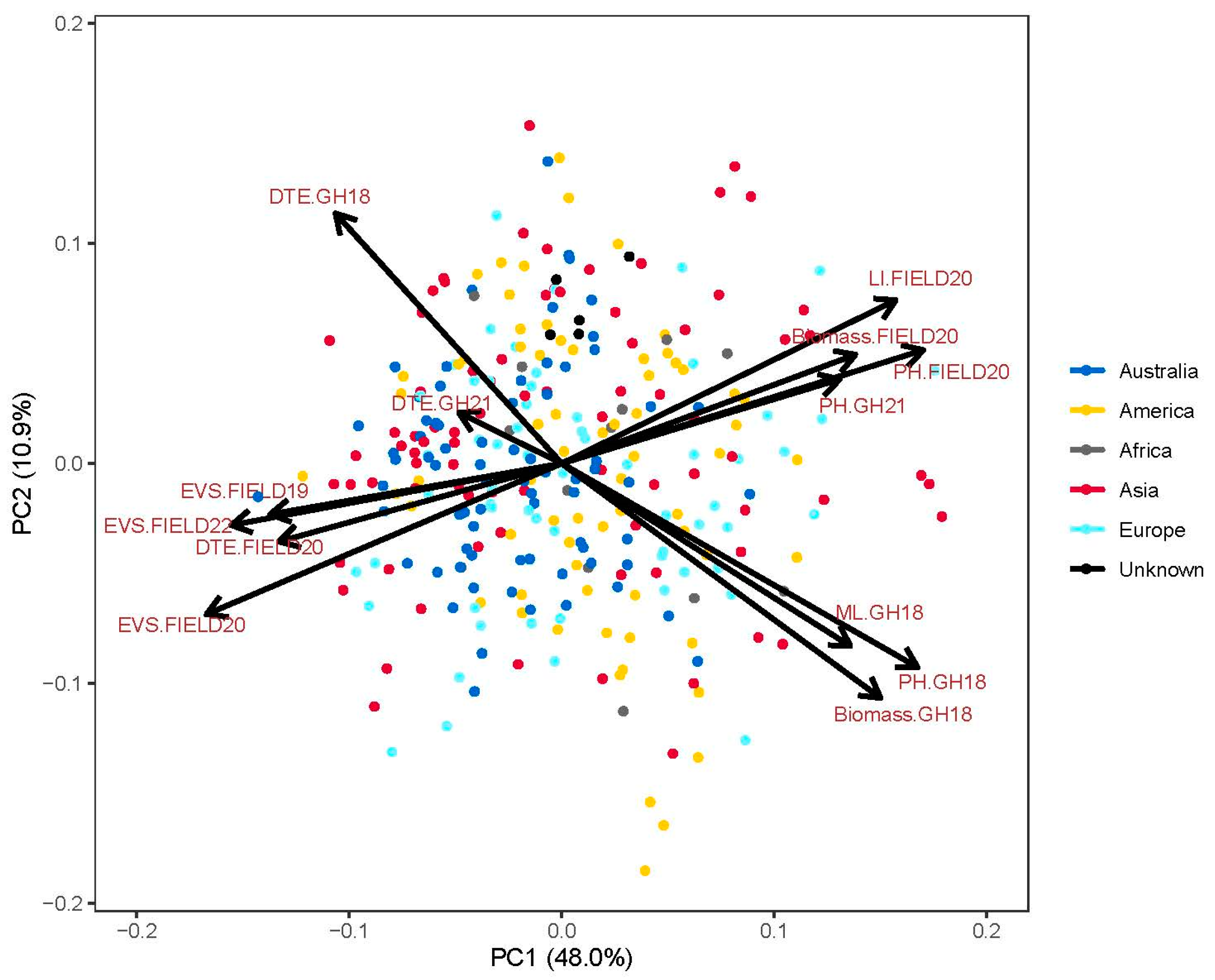
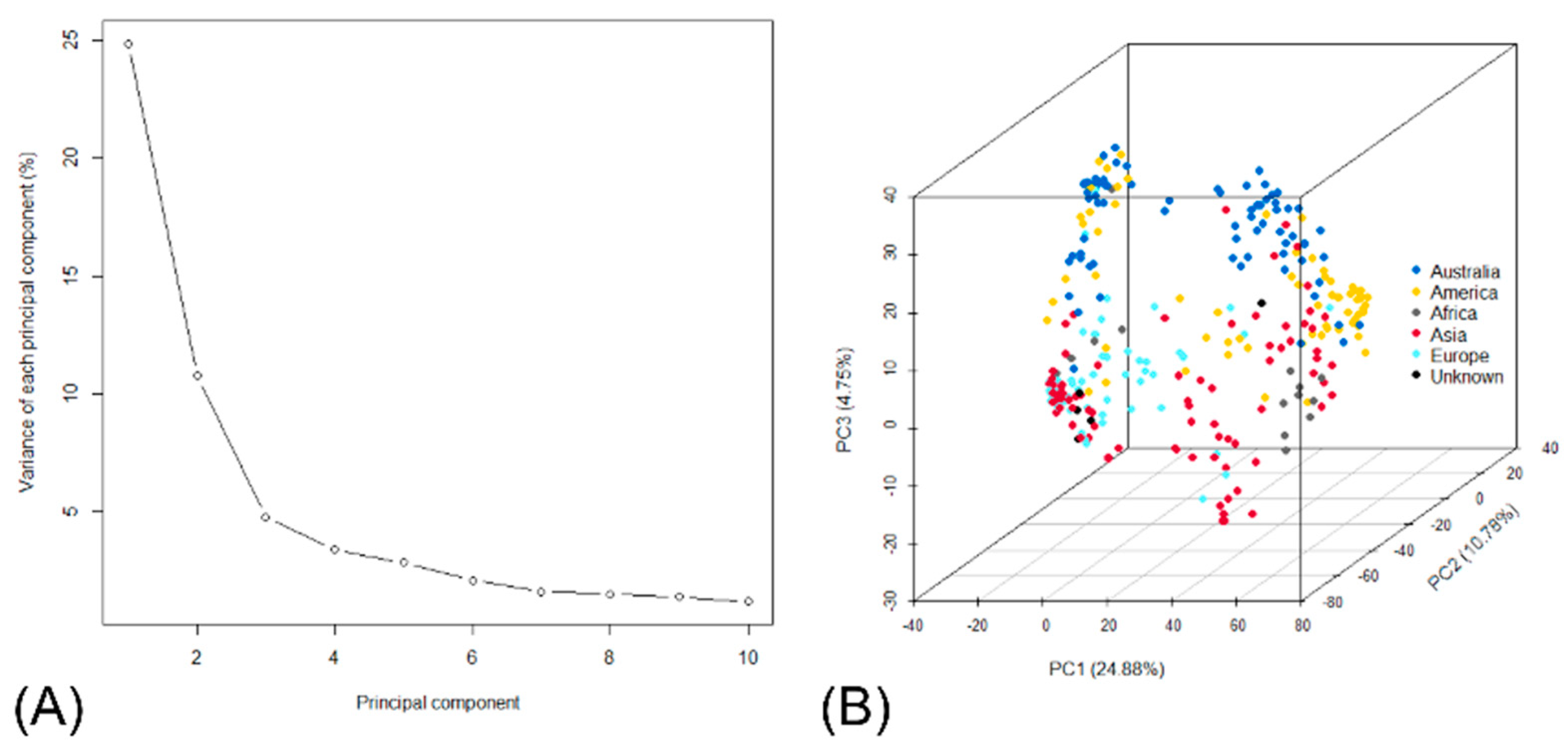
3.2. Population Structure
3.3. GWAS
3.4. Identification of Candidate Genes
4. Discussion
4.1. Genotypic Variation and Relationship of Traits
4.2. GWAS Analysis
4.3. Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunn, B.; Dunn, T.; Mitchell, J.; Brinkhoff, J. Effects of plant population and row spacing on grain yield of aerial-sown and drill-sown rice. Crop Pasture Sci. 2020, 71, 219–228. [Google Scholar] [CrossRef]
- Lal, B.; Nayak, A.K.; Gautam, P.; Tripathi, R.; Singh, T.; Katara, J. Aerobic rice: A water saving approach for rice production. Int. J. Res. BioSci. 2012, 1, 1–6. [Google Scholar]
- Chamara, B.S.; Marambe, B.; Kumar, V.; Ismail, A.M.; Septiningsih, E.M.; Chauhan, B.S. Optimizing sowing and flooding depth for anaerobic germination-tolerant genotypes to enhance crop establishment, early growth, and weed management in dry-seeded rice (Oryza sativa L.). Front. Plant Sci. 2018, 9, e1654. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhan, J.; Wang, J.; Li, X.; Wei, Y.; Wu, H.; Dai, H. Development Status of Direct Seeding Rice and Study on Response Mechanism of Submergence. Open Access Libr. J. 2022, 9, 1–13. [Google Scholar] [CrossRef]
- Rao, A.; Johnson, D.; Sivaprasad, B.; Ladha, J.; Mortimer, A. Weed management in direct-seeded rice. Adv. Agron. 2007, 93, 153–255. [Google Scholar]
