Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Samples
2.2. Measurement of the Physical and Chemical Properties of Soil
2.3. Microbial Isolation and Counting
2.4. DNA Extraction and 16S Sequencing
2.5. Data Analysis
3. Results and Analysis
3.1. Physical and Chemical Properties of Sandy Soil in the Taklimakan Desert
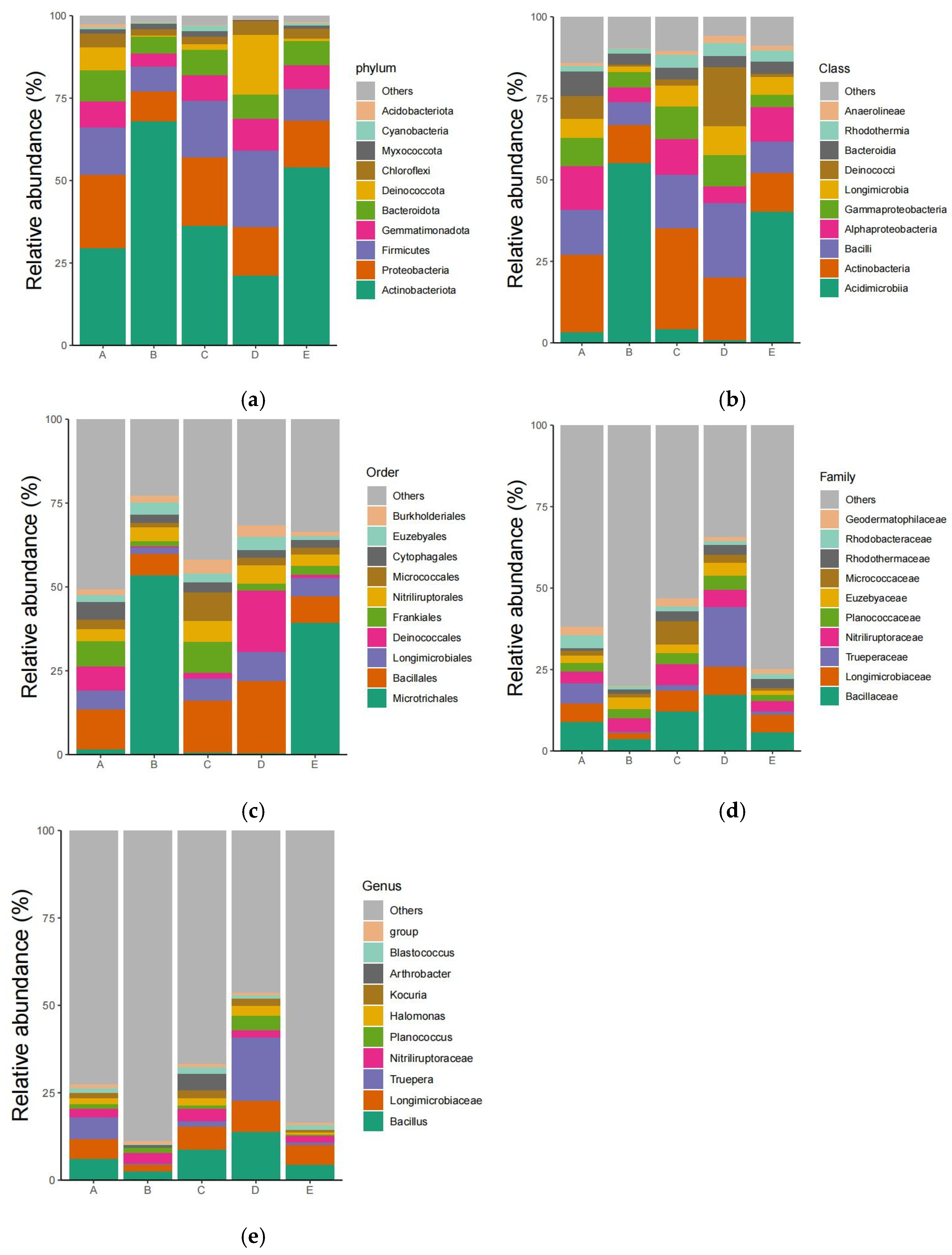
3.2. Species Composition of Different Samples Is Different
3.3. High-Throughput Determination of Bacterial Composition in Soil Samples from the Taklimakan Desert
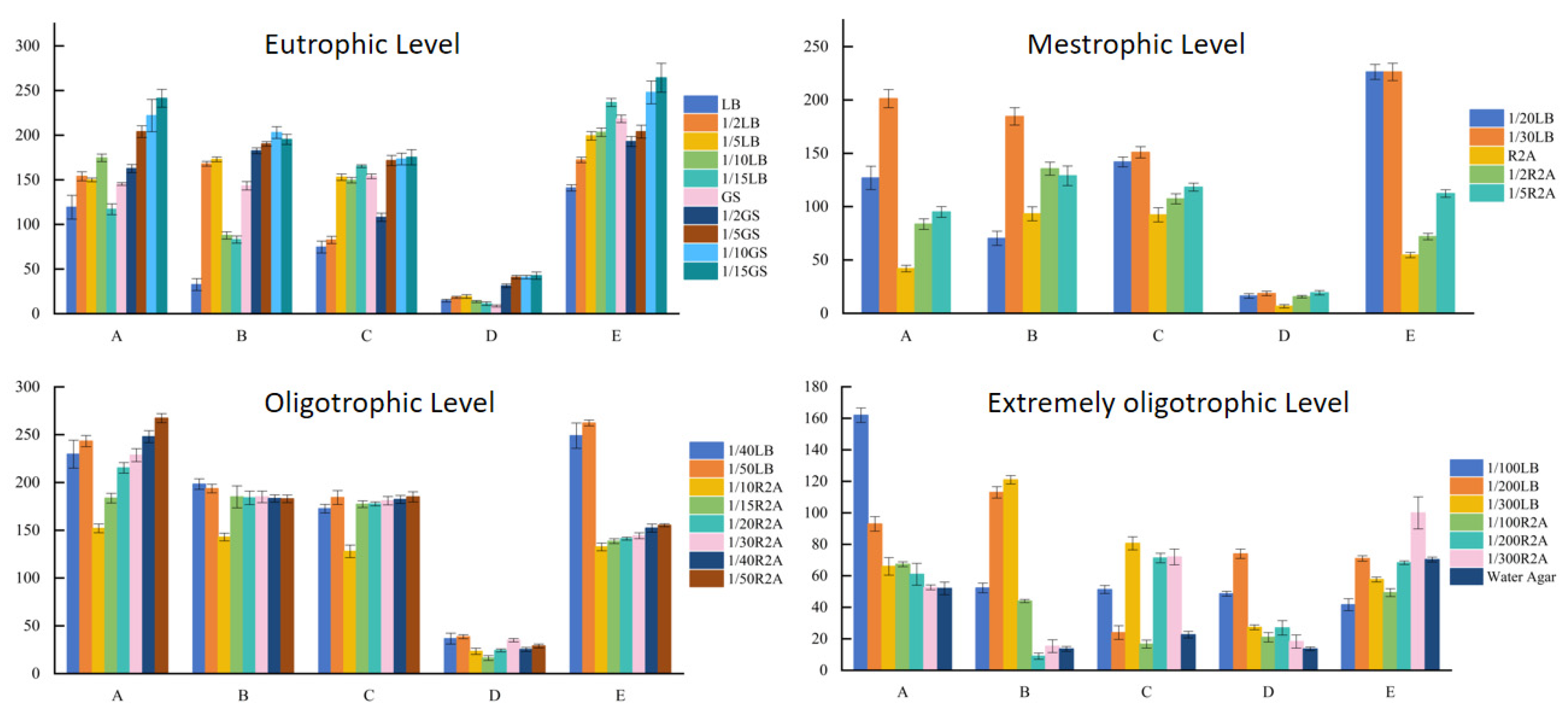
3.4. The Number of Colonies Isolated from Different Nutrient Levels of Medium
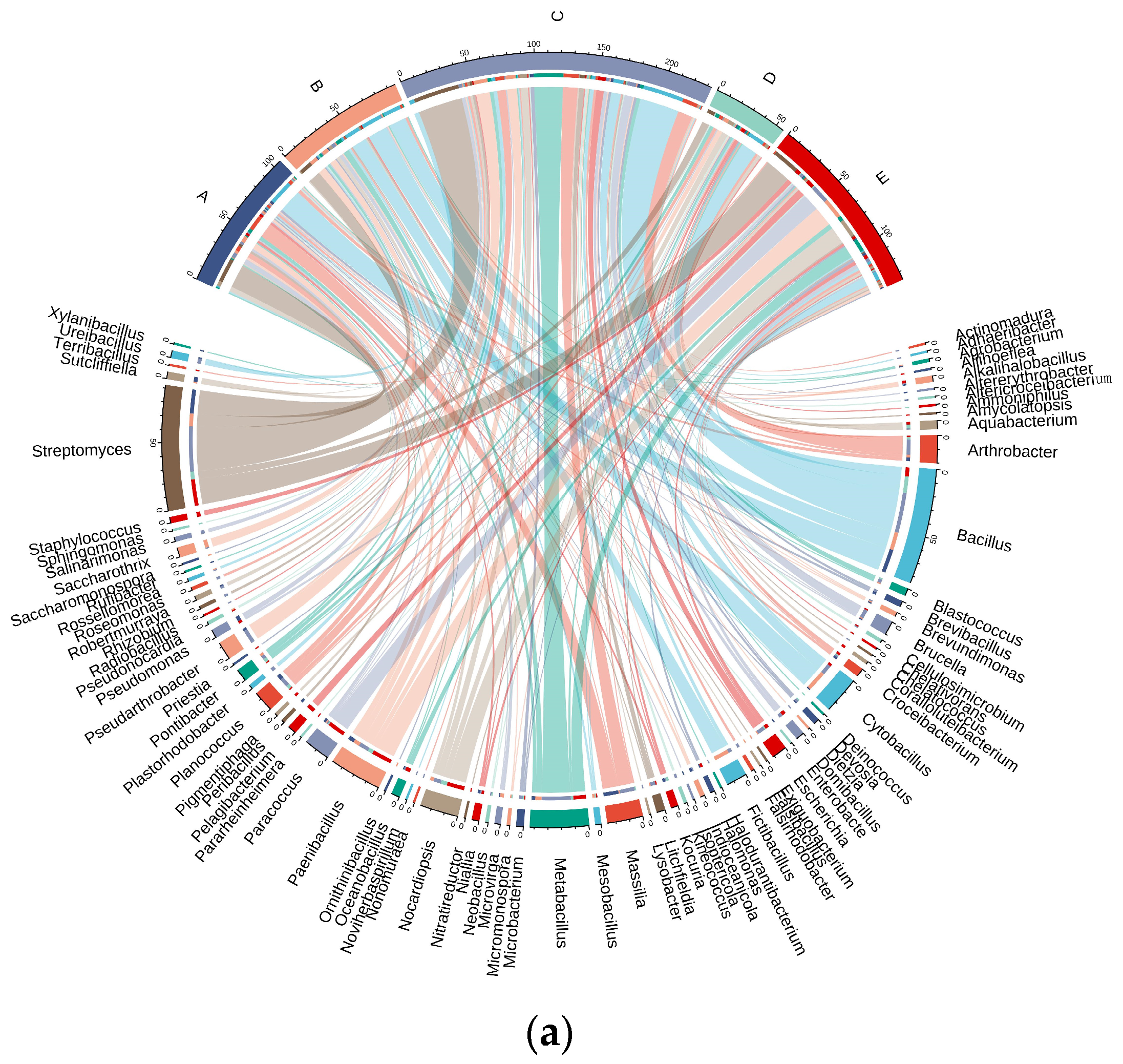
3.5. Distribution of Culturable Microbial Species in the Taklimakan Desert
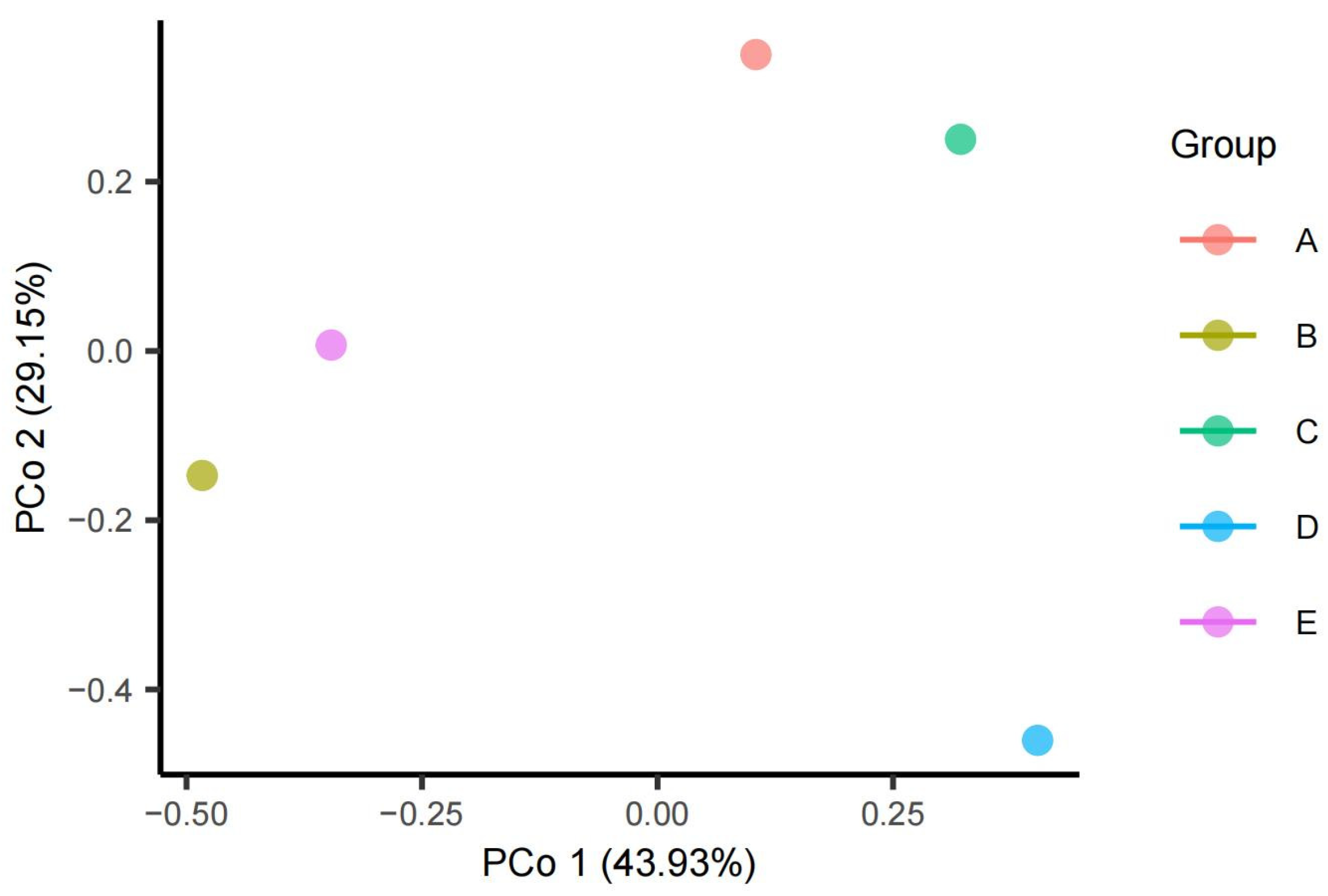
3.6. Microbial Community Structure and Diversity Analysis Based on 16S rRNA Bacterial Species Analysis
3.6.1. Culturable Microbial Community Structure of the Taklimakan Desert
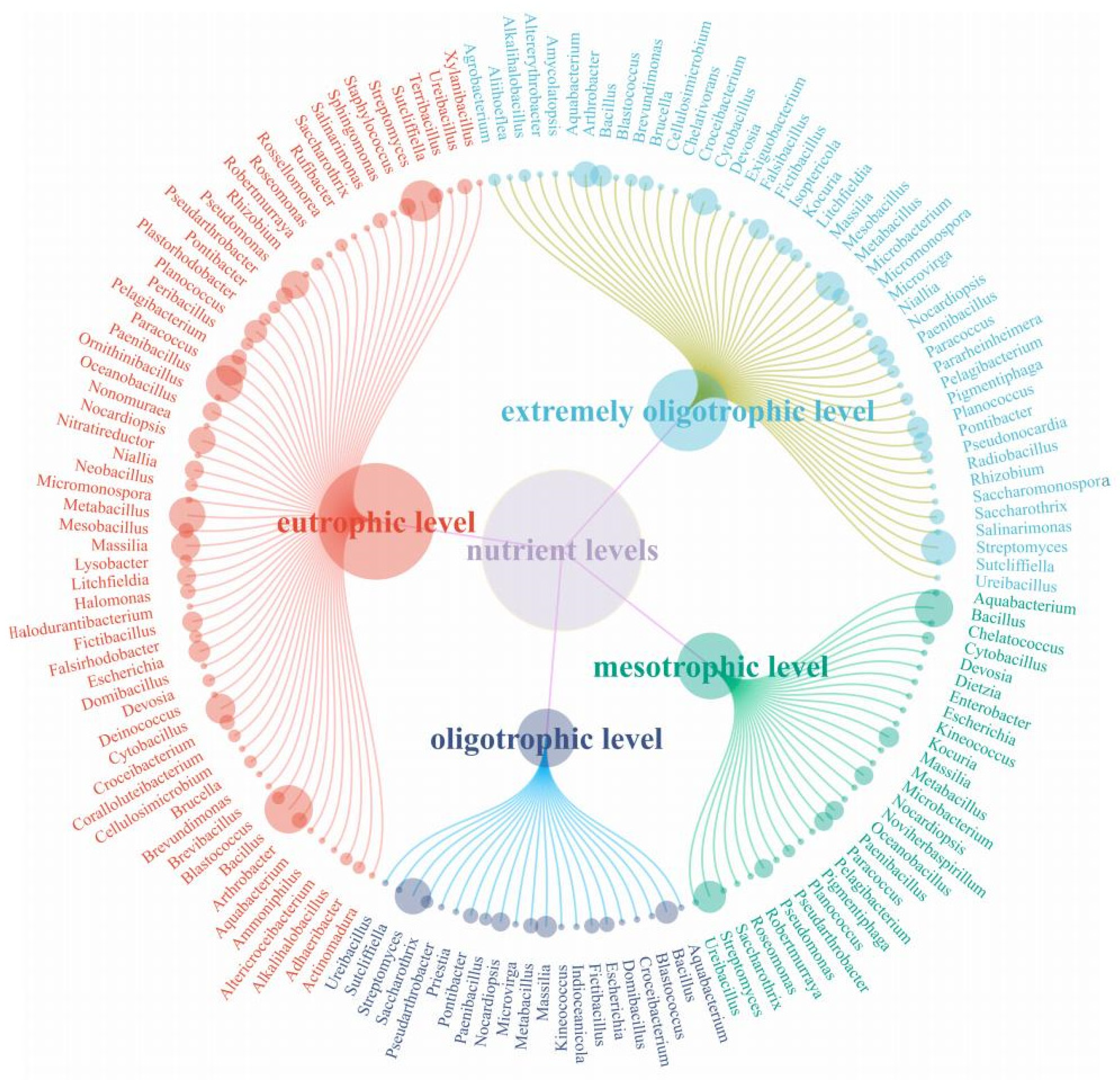
3.6.2. Community Structure of Microorganisms in the Taklimakan Desert under Different Nutrient Levels of Medium
3.6.3. Rare Microorganisms Isolated from Different Medium Nutrient Levels
3.7. Diversity of Culture-Free and Culturable Bacterial Communities in the Taklimakan Desert
4. Discussion
4.1. Bacterial Diversity in the Taklimakan Desert
4.2. Influence of Nutrient Level on Bacterial Isolation and Culture
4.3. Simple Culture-Based Strategy Can Obtain Numerous Unknown and Rare Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koch, A.L. Oligotrophs versus copiotrophs. Bioessays 2001, 23, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Hao, Z.; Gao, G.; Wang, Y. Ecological function of oligotrophic bacteria and their Applications in the environment. Microbiol. China 2012, 39, 0526–0535. [Google Scholar] [CrossRef]
