Smad4 Heterozygous Knockout Effect on Pancreatic and Body Weight in F1 Population Using Collaborative Cross Lines
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics and Animal Welfare Considerations
2.2. Crossbreeding to Generate F1 Offspring
2.3. Animal Housing and Nutritional Care
2.4. Extraction of Genomic DNA
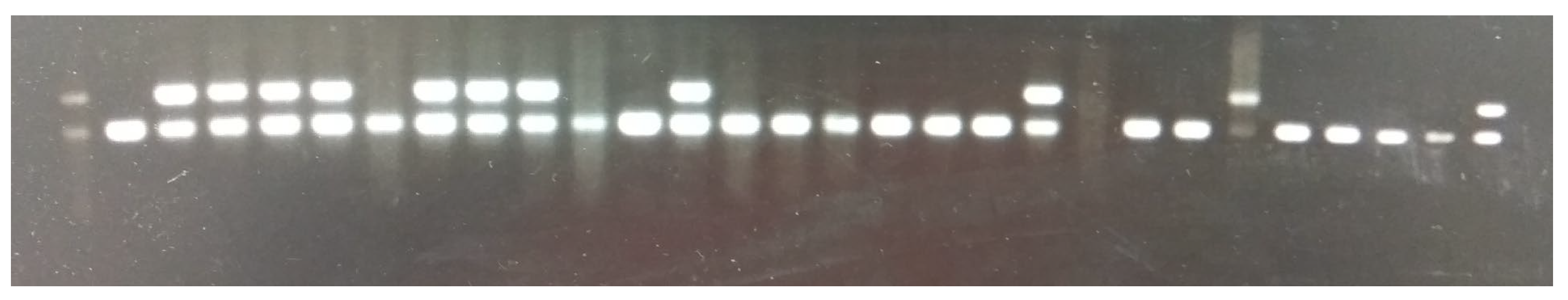
2.5. F1 Mouse PCR Genotyping
- Primer 30403 (5′-TGT AGT TCT GTC TTT CCT TCC TG-3′)
- Primer 30404 (5′-ACT GAC CTT TAT ATA CGC GCT TG-3′)
- Primer oIMR2088 (5′-AGA CTG CCT TGG GAA AAG CG-3′)
2.6. Tissue Harvesting
2.7. Data Analysis
3. Results
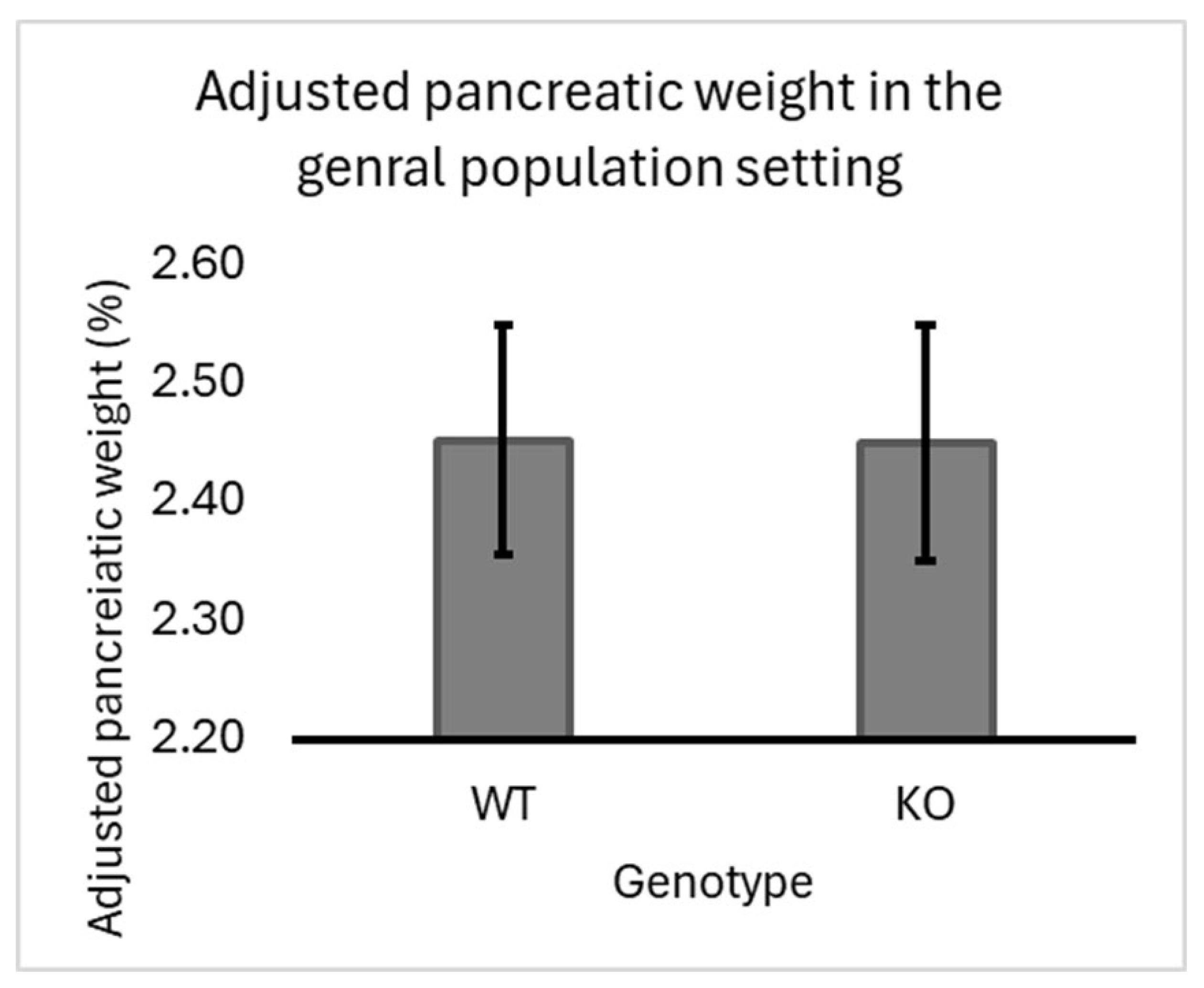
3.1. The Effect of Smad4 Kock out in the General Population of F1 Mice
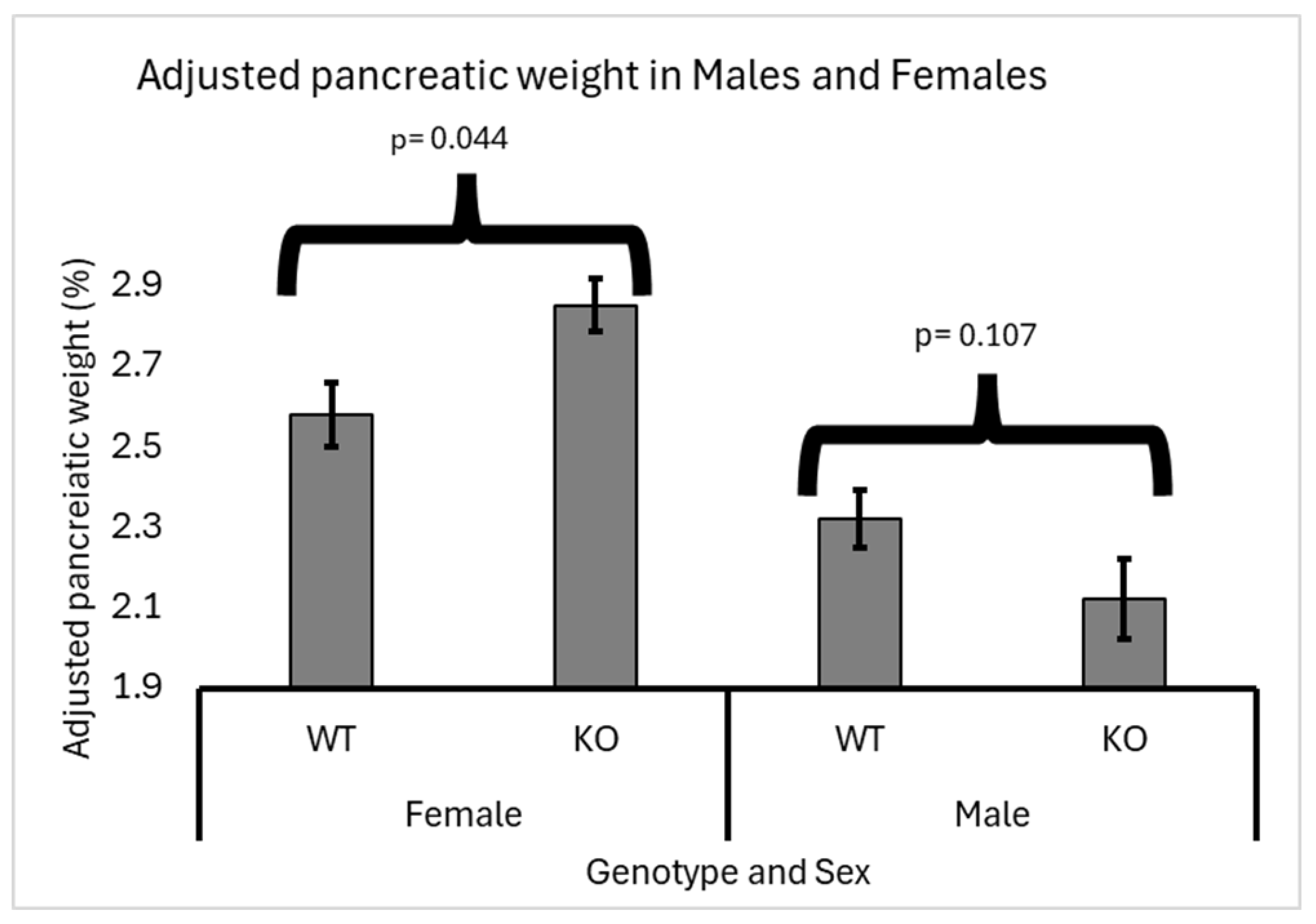
3.2. Sex Effect
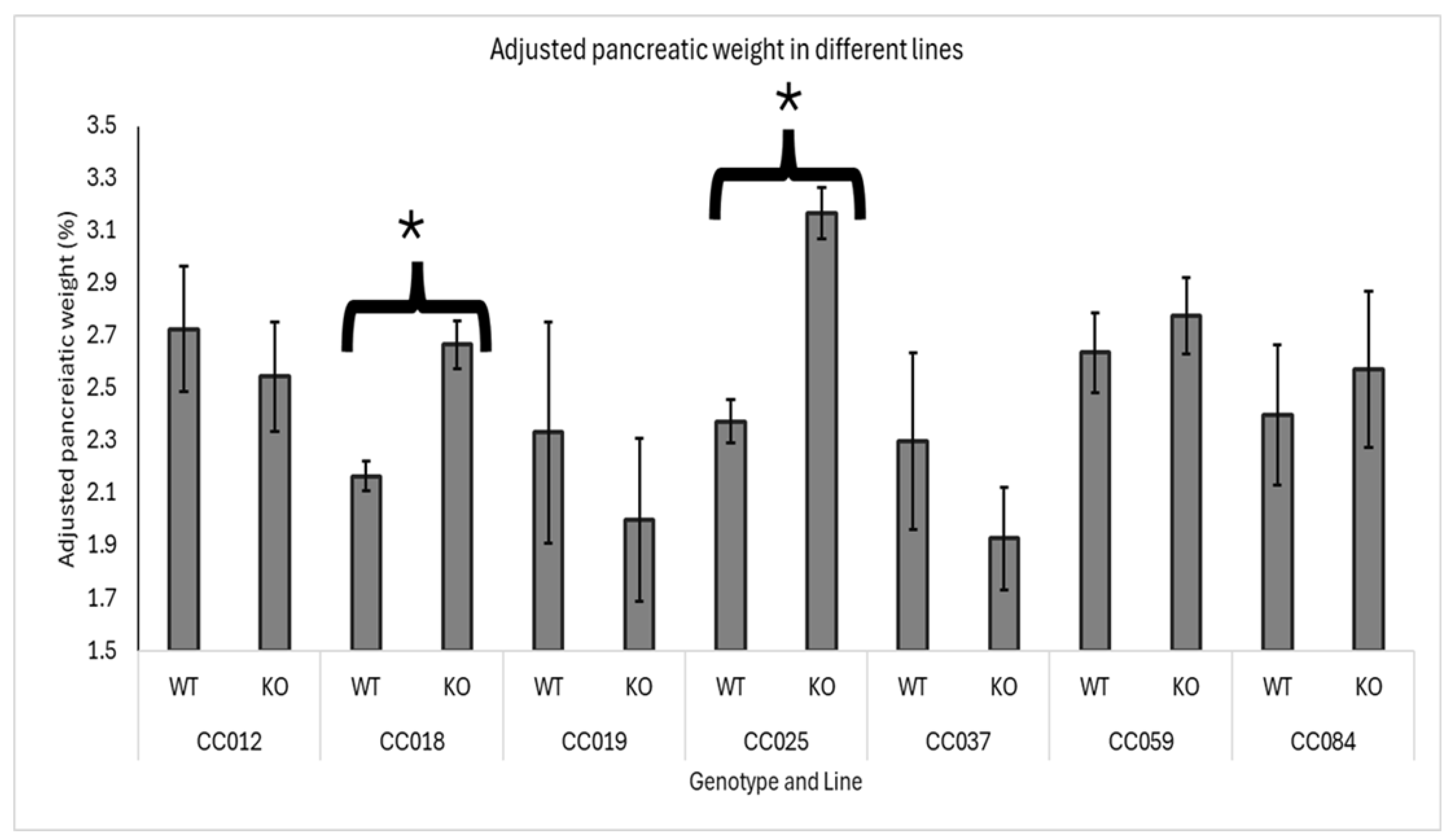
3.3. Line Genetic Effect
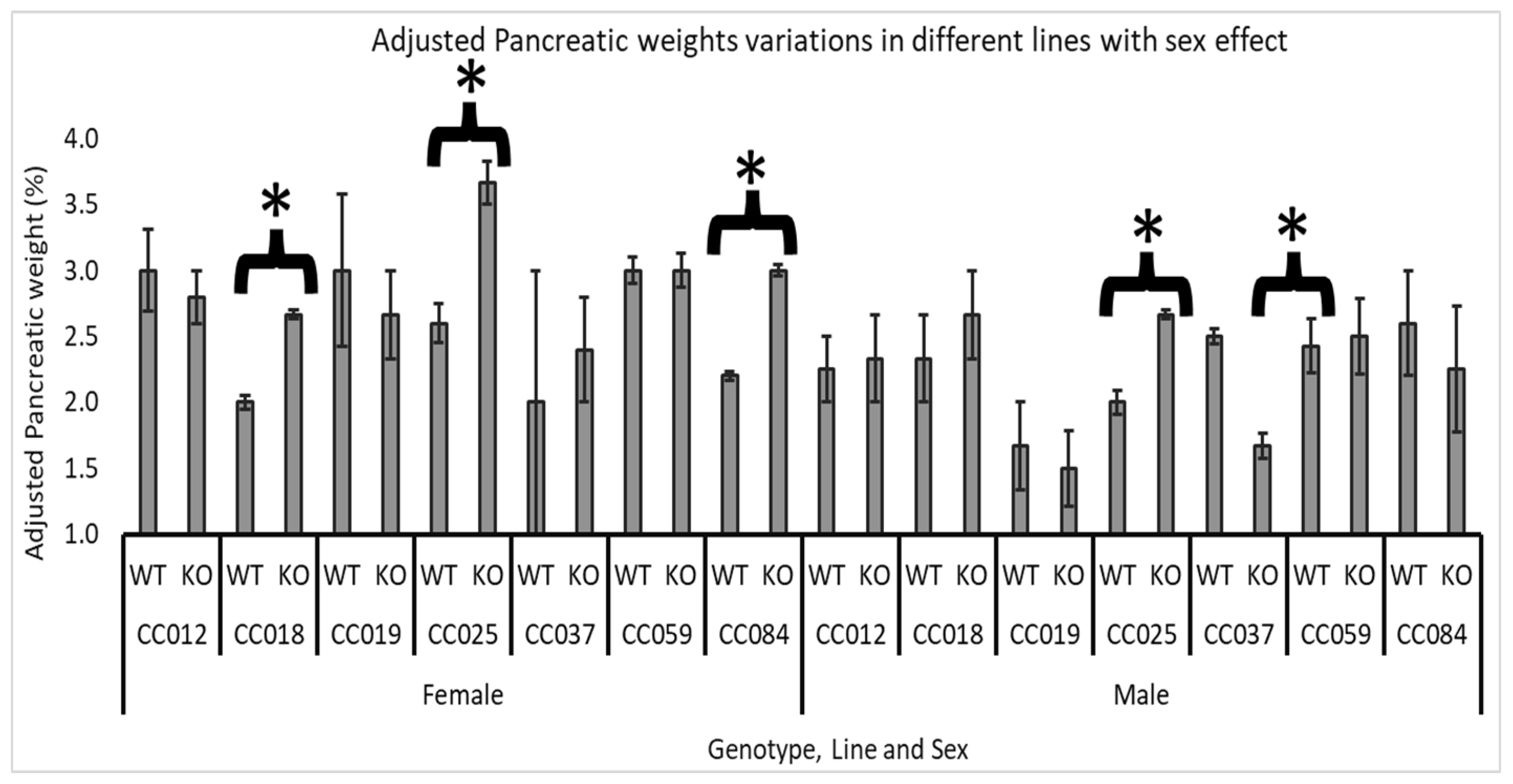
3.4. Line and Sex Effect
3.5. Heritability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Paredes, J.L.; Orabi, A.I.; Ahmad, T.; Benbourenane, I.; Tobita, K.; Tadros, S.; Bae, K.T.; Husain, S.Z. A Non-Invasive Method of Quantifying Pancreatic Volume in Mice Using Micro-MRI. PLoS ONE 2014, 9, e92263. [Google Scholar] [CrossRef] [PubMed]
- Nathan, J.D.; Romac, J.; Peng, R.Y.; Peyton, M.; Macdonald, R.J.; Liddle, R.A. Transgenic Expression of Pancreatic Secretory Trypsin Inhibitor-I Ameliorates Secretagogue-Induced Pancreatitis in Mice. Gastroenterology 2005, 128, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, K.; Fujimoto, T.; Kawanami, T.; Takiguchi, S.; Jimi, A.; Funakoshi, A.; Shirasawa, S. Pancreatic Hypertrophy in Ki-Ras-Induced Actin-Interacting Protein Gene Knockout Mice. Pancreas 2011, 40, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Gelbart, W.M.; Harland, R.M.; Heldin, C.H.; Kern, S.E.; Massagué, J.; Melton, D.A.; Mlodzik, M.; Padgett, R.W.; Roberts, A.B.; et al. Nomenclature: Vertebrate Mediators of TGF beta Family Signals. Cell 1996, 87, 173. [Google Scholar] [CrossRef]
- Wrana, J.L. The Secret Life of Smad4. Cell 2009, 136, 13–14. [Google Scholar] [CrossRef]
- Hahn, S.A.; Schutte, M.; Shamsul Hoque, A.T.M.; Moskaluk, C.A.; Da Costa, L.T.; Rozenblum, E.; Weinstein, C.L.; Fischer, A.; Yeo, C.J.; Hruban, R.H.; et al. DPC4, A Candidate Tumor Suppressor Gene at Human Chromosome 18q21.1. Science 1996, 271, 350–353. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.; Huang, C.; Cen, G.; Jiang, T.; Cao, J.; Huang, K.; Qiu, Z. SMAD4 and Its Role in Pancreatic Cancer. Tumor Biol. 2015, 36, 111–119. [Google Scholar] [CrossRef]
- Liu, F.; Pouponnot, C.; Massagué, J. Dual Role of the Smad4/DPC4 Tumor Suppressor in TGF-Inducible Transcriptional Complexes. Genes Dev. 1997, 11, 3157–3167. [Google Scholar] [CrossRef]
- Gu, Y.; Ji, Y.; Jiang, H.; Qiu, G. Clinical Effect of Driver Mutations of KRAS, CDKN2A/P16, TP53, and SMAD4 in Pancreatic Cancer: A Meta-Analysis. Genet. Test. Mol. Biomark. 2020, 24, 777–788. [Google Scholar] [CrossRef]