- Mahender, A.; Anandan, A.; Pradhan, S.K. Early seedling vigour, an imperative trait for direct-seeded rice: An overview on physio-morphological parameters and molecular markers. Planta 2015, 241, 1027–1050. [Google Scholar] [CrossRef]
- Namuco, O.S.; Cairns, J.E.; Johnson, D.E. Investigating early vigour in upland rice (Oryza sativa L.): Part I. Seedling growth and grain yield in competition with weeds. Field Crops Res. 2009, 113, 197–206. [Google Scholar] [CrossRef]
- Yang, J.; Yang, G.; Yang, M.; Su, L.; Xia, A.; Li, D.; Huang, C.; Zhou, D.; Liu, Y.; Wang, H.; et al. Quantitative trait locus analysis of seed germination and early seedling growth in rice. Front. Plant Sci. 2019, 10, e1582. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhang, R.; Chen, J.; Cao, F.; Jiang, L.; Zou, Y. Morphological and physiological traits of seeds and seedlings in two rice cultivars with contrasting early vigor. Plant Prod. Sci. 2017, 20, 95–101. [Google Scholar] [CrossRef]
- Ros, C.; Bell, R.; White, P. Seedling vigour and the early growth of transplanted rice (Oryza sativa). Plant Soil 2003, 252, 325–337. [Google Scholar] [CrossRef]
- Shi, Z.; Chang, T.-G.; Chen, F.; Zhao, H.; Song, Q.; Wang, M.; Wang, Y.; Zhou, Z.; Wang, C.; Zhou, S.-C.; et al. Morphological and physiological factors contributing to early vigor in the elite rice cultivar 9311. Sci. Rep. 2020, 10, 14813. [Google Scholar] [CrossRef] [PubMed]
- Ward, R. Rice Growing Guide; NSW Department of Primary Industries: Orange, NSW, Australia, 2021.