- Kuznetsov, S.I.; Dubinina, G.A.; Lapteva, N.A. Biology of oligotrophic bacteria. Annu. Rev. Microbiol. 1979, 33, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Peng, Z.; Tu, H.; Qiu, Y.; Liu, Y.; Li, X.; Gao, H.; Pan, H.; Chen, B.; Liang, C.; et al. Oligotrophic microbes are recruited to resist multiple global change factors in agricultural subsoils. Environ. Int. 2024, 183, 108429. [Google Scholar] [CrossRef] [PubMed]
- Noell, S.E.; Brennan, E.; Washburn, Q.; Davis, E.W.; Hellweger, F.L.; Giovannoni, S.J. Differences in the regulatory strategies of marine oligotrophs and copiotrophs reflect differences in motility. Environ. Microbiol. 2023, 25, 1265–1280. [Google Scholar] [CrossRef] [PubMed]
- Yongjian, C.; W, N.J.; Priyanka, K.; M, M.R.; Albert, B. Life-history strategies of soil microbial communities in an arid ecosystem. Isme J. 2021, 15, 649–657. [Google Scholar] [CrossRef]
- Xie, G.; Zhang, Y.; Gong, Y.; Luo, W.; Tang, X. Extreme trophic tales: Deciphering bacterial diversity and potential functions in oligotrophic and hypereutrophic lakes. Bmc Microbiol. 2024, 24, 348. [Google Scholar] [CrossRef] [PubMed]
- Panova-Karadzhova, A.; Dimkov, R. Ecological Adaptations of Heterotrophic and Oligotrophic Bacteria in Iskar Reservoir. Biotechnol. Biotec. Eq. 2009, 23, 158–162. [Google Scholar] [CrossRef][Green Version]
- Hayakawa, D.H.; Huggett, M.J.; Rappé, M.S. Ecology and Cultivation of Marine Oligotrophic Bacteria. In Extremophiles Handbook; Springer: Tokyo, Japan, 2011. [Google Scholar] [CrossRef]
- Pan, H.; Cheng, Z.; Zhang, X.; Mu, S.; Qi, X.; Wang, F. A study on an oligotrophic bacteria and its ecological characteristics in an arid desert area. Sci. China Ser. D Earth Sci. 2007, 50, 128–134. [Google Scholar] [CrossRef]
- Button, D.K.; Schut, F.; Quang, P.; Martin, R.; Robertson, B.R. Viability and isolation of marine bacteria by dilution culture: Theory, procedures, and initial results. Appl. Environ. Microb. 1993, 59, 881–891. [Google Scholar] [CrossRef]