- Zhao, M.; Mishra, L.; Deng, C.X. The Role of TGF-β/SMAD4 Signaling in Cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef]
- Brown, S.D.M. Advances in Mouse Genetics for the Study of Human Disease. Hum. Mol. Genet. 2021, 30, R274–R284. [Google Scholar] [CrossRef] [PubMed]
- Noll, K.E.; Ferris, M.T.; Heise, M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe 2019, 25, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Churchill, G.A.; Airey, D.C.; Allayee, H.; Angel, J.M.; Attie, A.D.; Beatty, J.; Beavis, W.D.; Belknap, J.K.; Bennett, B.; Berrettini, W.; et al. The Collaborative Cross, a Community Resource for the Genetic Analysis of Complex Traits. Nat. Genet. 2004, 36, 1133–1137. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.; Gibson, H.; Frelinger, J.; Buntzman, A. Using the Collaborative Cross and Diversity Outbred Mice in Immunology. Curr. Protoc. 2022, 2, e547. [Google Scholar] [CrossRef]
- Dorman, A.; Binenbaum, I.; Abu-Toamih Atamni, H.J.; Chatziioannou, A.; Tomlinson, I.; Mott, R.; Iraqi, F.A. Genetic Mapping of Novel Modifiers for Apc Min Induced Intestinal Polyps’ Development Using the Genetic Architecture Power of the Collaborative Cross Mice. BMC Genom. 2021, 22, 566. [Google Scholar] [CrossRef]