- Zhan, J.; Lu, X.; Liu, H.; Zhao, Q.; Ye, G. Mesocotyl elongation, an essential trait for dry-seeded rice (Oryza sativa L.): A review of physiological and genetic basis. Planta 2019, 251, 27. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Sasaki, K.; Kang, J.-W.; Sato, T.; Song, W.-Y.; Ahn, S.-N. Mesocotyl elongation is essential for seedling emergence under deep-seeding condition in rice. Rice 2017, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Banayo, N.P.M.C.; Bueno, C.S.; Kashiwagi, J.-i.; Nakashima, T.; Corales, A.M.; Garcia, R.; Sandhu, N.; Kumar, A.; Kato, Y. Longer mesocotyl contributes to quick seedling establishment, improved root anchorage, and early vigor of deep-sown rice. Field Crops Res. 2018, 228, 84–92. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Yu, S.-B.; Yu, T.; Huang, Z.; Zhu, Y.-G. Mapping quantitative trait loci (QTLs) for seedling-vigor using recombinant inbred lines of rice (Oryza sativa L.). Field Crops Res. 2005, 91, 161–170. [Google Scholar] [CrossRef]
- Cairns, J.; Namuco, O.; Torres, R.; Simborio, F.; Courtois, B.; Aquino, G.; Johnson, D. Investigating early vigour in upland rice (Oryza Sativa L.): Part ii. Identification of QTLs controlling early vigour under greenhouse and field conditions. Field Crops Res. 2009, 113, 207–217. [Google Scholar] [CrossRef]
- Singh, U.M.; Yadav, S.; Dixit, S.; Ramayya, P.J.; Devi, M.N.; Raman, K.A.; Kumar, A. QTL hotspots for early vigor and related traits under dry direct-seeded system in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, e286. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Niu, A.; Cai, H.; Zhao, Y.; Liu, J.; Zhu, Y.; Zhang, Z. Genetic dissection of seedling and early vigor in a recombinant inbred line population of rice. Plant Sci. 2007, 172, 212–220. [Google Scholar] [CrossRef]
- Lee, H.-S.; Sasaki, K.; Higashitani, A.; Ahn, S.-N.; Sato, T. Mapping and characterization of quantitative trait loci for mesocotyl elongation in rice (Oryza sativa L.). Rice 2012, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Jiang, S.-K.; Feng, L.-L.; Xu, Z.-J.; Chen, W.-F. Analysis of QTLs for mesocotyl length in rice (Oryza Sativa L.). Acta Agron. Sin. 2010, 36, 1108–1113. [Google Scholar] [CrossRef]
- Zeng, M.; Yang, J.; Wu, K.; Wang, H.; Sun, K.; Chen, Z.; Guo, T.; Chen, C. Genome-wide association study reveals early seedling vigour-associated quantitative trait loci in indica rice. Euphytica 2021, 217, 141. [Google Scholar] [CrossRef]
- Wang, F.; Longkumer, T.; Catausan, S.C.; Calumpang, C.L.F.; Tarun, J.A.; Cattin-Ortola, J.; Ishizaki, T.; Pariasca Tanaka, J.; Rose, T.; Wissuwa, M.; et al. Genome-wide association and gene validation studies for early root vigour to improve direct seeding of rice. Plant Cell Environ. 2018, 41, 2731–2743. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, Q.; Wang, C.-C.; Liu, Z.-X.; Jiang, Y.-J.; Zhai, L.-Y.; Zheng, T.-Q.; Xu, J.-L.; Li, Z.-K. Genetic dissection of seedling vigour in a diverse panel from the 3000 rice (Oryza sativa L.) genome project. Sci. Rep. 2019, 9, 4804. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Feng, F.; Lian, X.; Teng, X.; Wei, H.; Yu, H.; Xie, W.; Yan, M.; Fan, P.; Li, Y.; et al. Genome-wide association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 2015, 15, 218. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhan, J.; Li, J.; Lu, X.; Liu, J.; Wang, Y.; Zhao, Q.; Ye, G. Genome-wide association study (GWAS) for mesocotyl elongation in rice (Oryza sativa L.) under multiple culture conditions. Genes 2020, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, W.; Jiang, C.; Wang, X.; Xiong, H.; Todorovska, E.G.; Yin, Z.; Chen, Y.; Wang, X.; Xie, J.; et al. Genetic architecture and candidate genes for deep-sowing tolerance in rice revealed by Non-syn GWAS. Front. Plant Sci. 2018, 9, e332. [Google Scholar] [CrossRef] [PubMed]
- Coombes, N. DiGGer: DiGGer Design Generator under Correlation and Blocking; NSW DPI Biometrics: Sydney, NSW, Australia, 2009.
- Butler, D.; Cullis, B. On Model Based Design of Comparative Experiments in R; National Institute for Applied Statistics Research Australia, University of Wollongong: Wollongong, NSW, Australia, 2022. [Google Scholar]
- Isbell, R.; National Committee on Soil and Terrain. The Australian Soil Classification; CSIRO Publishing: Clayton, VIC, USA, 2021.
- Buxton, J.W.; Jia, W. A controlled water table irrigation system for hydroponic lettuce production. In Proceedings of the International Symposium on Growing Media and Hydroponics, Kassandra, Greece, 31 August–6 September 1999; pp. 281–288. [Google Scholar]
- Hunter, M.; Mitchell, J.; Dieters, M. Semi-automated, non-weighing, pot-in-bucket (PIB), water management in pot plant culture. In Proceedings of the 16th Australian Agronomy Conference, Armidale, NSW, Australia, 14–18 October 2012. [Google Scholar]
- IRRI. Standard Evaluation System for Rice; International Rice Research Institute: Los Baños, Laguna, Philippines, 2002. [Google Scholar]
- Butler, D.; Cullis, B.R.; Gilmour, A.; Gogel, B. ASReml-R Reference Manual; The State of Queensland, Department of Primary Industries Fisheries: Brisbane, QLD, Australia, 2009.
- Isik, F.; Holland, J.; Maltecca, C. Genetic Data Analysis for Plant and Animal Breeding; Springer: Berlin/Heidelberg, Germany, 2017; Volume 400. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. tidyverse: Easily install and load the ‘tidyverse’. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Slowikowski, K. ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. 2020. Available online: https://cran.r-project.org/web/packages/ggrepel/index.html (accessed on 1 April 2023).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2020. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 1 April 2023).
- Vinarao, R.; Proud, C.; Zhang, X.; Snell, P.; Fukai, S.; Mitchell, J. Stable and Novel Quantitative Trait Loci (QTL) Confer Narrow Root Cone Angle in an Aerobic Rice (Oryza sativa L.) Production System. Rice 2021, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Stekhoven, D. missForest: Nonparametric Missing Value Imputation Using Random Forest, R Package Version 1.5. 2022. Available online: https://cran.r-project.org/web/packages/missForest/index.html (accessed on 1 April 2023).