- Giovannoni, S.J. Cultivation and Growth Characteristics of a Diverse Group of Oligotrophic Marine Gammaproteobacteria. Appl. Environ. Microbiol. 2004, 70, 432–440. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Mou, S.; Wang, F. Study on the Physiological and Biochemical Properties of Five Strands of Oligotrophic Bacteria in Arid Deserts in Xinjiang. Arid. Land Geogr. 2005, 6, 831–835. [Google Scholar] [CrossRef]
- Schmidt, T.M.; Schaechter, M. Topics in Ecological and Environmental Microbiology; Academic Press: Cambridge, MA, USA, 2011; ISBN 0123838789. [Google Scholar]
- Leadbetter, J.R. Cultivation of recalcitrant microbes: Cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 2003, 6, 274–281. [Google Scholar] [CrossRef]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microb. 2002, 68, 2391–2396. [Google Scholar] [CrossRef]
- Zhao, S.; Yu, Y.; Xia, D.; Yin, D.; He, J.; Liu, N.; Li, F. Urban particle size distributions during two contrasting dust events originating from Taklimakan and Gobi Deserts. Environ. Pollut. 2015, 207, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, T. Exploration of the dynamic water resource carrying capacity of the Keriya River Basin on the southern margin of the Taklimakan Desert, China. Reg. Sustain. 2021, 2, 73–82. [Google Scholar] [CrossRef]
- Mclean, E.O. Soil pH and lime requirement. Methods Soil Anal. Part 1982, 12, 17–44. [Google Scholar] [CrossRef]
- Liebner, S.; Rublack, K.; Stuehrmann, T.; Wagner, D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the Lena Delta, Siberia. Microb. Ecol. 2009, 57, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Bremner, J.M.; Mulvaney, C.S. Nitrogen—Total. Methods Soil Anal. Part 2 Chem. Microbiol. Prop. 1982, 9, 595–624. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. Methods Soil Anal. Part 2 Chem. Microbiol. Prop. 1982, 9, 539–579. [Google Scholar]