- Iraqi, F.A.; Mahajne, M.; Salaymah, Y.; Sandovski, H.; Tayem, H.; Vered, K.; Balmer, L.; Hall, M.; Manship, G.; Morahan, G.; et al. The Genome Architecture of the Collaborative Cross Mouse Genetic Reference Population. Genetics 2012, 190, 389–401. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-Quality Mouse Genomic Dna with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Zohud, O.; Midlej, K.; Lone, I.M.; Nashef, A.; Abu-Elnaaj, I.; Iraqi, F.A. Studying the Effect of the Host Genetic Background of Juvenile Polyposis Development Using Collaborative Cross and Smad4 Knock-Out Mouse Models. Int. J. Mol. Sci. 2024, 25, 5812. [Google Scholar] [CrossRef]
- Veite-Schmahl, M.J.; Regan, D.P.; Rivers, A.C.; Nowatzke, J.F.; Kennedy, M.A. Dissection of the Mouse Pancreas for Histological Analysis and Metabolic Profiling. J. Vis. Exp. 2017, 19, 55647. [Google Scholar] [CrossRef]
- Chang, W.; Renaut, P.; Pretorius, C. SMAD4 Juvenile Polyposis Syndrome and Hereditary Haemorrhagic Telangiectasia Presenting in a Middle-Aged Man as a Large Fungating Gastric Mass, Polyposis in Both Upper and Lower GI Tract and Iron Deficiency Anaemia, with No Known Family History. BMJ Case Rep. 2020, 13, e236855. [Google Scholar] [CrossRef]
- Bardeesy, N.; Cheng, K.H.; Berger, J.H.; Chu, G.C.; Pahler, J.; Olson, P.; Hezel, A.F.; Horner, J.; Lauwers, G.Y.; Hanahan, D.; et al. Smad4 Is Dispensable for Normal Pancreas Development yet Critical in Progression and Tumor Biology of Pancreas Cancer. Genes Dev. 2006, 20, 3130–3146. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Tanaka, S.; Umemori, H.; Minowa, O.; Usui, M.; Ikematsu, N.; Hosoda, E.; Imamura, T.; Kuno, J.; Yamashita, T.; et al. Negative Regulation of BMP/Smad Signaling by Tob in Osteoblasts. Cell 2000, 103, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Mamot, C.; Mild, G.; Reuter, J.; Laffer, U.; Metzger, U.; Terracciano, L.; Boulay, J.L.; Herrmann, R.; Rochlitz, C. Infrequent Mutation of the Tumour-Suppressor Gene Smad4 in Early-Stage Colorectal Cancer. Br. J. Cancer 2003, 88, 420–423. [Google Scholar] [CrossRef][Green Version]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.H.; Pointu, H.; Olsen, B.B.; Cochet, C.; Issinger, O.-G.; Boldyreff, B. Disruption of the Regulatory β Subunit of Protein Kinase CK2 in Mice Leads to a Cell-Autonomous Defect and Early Embryonic Lethality. Mol. Cell. Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef]
- Shimada, S.; Yoshizawa, T.; Takahashi, Y.; Nitahara-Kasahara, Y.; Okada, T.; Nomura, Y.; Yamanaka, H.; Kosho, T.; Matsumoto, K. Backcrossing to an Appropriate Genetic Background Improves the Birth Rate of Carbohydrate Sulfotransferase 14 Gene-Deleted Mice. Exp. Anim. 2020, 69, 407–413. [Google Scholar] [CrossRef]
- Qahaz, N.; Lone, I.M.; Khadija, A.; Ghnaim, A.; Zohud, O.; Nun, N.B.; Nashef, A.; Abu El-Naaj, I.; Iraqi, F.A. Host Genetic Background Effect on Body Weight Changes Influenced by Heterozygous Smad4 Knockout Using Collaborative Cross Mouse Population. Int. J. Mol. Sci. 2023, 24, 16136. [Google Scholar] [CrossRef]