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, S.S.; Bayissa, K.N.; Matros, A.; Pillen, K.; Ordon, F.; Wehner, G. Genome wide association study of Ethiopian barley for terminal drought stress tolerance under field and climate chamber conditions. Cereal Res. Commun. 2023. [Google Scholar] [CrossRef]
- Volante, A.; Tondelli, A.; Desiderio, F.; Abbruscato, P.; Menin, B.; Biselli, C.; Casella, L.; Singh, N.; McCouch, S.R.; Tharreau, D.; et al. Genome wide association studies for japonica rice resistance to blast in field and controlled conditions. Rice 2020, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Ming, L.; Fu, D.; Wu, Z.; Zhao, H.; Xu, X.; Xu, T.; Xiong, X.; Li, M.; Zheng, Y.; Li, G.; et al. Transcriptome-wide association analyses reveal the impact of regulatory variants on rice panicle architecture and causal gene regulatory networks. Nat. Commun. 2023, 14, 7501. [Google Scholar] [CrossRef] [PubMed]
- Norton, G.J.; Douglas, A.; Lahner, B.; Yakubova, E.; Guerinot, M.L.; Pinson, S.R.M.; Tarpley, L.; Eizenga, G.C.; McGrath, S.P.; Zhao, F.-J.; et al. Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at four international field sites. PLoS ONE 2014, 9, e89685. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.-Y.; Li, F.-P.; Choi, B.; Heo, E.-B.; Kim, K.-W.; Park, Y.-J. A genome-wide association study reveals candidate genes related to salt tolerance in rice (Oryza Sativa) at the germination stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, W.; Zhang, S.; Yang, T.; Liu, Q.; Dong, J.; Fu, H.; Mao, X.; Liu, B. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice 2018, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R. Gene Nomenclature System for Rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef]
- Chen, Y.; Xiang, Z.; Liu, M.; Wang, S.; Zhang, L.; Cai, D.; Huang, Y.; Mao, D.; Fu, J.; Chen, L. ABA biosynthesis gene OsNCED3 contributes to preharvest sprouting resistance and grain development in rice. Plant Cell Environ. 2022, 46, 1384–1401. [Google Scholar] [CrossRef] [PubMed]
- Lar, S.M.; Seo, J.; Jang, S.-G.; Zhang, H.; Lee, A.-R.; Cao, F.-Y.; Lee, J.-H.; Kim, N.-E.; Lee, Y.; Park, Y.-J.; et al. Genome-wide association study for detecting salt-tolerance loci and candidate genes in rice. Agriculture 2021, 11, 1174. [Google Scholar] [CrossRef]
- Babu, P.M.; Neeraja, C.N.; Rathod, S.; Suman, K.; Uttam, G.A.; Chakravartty, N.; Lachagari, V.B.R.; Chaitanya, U.; Rao, L.V.S.; Voleti, S.R. Stable SNP allele associations with high hrain zinc content in polished rice (Oryza sativa L.) identified based on ddRAD sequencing. Front. Genet. 2020, 11, e763. [Google Scholar] [CrossRef] [PubMed]
- RGAP. Rice Genome Annotation Project. Available online: https://rice.uga.edu/cgi-bin/gbrowse/rice (accessed on 5 December 2022).
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 zinc finger proteins response to abiotic stress in plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Ramkumar, M.K.; Dutta, B.; Kumar, A.; Pandey, R.; Jain, P.K.; Gaikwad, K.; Mishra, D.C.; Chaturvedi, K.K.; Rai, A.; et al. Integration of miRNA dynamics and drought tolerant QTLs in rice reveals the role of miR2919 in drought stress response. BMC Genom. 2023, 24, 526. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Howell, K.A.; Heazlewood, J.L.; Tan, T.Y.W.; Narsai, R.; Huang, S.; Whelan, J.; Millar, A.H. Analysis of the Rice Mitochondrial Carrier Family Reveals Anaerobic Accumulation of a Basic Amino Acid Carrier Involved in Arginine Metabolism during Seed Germination. Plant Physiol. 2010, 154, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qiu, Y.; Hu, Y.; Yu, D. Heterologous expression of AtWRKY57 confers drought tolerance in Oryza sativa. Front. Plant Sci. 2016, 7, e145. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, S.; Krishnan, G.S.; Mohapatra, T. Molecular basis of genetic plasticity to varying environmental conditions on growing rice by dry/direct-sowing and exposure to drought stress: Insights for DSR varietal development. Front. Plant Sci. 2022, 13, e1013207. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Kumar, A.; Grover, N.; Ellur, R.K.; Bollinedi, H.; Krishnan, S.G.; Bhowmick, P.K.; Vinod, K.K.; Nagarajan, M.; Singh, A.K. Genome-wide association study reveals marker–trait associations for early vegetative stage salinity tolerance in rice. Plants 2021, 10, 559. [Google Scholar] [CrossRef] [PubMed]
- Karnatam, K.S.; Jaganathan, D.; Dilip, K.R.; N, M.B.; Muthurajan, R. Shortlisting putative candidate genes underlying qDTY1.1, a major effect drought tolerant QTL in rice Oryza sativa L. Electron. J. Plant Breed. 2020, 11, 916–924. Available online: https://www.ejplantbreeding.org/index.php/EJPB/article/view/3651 (accessed on 1 April 2023).
- Li, H.; Yue, H.; Xie, J.; Bu, J.; Li, L.; Xin, X.; Zhao, Y.; Zhang, H.; Yang, L.; Wang, J.; et al. Transcriptomic profiling of the high-vigour maize (Zea mays L.) hybrid variety response to cold and drought stresses during seed germination. Sci. Rep. 2021, 11, 19345. [Google Scholar] [CrossRef] [PubMed]
- Catusse, J.; Meinhard, J.; Job, C.; Strub, J.-M.; Fischer, U.; Pestsova, E.; Westhoff, P.; Van Dorsselaer, A.; Job, D. Proteomics reveals potential biomarkers of seed vigor in sugarbeet. Proteomics 2011, 11, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.S.; Muthusamy, V.; Rashmi, T.; Basu, S.; Anand, A.; Mehta, B.K.; Gain, N.; Zunjare, R.U.; Singh, A.K.; Gupta, H.S.; et al. Characterization of crtRB1- and vte4-based biofortified sweet corn inbreds for seed vigour and physico-biochemical traits. J. Appl. Genet. 2022, 63, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Misra, G.; Joshi-Saha, A. Genetic mapping and transcriptome profiling of a chickpea (Cicer arietinum L.) mutant identifies a novel locus (CaEl) regulating organ size and early vigor. Plant J. 2023, 116, 1401–1420. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mustafiz, A.; Sahoo, K.K.; Sharma, V.; Samanta, S.; Sopory, S.K.; Pareek, A.; Singla-Pareek, S.L. Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 2012, 79, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Sircar, S.; Parekh, N. Meta-analysis of drought-tolerant genotypes in Oryza sativa: A network-based approach. PLoS ONE 2019, 14, e0216068. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.-H.; Nalini Chandran, A.K.; Park, J.-C.; Gho, Y.-S.; Lee, S.-W.; An, G.; Jung, K.-H. OsPhyB-mediating novel regulatory pathway for drought tolerance in rice root identified by a global RNA-Seq transcriptome analysis of rice genes in response to water deficiencies. Front. Plant Sci. 2017, 8, e580. [Google Scholar] [CrossRef] [PubMed]
- Durand, T.C.; Cueff, G.; Godin, B.; Valot, B.; Clément, G.; Gaude, T.; Rajjou, L. Combined proteomic and metabolomic profiling of the Arabidopsis thaliana vps29 mutant reveals pleiotropic functions of the retromer in seed development. Int. J. Mol. Sci. 2019, 20, 362. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Peng, W.; Huang, J.; Yan, X.; Yao, W.; Ouyang, J.; Li, S. Inositolphosphorylceramide synthases, OsIPCSs, regulate plant height in rice. Plant Sci. 2023, 335, 111798. [Google Scholar] [CrossRef]
- Ren, D.; Ding, C.; Qian, Q. Molecular bases of rice grain size and quality for optimized productivity. Sci. Bull. 2023, 68, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Mukherji, S.; Sircar, S.M. High temperature—Induced changes in germination, seedling vigour and the metabolic activities in rice seeds. Biol. Plant. 1973, 15, 65–71. [Google Scholar] [CrossRef]