- Foster, A.W.; Clough, S.E.; Aki, Z.; Young, T.R.; Clarke, A.R.; Robinson, N.J. Metalation calculators for E. coli strain JM109 (DE3): Aerobic, anaerobic, and hydrogen peroxide exposed cells cultured in LB media. Metallomics 2022, 14, mfac58. [Google Scholar] [CrossRef]
- Ahn, Y.; Kim, J.M.; Ahn, H.; Lee, Y.; LiPuma, J.J.; Hussong, D.; Cerniglia, C.E. Evaluation of liquid and solid culture media for the recovery and enrichment of Burkholderia cenocepacia from distilled water. J. Ind. Microbiol. Biotechnol. 2014, 41, 1109–1118. [Google Scholar] [CrossRef]
- Ludwig, W. Nucleic acid techniques in bacterial systematics and identification. Int. J. Food Microbiol. 2007, 120, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Seok-Hwan, Y.; Sung-Min, H.; Soonjae, K.; Jeongmin, L.; Yeseul, K.; Hyungseok, S.; Jongsik, C. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Nishimaki, T.; Sato, K. An extension of the Kimura two-parameter model to the natural evolutionary process. J. Mol. Evol. 2019, 87, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Bahadur, A.; Zhang, W.; Sajjad, W.; Nasir, F.; Zhang, G.; Liu, G.; Chen, T. Bacterial diversity patterns of desert dunes in the northeastern Qinghai-Tibet Plateau, China. Arch. Microbiol. 2021, 203, 2809–2823. [Google Scholar] [CrossRef]
- An, S.; Sin, H.H.; DuBow, M.S. Modification of atmospheric sand-associated bacterial communities during Asian sandstorms in China and South Korea. Heredity 2015, 114, 460–467. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb. Ecol. 2013, 66, 850–860. [Google Scholar] [CrossRef]
- Straub, D.; Blackwell, N.; Langarica-Fuentes, A.; Peltzer, A.; Kleindienst, S. Interpretations of Environmental Microbial Community Studies Are Biased by the Selected 16S rRNA (Gene) Amplicon Sequencing Pipeline. Front. Microbiol. 2020, 11, 550420. [Google Scholar] [CrossRef]