- Garcia-Carracedo, D.; Yu, C.-C.; Akhavan, N.; Fine, S.A.; Schönleben, F.; Maehara, N.; Karg, D.C.; Xie, C.; Qiu, W.; Fine, R.L.; et al. Smad4 Loss Synergizes with TGFα Overexpression in Promoting Pancreatic Metaplasia, PanIN Development, and Fibrosis. PLoS ONE 2015, 10, e0120851. [Google Scholar] [CrossRef]
- Ahmed, S.; Bradshaw, A.D.; Gera, S.; Zahidunnabi Dewan, M.; Xu, R. The TGF-β/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. J. Clin. Med. 2017, 6, 5. [Google Scholar] [CrossRef]
- Simeone, D.M.; Zhang, L.; Treutelaar, M.K.; Zhang, L.; Graziano, K.; Logsdon, C.D.; Burant, C.F. Islet Hypertrophy Following Pancreatic Disruption of Smad4 Signaling. Am. J. Physiol. Endocrinol. Metab. 2006, 291, 1305–1316. [Google Scholar] [CrossRef]
- Chen, Y.W.; Hsiao, P.J.; Weng, C.C.; Kuo, K.K.; Kuo, T.L.; Wu, D.C.; Hung, W.C.; Cheng, K.H. SMAD4 Loss Triggers the Phenotypic Changes of Pancreatic Ductal Adenocarcinoma Cells. BMC Cancer 2014, 14, 181. [Google Scholar] [CrossRef]
- Fullerton, P.T.; Creighton, C.J.; Matzuk, M.M. Insights into Smad4 Loss in Pancreatic Cancer from Inducible Restoration of TGF-β Signaling. Mol. Endocrinol. 2015, 29, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Lilly, A.C.; Astsaturov, I.; Golemis, E.A. Intrapancreatic Fat, Pancreatitis, and Pancreatic Cancer. Cell. Mol. Life Sci. 2023, 80, 206. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Wig, J.D.; Srinivasan, R. The Smad Family and Its Role in Pancreatic Cancer. Indian J. Cancer 2011, 48, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Yam, P.; Albright, J.; VerHague, M.; Gertz, E.R.; Pardo-Manuel de Villena, F.; Bennett, B.J. Genetic Background Shapes Phenotypic Response to Diet for Adiposity in the Collaborative Cross. Front. Genet. 2021, 11, 615012. [Google Scholar] [CrossRef]
- Vergara, M.M.; Labbé, J.; Tannous, J. Reflection on the Challenges, Accomplishments, and New Frontiers of Gene Drives. BioDesign Res. 2022, 2022, 9853416. [Google Scholar] [CrossRef]
- Gu, A.; Li, J.; Qiu, S.; Hao, S.; Yue, Z.Y.; Zhai, S.; Li, M.Y.; Liu, Y. Pancreatic Cancer Environment: From Patient-Derived Models to Single-Cell Omics. Mol. Omics 2024, 20, 220–233. [Google Scholar] [CrossRef]
- Guo, J.; Wang, M.F.; Zhu, Y.; Watari, F.; Xu, Y.H.; Chen, X. Exploitation of Platelets for Antitumor Drug Delivery and Modulation of the Tumor Immune Microenvironment. Acta Mater. Med. 2023, 2, 172–190. [Google Scholar] [CrossRef]
| Line | ||||||||
|---|---|---|---|---|---|---|---|---|
| CC012 | CC018 | CC019 | CC025 | CC037 | CC059 | CC084 | ||
| Female | WT | 7 | 3 | 3 | 5 | 4 | 4 | 5 |
| KO | 5 | 3 | 3 | 3 | 5 | 5 | 3 | |
| Male | WT | 4 | 3 | 3 | 3 | 6 | 7 | 5 |
| KO | 6 | 3 | 4 | 3 | 9 | 4 | 4 | |
| Sex | Genotype | Trait | VG | H2 | CVg |
|---|---|---|---|---|---|
| Female | WT | Adjusted Pan Weight % | 0.284903 | 0.477302 | 0.214363 |
| Female | WT | Delta_BW | 14.5507 | 0.153954 | 0.062006 |