- Punchkhon, C.; Plaimas, K.; Buaboocha, T.; Siangliw, J.L.; Toojinda, T.; Comai, L.; De Diego, N.; Spíchal, L.; Chadchawan, S. Drought-tolerance gene identification using genome comparison and co-expression network analysis of chromosome substitution lines in rice. Genes 2020, 11, 1197. [Google Scholar] [CrossRef]
- Redona, E.; Mackill, D. Genetic variation for seedling vigor traits in rice. Crop Sci. 1996, 36, 285–290. [Google Scholar] [CrossRef]
- Krishna, G.R.; Rani, A.R.; Reddy, B.V. Genetic variability for early rice seedling vigour in AF 3 population of BPT5204/IR88633-1-136-B2 under dry direct seeded condition. Int. J. Agric. Sci. Res. 2015, 5, 337–346. [Google Scholar]
- Yang, H.; Yang, Q.; Kang, Y.; Zhang, M.; Zhan, X.; Cao, L.; Cheng, S.; Wu, W.; Zhang, Y. Finding stable QTL for plant height in super hybrid rice. Agriculture 2022, 12, 165. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), green revolution rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.; Han, J.-J.; Han, M.-J.; An, G. Functional analyses of the flowering time gene OSMADS50, the putative suppressor of overexpression of CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef]
- Xiong, Q.; Ma, B.; Lu, X.; Huang, Y.-H.; He, S.-J.; Yang, C.; Yin, C.-C.; Zhao, H.; Zhou, Y.; Zhang, W.-K. Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell 2017, 29, 1053–1072. [Google Scholar] [CrossRef]
- Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.; Kitano, H.; Matsuoka, M. Green revolution: A mutant gibberellin-synthesis gene in rice. Nat. Commun. 2002, 416, 701702. [Google Scholar]
- Yoshida, S.; Bhattacharjee, D.; Cabuslay, G. Relationship between plant type and root growth in rice. Soil Sci. Plant Nutr. 1982, 28, 473–482. [Google Scholar] [CrossRef]
- Mathan, J.; Bhattacharya, J.; Ranjan, A. Enhancing crop yield by optimizing plant developmental features. Development 2016, 143, 3283–3294. [Google Scholar] [CrossRef] [PubMed]
- Dass, A.; Shekhawat, K.; Choudhary, A.K.; Sepat, S.; Rathore, S.S.; Mahajan, G.; Chauhan, B.S. Weed management in rice using crop competition—A review. Crop Prot. 2017, 95, 45–52. [Google Scholar] [CrossRef]
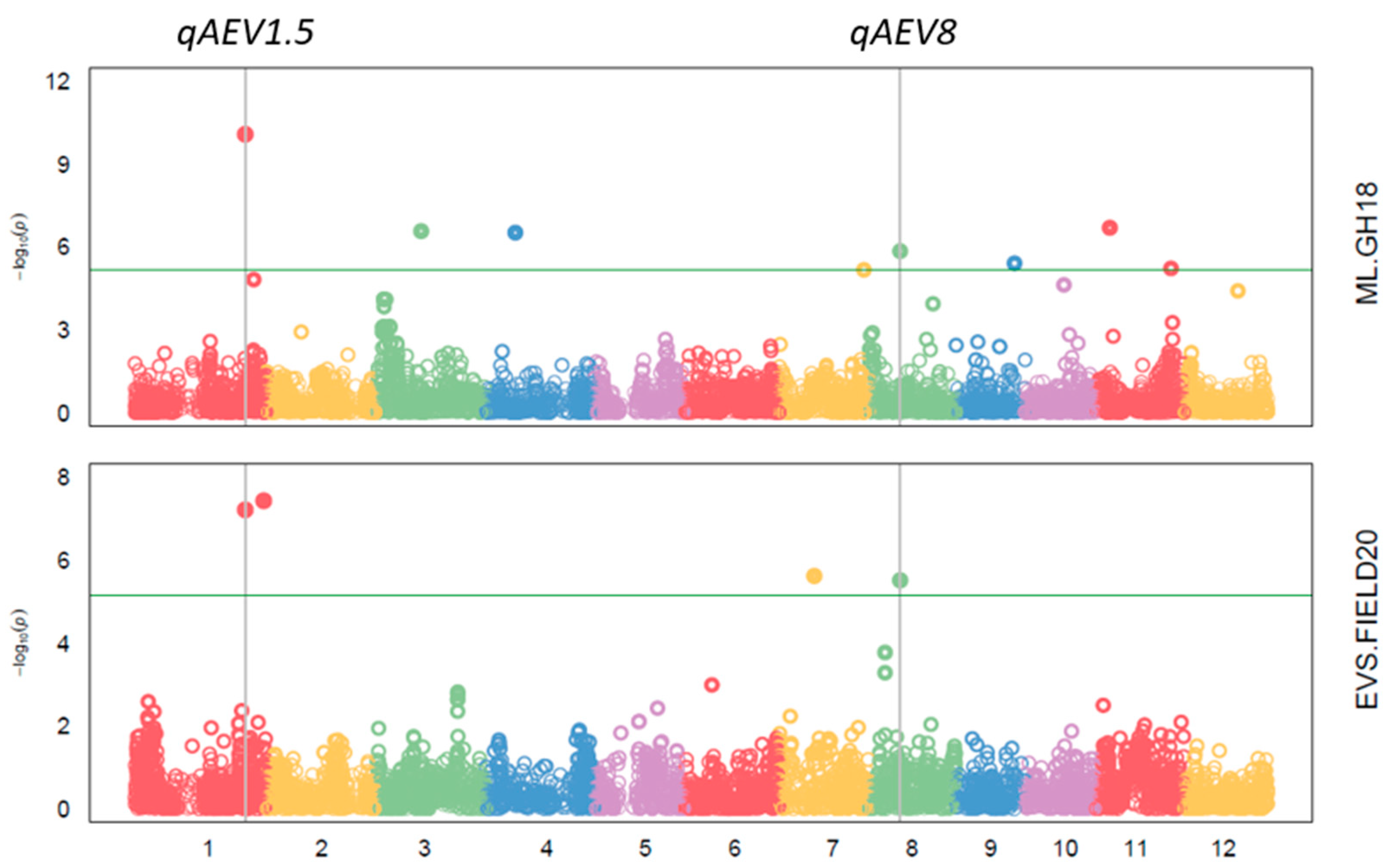
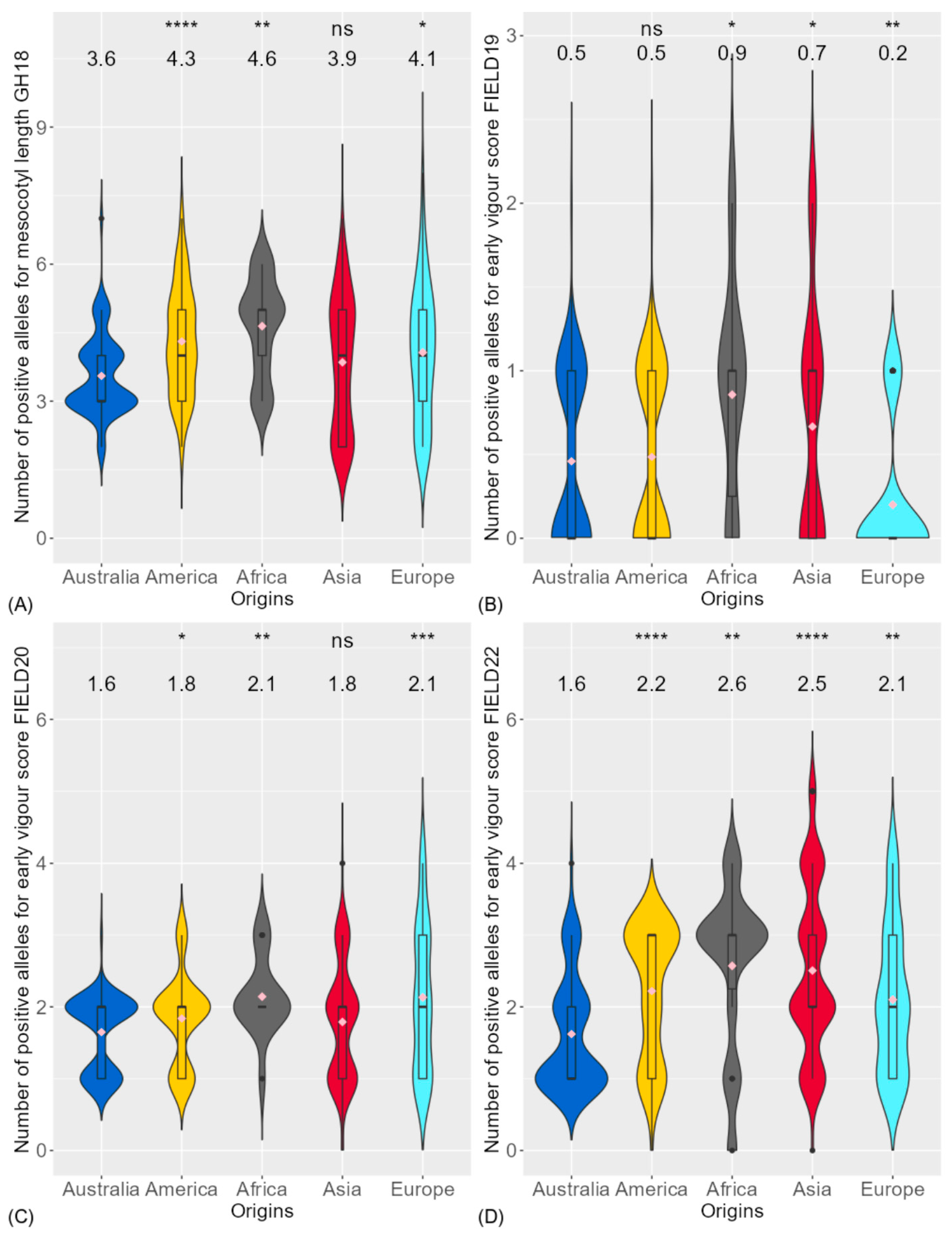
| Trait | Experiment | Mean | Maximum | Minimum | Heritability | LSD |
|---|---|---|---|---|---|---|
| Mesocotyl length (mm) | GH18 | 9.7 | 37.2 | 1.6 | 0.82 | 24.7 |
| Early vigour score | FIELD19 | 4.8 | 9.2 | 0.9 | 0.57 | 2 |
| FIELD20 | 5.8 | 9.9 | 1.7 | 0.75 | 2.1 | |
| FIELD22 | 4.7 | 8.2 | 0.8 | 0.65 | 2 | |
| Plant height (cm) | GH18 | 14.3 | 34.8 | 1.4 | 0.80 | 11.2 |
| FIELD20 | 29.6 | 47.0 | 12.8 | 0.82 | 6.7 | |
| GH21 | 44.1 | 60.5 | 24.3 | 0.75 | 8.5 | |
| Days to emergence | GH18 | 10 | 16 | 4 | 0.42 | 5 |
| FIELD20 | 10 | 16 | 8 | 0.78 | 2 | |
| GH21 | 5 | 10 | 3 | 0.40 | 2 | |
| Biomass per plant (mg) | GH18 | 12.1 | 32.5 | 0.7 | 0.75 | 10.0 |
| Biomass per m2 (g m−2) | FIELD20 | 70.5 | 196.4 | 13.5 | 0.64 | 47.3 |
| Light interception (%) | FIELD20 | 31 | 74 | 5 | 0.56 | 18 |
| QTL | Trait and Experiment | SNP | Chr. | Pos. | p Value | MAF | Effect | PVE |
|---|---|---|---|---|---|---|---|---|
| qAEV1.1 | EVS FIELD19 | 1_6304441 | 1 | 6,304,441 | 1.91 × 10−8 | 0.36 | −0.41 | 11.76 |
| qAEV1.2 | EVS FIELD22 | 1_32499722 | 1 | 32,499,722 | 9.70 × 10−8 | 0.13 | −0.51 | 8.65 |
| EVS FIELD19 | 1_32510801 | 1 | 32,510,801 | 9.94 × 10−7 | 0.14 | −0.42 | 21.44 | |
| PH GH21 | 1_32510801 | 1 | 32,510,801 | 5.99 × 10−9 | 0.14 | 2.20 | 7.61 | |
| qAEV1.3 | DTE FIELD20 | 1_34429355 | 1 | 34,429,355 | 5.06 × 10−6 | 0.14 | 0.62 | 9.73 |
| PH GH18 | 1_34600584 | 1 | 34,600,584 | 4.33 × 10−9 | 0.09 | 27.12 | 22.92 | |
| qAEV1.4 | LI FIELD20 | 1_35998551 | 1 | 35,998,551 | 3.73 × 10−8 | 0.11 | −0.06 | 7.29 |
| qAEV1.5 | EVS FIELD20 | 1_36358593 | 1 | 36,358,593 | 6.10 × 10−8 | 0.11 | 0.65 | 9.98 |
| ML GH18 | 1_36358593 | 1 | 36,358,593 | 8.48 × 10−11 | 0.11 | −3.04 | 8.59 | |