- Maillet, A.; Bouju-Albert, A.; Roblin, S.; Vaissié, P.; Leuillet, S.; Dousset, X.; Jaffrès, E.; Combrisson, J.; Prévost, H. Impact of DNA extraction and sampling methods on bacterial communities monitored by 16S rDNA metabarcoding in cold-smoked salmon and processing plant surfaces. Food Microbiol. 2021, 95, 103705. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, C.A.; Griffin, D.W.; Garrison, V.H.; Peak, K.K.; Royall, N.; Smith, R.R.; Shinn, E.A. Characterization of aerosolized bacteria and fungi from desert dust events in Mali, West Africa. Aerobiologia 2004, 20, 99–110. [Google Scholar] [CrossRef]
- Griffin, D.W.; Kubilay, N.; Koçak, M.; Gray, M.A.; Borden, T.C.; Shinn, E.A. Airborne desert dust and aeromicrobiology over the Turkish Mediterranean coastline. Atmos. Environ. 2007, 41, 4050–4062. [Google Scholar] [CrossRef]
- Lee, D.W.; Lee, S.D. Marmoricola scoriae sp. nov., isolated from volcanic ash. Int. J. Syst. Evol. Microbiol. 2010, 60, 2135–2139. [Google Scholar] [CrossRef]
- Urzì, C.; Salamone, P.; Schumann, P.; Stackebrandt, E. Marmoricola aurantiacus gen. nov., sp. nov., a coccoid member of the family Nocardioidaceae isolated from a marble statue. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 2, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Arocha-Garza, H.F.; Canales-Del Castillo, R.; Eguiarte, L.E.; Souza, V.; De la Torre-Zavala, S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. Peerj 2017, 5, e3247. [Google Scholar] [CrossRef] [PubMed]
- Contador, C.A.; Veas-Castillo, L.; Tapia, E.; Antipán, M.; Miranda, N.; Ruiz-Tagle, B.; García-Araya, J.; Andrews, B.A.; Marin, M.; Dorador, C. Atacama Database: A platform of the microbiome of the Atacama Desert. Antonie Van Leeuwenhoek 2020, 113, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Mandakovic, D.; Maldonado, J.; Pulgar, R.; Cabrera, P.; Gaete, A.; Urtuvia, V.; Seeger, M.; Cambiazo, V.; González, M. Microbiome