| Female | KO | Adjusted Pan Weight % | 0.257129 | 0.603407 | 0.201222 |
| Female | KO | Delta_BW | 4.166905 | 0.036585 | 0.032808 |
| Male | WT | Adjusted Pan Weight % | 0.536923 | 0.456597 | 0.319978 |
| Male | WT | Delta_BW | 6.331883 | 0.04312 | 0.04313 |
| Male | KO | Adjusted Pan Weight % | 0.243643 | 0.412505 | 0.196654 |
| Male | KO | Delta_BW | 4.073576 | 0.030492 | 0.03395 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohud, O.; Lone, I.M.; Midlej, K.; Nashef, A.; Iraqi, F.A. Smad4 Heterozygous Knockout Effect on Pancreatic and Body Weight in F1 Population Using Collaborative Cross Lines. Biology 2024, 13, 918. https://doi.org/10.3390/biology13110918
Zohud O, Lone IM, Midlej K, Nashef A, Iraqi FA. Smad4 Heterozygous Knockout Effect on Pancreatic and Body Weight in F1 Population Using Collaborative Cross Lines. Biology. 2024; 13(11):918. https://doi.org/10.3390/biology13110918
Chicago/Turabian StyleZohud, Osayd, Iqbal M. Lone, Kareem Midlej, Aysar Nashef, and Fuad A. Iraqi. 2024. "Smad4 Heterozygous Knockout Effect on Pancreatic and Body Weight in F1 Population Using Collaborative Cross Lines" Biology 13, no. 11: 918. https://doi.org/10.3390/biology13110918
APA StyleZohud, O., Lone, I. M., Midlej, K., Nashef, A., & Iraqi, F. A. (2024). Smad4 Heterozygous Knockout Effect on Pancreatic and Body Weight in F1 Population Using Collaborative Cross Lines. Biology, 13(11), 918. https://doi.org/10.3390/biology13110918