| qAEV1.6 | PH GH21 | 1_38847713 | 1 | 38,847,713 | 1.78 × 10−10 | 0.20 | −2.23 | 7.61 |
| PH FIELD20 | 1_38847713 | 1 | 38,847,713 | 7.90 × 10−8 | 0.21 | −18.71 | 5.01 | |
| qAEV1.7 | Biomass GH18 | 1_40490610 | 1 | 40,490,610 | 7.44 × 10−7 | 0.11 | −1.74 | 5.67 |
| Biomass GH18 | 1_40527461 | 1 | 40,527,461 | 1.96 × 10−7 | 0.41 | −1.27 | 3.23 | |
| qAEV1.8 | EVS FIELD20 | 1_42422994 | 1 | 42,422,994 | 3.68 × 10−8 | 0.06 | −0.88 | 17.41 |
| qAEV2.1 | Biomass GH18 | 2_2713635 | 2 | 2,713,635 | 1.46 × 10−6 | 0.11 | 1.49 | 4.42 |
| qAEV2.2 | PH GH21 | 2_6543528 | 2 | 6,543,528 | 3.20 × 10−7 | 0.12 | −1.99 | 6.01 |
| qAEV2.3 | DTE FIELD20 | 2_30871430 | 2 | 30,871,430 | 1.36 × 10−6 | 0.08 | 0.79 | 22.37 |
| qAEV3.1 | PH GH18 | 3_1248008 | 3 | 1,248,008 | 8.33 × 10−11 | 0.44 | 17.78 | 7.35 |
| Biomass GH18 | 3_1248008 | 3 | 1,248,008 | 5.00 × 10−9 | 0.44 | 1.30 | 3.42 | |
| qAEV3.2 | DTE GH18 | 3_13599473 | 3 | 13,599,473 | 1.24 × 10−8 | 0.43 | −0.66 | 8.74 |
| qAEV3.3 | ML GH18 | 3_15001141 | 3 | 15,001,141 | 2.72 × 10−7 | 0.47 | 1.59 | 1.77 |
| qAEV4.1 | Biomass GH18 | 4_4418061 | 4 | 4,418,061 | 7.89 × 10−7 | 0.39 | 1.17 | 2.71 |
| qAEV4.2 | ML GH18 | 4_9669965 | 4 | 9,669,965 | 3.08 × 10−7 | 0.07 | 3.75 | 5.82 |
| qAEV4.3 | LI FIELD20 | 4_12046452 | 4 | 12,046,452 | 5.27 × 10−8 | 0.48 | 0.04 | 8.94 |
| PH GH21 | 4_12046452 | 4 | 12,046,452 | 1.27 × 10−8 | 0.47 | 1.42 | 3.12 | |
| qAEV4.4 | DTE GH18 | 4_21145795 | 4 | 21,145,795 | 3.79 × 10−8 | 0.17 | 0.94 | 9.23 |
| qAEV4.5 | EVS FIELD22 | 4_33718024 | 4 | 33,718,024 | 9.43 × 10−7 | 0.35 | 0.31 | 3.53 |
| qAEV5.1 | EVS FIELD22 | 5_14202497 | 5 | 14,202,497 | 9.73 × 10−7 | 0.48 | −0.29 | 1.92 |
| qAEV5.2 | PH GH18 | 5_27968982 | 5 | 27,968,982 | 8.96 × 10−11 | 0.18 | −23.12 | 1.44 |
| qAEV6.1 | PH FIELD20 | 6_21702460 | 6 | 21,702,460 | 7.16 × 10−8 | 0.07 | 33.18 | 36.33 |
| EVS FIELD22 | 6_21702460 | 6 | 21,702,460 | 1.10 × 10−8 | 0.07 | −0.74 | 16.69 | |
| qAEV6.2 | EVS FIELD22 | 6_23451697 | 6 | 23,451,697 | 1.01 × 10−6 | 0.19 | 0.38 | 4.19 |
| qAEV6.3 | LI FIELD20 | 6_27996796 | 6 | 27,996,796 | 3.48 × 10−7 | 0.06 | 0.07 | 34.84 |
| qAEV7.1 | EVS FIELD20 | 7_11559181 | 7 | 11,559,181 | 2.40 × 10−6 | 0.22 | −0.38 | 2.42 |
| qAEV7.2 | DTE GH21 | 7_24657049 | 7 | 24,657,049 | 2.43 × 10−7 | 0.06 | 0.44 | 42.02 |
| qAEV7.3 | ML GH18 | 7_27838498 | 7 | 27,838,498 | 6.83 × 10−6 | 0.40 | 1.27 | 1.84 |
| qAEV8 | Biomass m2 FIELD20 | 8_10101994 | 8 | 10,101,994 | 5.07 × 10−6 | 0.33 | −7.03 | 22.05 |
| EVS FIELD20 | 8_10101994 | 8 | 10,101,994 | 3.06 × 10−6 | 0.33 | 0.46 | 7.91 | |
| ML GH18 | 8_10101994 | 8 | 10,101,994 | 1.44 × 10−6 | 0.32 | −1.63 | 1.53 | |
| qAEV9 | ML GH18 | 9_19288810 | 9 | 19,288,810 | 3.96 × 10−6 | 0.08 | −2.40 | 7.19 |
| qAEV11.1 | ML GH18 | 11_4596939 | 11 | 4,596,939 | 2.05 × 10−7 | 0.35 | 1.38 | 1.33 |
| qAEV11.2 | PH FIELD20 | 11_21913577 | 11 | 21,913,577 | 4.17 × 10−7 | 0.06 | −24.11 | 2.67 |
| qAEV11.3 | ML GH18 | 11_24666757 | 11 | 24,666,757 | 6.07 × 10−6 | 0.21 | 1.24 | 2.70 |
| QTL | Gene | Gene Identification | Gene Ontology Classification |
|---|---|---|---|
| qAEV1.5 | LOC_Os01g62460 | ZOS1-16-C2H2 zinc finger protein | Sequence-specific DNA-binding transcription factor activity; C2H2 zinc finger proteins have been shown to be involved in plant growth and development [57] |
| LOC_Os01g62480 | Laccase precursor protein | Drought tolerance, xylem structure, cell wall, cell length [58] | |
| LOC_Os01g62500 | OsFtsH3 FtsH protease, homologue of AtFtsH3/10 | Targets mitochondria and is involved in arginine metabolism during rice seed germination [59] | |
| LOC_Os01g62514 | WRKY56 | Sequence-specific DNA-binding transcription factor activity; WRKY gene family is involved in drought tolerance and root thickness [60] | |