analysis and bacterial isolation from Lejía Lake soil in Atacama Desert. Extremophiles 2018, 22, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Molina-Menor, E.; Gimeno-Valero, H.; Pascual, J.; Peretó, J.; Porcar, M. High culturable bacterial diversity from a European desert: The Tabernas desert. Front. Microbiol. 2021, 11, 583120. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Schmid, M.; Hartmann, A.; Tripathi, A.K. Identification of diazotrophs in the culturable bacterial community associated with roots of Lasiurus sindicus, a perennial grass of Thar Desert, India. Microb. Ecol. 2007, 54, 82–90. [Google Scholar] [CrossRef]
- Yu, L.Z.H.; Luo, X.S.; Liu, M.; Huang, Q. Diversity of ionizing radiation-resistant bacteria obtained from the Taklimakan Desert. J. Basic Microb. 2015, 55, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Jolivet, E.; Corre, E.; L Haridon, S.; Forterre, P.; Prieur, D. Thermococcus marinus sp. nov. and Thermococcus radiotolerans sp. nov., two hyperthermophilic archaea from deep-sea hydrothermal vents that resist ionizing radiation. Extremophiles 2004, 8, 219–227. [Google Scholar] [CrossRef]
- Andam, C.P.; Doroghazi, J.R.; Campbell, A.N.; Kelly, P.J.; Choudoir, M.J.; Buckley, D.H. A latitudinal diversity gradient in terrestrial bacteria of the genus Streptomyces. Mbio 2016, 7, 10–1128. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Stach, J.E.; Goodfellow, M. Genetic and phenotypic evidence for Streptomyces griseus ecovars isolated from a beach and dune sand system. Antonie Van Leeuwenhoek 2008, 94, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Davelos, A.L.; Xiao, K.; Samac, D.A.; Martin, A.P.; Kinkel, L.L. Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microb. Ecol. 2004, 48, 601–612. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kinkel, L.L.; Schlatter, D.C.; Xiao, K.; Baines, A.D. Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. Isme J. 2014, 8, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Souza, V.; Eguiarte, L.E.; Travisano, M.; Elser, J.J.; Rooks, C.; Siefert, J.L. Travel, sex, and food: What’s speciation got to do with it? Astrobiology 2012, 12, 634–640. [Google Scholar] [CrossRef]
- Zhou, N.; Jiang, C.; Liu, S. Cultivation of microorganisms from environments: Nutrient level of the culture medium is of great importance. Microbiol. China 2016, 43, 1075–1081. [Google Scholar] [CrossRef]
- Cao, X.; Xiong, H.; Fan, Y.; Xiong, L. Comparing the Effects of Two Culture Methods to Determine the Total Heterotrophic Bacterial Colony Count in Hospital Purified Water. J. Epidemiol. Glob. Health 2024, 14, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Tianjiao, D.; Donghui, W.; Colin, T.B.; Linwei, W.; Xue, G.; Suo, L.; Yifan, S.; Jiesi, L.; Jizhong, Z.; Yunfeng, Y. Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 2022, 13, 175. [Google Scholar] [CrossRef]
- Dahal, R.H.; Kim, J. Microvirga soli sp. nov., an alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 127–132. [Google Scholar] [CrossRef]
- Huq, M.A. Microvirga rosea sp. nov.: A nanoparticle producing bacterium isolated from soil of rose garden. Arch. Microbiol. 2018, 200, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
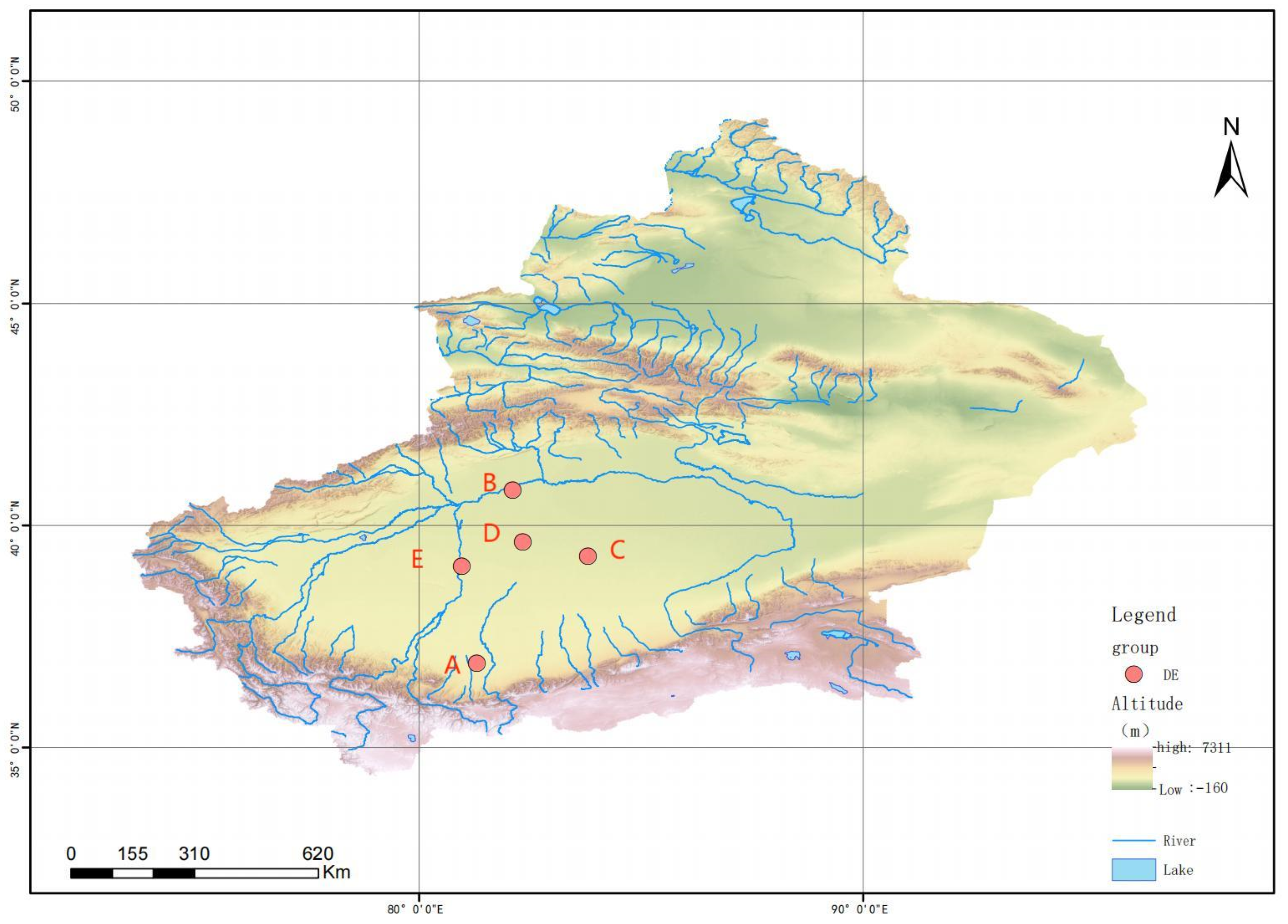


| No. | A | B | C | D | E |
|---|---|---|---|---|---|
| Direction | South | North | West | East | Central |
| Sampling area | Yutian County County | Alar City | Qiemo County | Shaya County | Lop County |
| Sample types | Sandy | Sandy | Sandy | Sandy | Sandy |
| Longitude | 81°17′40″ E | 82°5′59″ E | 83°48′0″ E | 82°19′41″ E | 80°57′25″ E |
| Latitude | 36°54′9″ N | 40°48′0″ N | 39°18′50″ N | 39°37′52″ N | 39°4′59″ N |
| Altitude (m) | 1364 | 926 | 990 | 988 | 1128.5 |
| No. | A | B | C | D | E |
|---|---|---|---|---|---|