| LOC_Os01g62570 | ATP/GTP/Ca++ binding protein | Cell growth; post-embryonic development | |
| LOC_Os01g62600 | Laccase precursor protein | Cell; response to stress; response to abiotic stimulus; leaf development under direct-sown and drought conditions [61] | |
| LOC_Os01g62610 | Peptidyl-prolyl cis–trans isomerase, FKBP-type | FKBPs gene family regulates hormone signalling in plant growth and development, stress response and seed germination [62] | |
| LOC_Os01g62630 | Aspartic proteinase nepenthesin precursor | Cell death; post-embryonic development; embryo development; drought response [63] | |
| LOC_Os01g62660 | MYB family transcription factor | Sequence-specific DNA-binding transcription factor activity; drought response [64] | |
| LOC_Os01g62760 | Protein phosphatase 2C | Drought response; protein phosphatase 2C involved in ABA metabolism [65] | |
| LOC_Os01g62800 | Methyltransferase | Methyltransferase gene family is related to the seed vigour index [66] | |
| LOC_Os01g62810 | Regulator of chromosome condensation | Regulation of plant organ elongation [67] | |
| LOC_Os01g62840 | Mannose-1-phosphate guanyltransferase | Drought response [63]; mannose-1-phosphate guanyltransferase regulates chlorophyll retention and seedling growth [68] | |
| LOC_Os01g62900 | Amino acid kinase | Drought response [69] | |
| LOC_Os01g62920 | Homeodomain protein | Sequence-specific DNA-binding transcription factor activity; homeodomain protein involved in plant hormone regulation including auxin, ABA, cytokinin and GA [23] | |
| LOC_Os01g62950 | RAS-related protein | Drought response [70]; cellular component; RAS-related protein responses to ABA, lipid metabolic processes and carbohydrate metabolic processes [71] | |
| LOC_Os01g63010 | Universal stress protein domain containing protein | Regulates genes under direct-sown and drought stress conditions [61] | |
| LOC_Os01g63060 | Phosphatidic acid phosphatase-related | Regulates the plant height, growth and development of rice [72] | |
| qAEV8 | LOC_Os08g16260 | Cytochrome P450 protein | Cytochrome P450 protein regulates cell size in the embryo and apoptosis in the endosperm [73] |
| LOC_Os08g16320 | Cytochrome P450 protein | Cytochrome P450 protein regulates cell size in the embryo and apoptosis in the endosperm [73] | |
| LOC_Os08g16480 | ATPase, AFG1 family domain-containing protein | ATPase is involved in germination percentage, seedling growth in terms of root and shoot lengths [74] | |
| LOC_Os08g16570 | Expressed protein | Drought resistance [75] | |
| LOC_Os08g16600 | WD-40 repeat protein family, expressed | WD-40 gene family is involved in signal transduction and hormone-controlled plant cell division [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, W.; Proud, C.; Vinarao, R.; Fukai, S.; Mitchell, J. Genome-Wide Association Study of Early Vigour-Related Traits for a Rice (Oryza sativa L.) japonica Diversity Set Grown in Aerobic Conditions. Biology 2024, 13, 261. https://doi.org/10.3390/biology13040261
Gong W, Proud C, Vinarao R, Fukai S, Mitchell J. Genome-Wide Association Study of Early Vigour-Related Traits for a Rice (Oryza sativa L.) japonica Diversity Set Grown in Aerobic Conditions. Biology. 2024; 13(4):261. https://doi.org/10.3390/biology13040261
Chicago/Turabian StyleGong, Wenliu, Christopher Proud, Ricky Vinarao, Shu Fukai, and Jaquie Mitchell. 2024. "Genome-Wide Association Study of Early Vigour-Related Traits for a Rice (Oryza sativa L.) japonica Diversity Set Grown in Aerobic Conditions" Biology 13, no. 4: 261. https://doi.org/10.3390/biology13040261
APA StyleGong, W., Proud, C., Vinarao, R., Fukai, S., & Mitchell, J. (2024). Genome-Wide Association Study of Early Vigour-Related Traits for a Rice (Oryza sativa L.) japonica Diversity Set Grown in Aerobic Conditions. Biology, 13(4), 261. https://doi.org/10.3390/biology13040261