| TC (g/kg) | (0.5033 ± 0.0120) c | (0.5465 ± 0.0014) a | (0.5292 ± 0.0040) b | (0.3596 ± 0.0042) d | (0.5341 ± 0.0092) ab |
| IP (g/kg) | (0.4805 ± 0.4810) b | (0.5155 ± 0.5125) a | (0.4828 ± 0.4842) b | (0.3334 ± 0.3341) c | (0.4923 ± 0.4848) b |
| OP (g/kg) | (0.0168 ± 0.0036) c | (0.0340 ± 0.0026) b | (0.0459 ± 0.0043) a | (0.0287 ± 0.0018) b | (0.0448 ± 0.0053) a |
| TN (g/kg) | (0.2611 ± 0.0064) b | (0.1272 ± 0.0041) d | (0.1431 ± 0.0011) c | (0.0758 ± 0.0013) e | (0.2754 ± 0.0096) a |
| TOC (g/kg) | (2.9835 ± 0.1152) a | (1.3887 ± 0.0921) c | (1.7306 ± 0.0447) b | (0.9425 ± 0.0298) d | (2.8418 ± 0.0900) a |
| NH4-N (mg/kg) | (1.0906 ± 0.0505) b | (0.8377 ± 0.1107) c | (1.4432 ± 0.1326) a | (1.5658 ± 0.0611) a | (1.6100 ± 0.14730) a |
| NO3-N (mg/kg) | (60.9560 ± 1.5623) a | (19.8417 ± 0.4272) c | (36.2060 ± 0.2706) b | (7.5463 ± 0.1370) d | (6.1933 ± 0.2146) e |
| TK (g/kg) | (15.8620 ± 0.0856) c | (15.6539 ± 0.0754) c | (18.4685 ± 0.2643) a | (16.6382 ± 0.2175) b | (16.3900 ± 0.1528) b |
| DOC (mg/kg) | (54.4943 ± 3.0400) a | (26.2365 ± 11.9005) b | (22.5329 ± 0.2920) b | (7.4250 ± 0.3521) c | (18.6680 ± 1.3207) b |
| DON (mg/kg) | (7.7808 ± 0.7826) a | (3.9640 ± 0.2978) b | (1.9260 ± 0.2446) c | (1.4253 ± 0.0494) c | (3.6013 ± 0.3176) b |
| pH | (8.9367 ± 0.0651) c | (9.800 ± 0.1127) a | (9.0600 ± 0.0520) bc | (8.7200 ± 0.0624) b | (9.0933 ± 0.0503) d |
| EC (S/m) | (1986.9233 ± 3.8618) a | (532.3300 ± 8.5440) c | (1351.6667 ± 7.0946) b | (115.2967 ± 2.5891) e | (169.233 ± 1.1930) d |
| ASL (m) | 1364 | 1128.5 | 926 | 990 | 998 |
| Sample | Observed | Chao1 | se.chao1 | ACE | se.ACE | Shannon | Simpson | InvSimpson | Fisher | Coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 1489 | 1504.068 | 5.952 | 1503.83 | 19.21 | 6.348 | 0.995 | 188.001 | 306.763 | 0.998 |
| B | 772 | 774.727 | 2.209 | 775.92 | 13.786 | 3.302 | 0.717 | 3.528 | 125.895 | 0.999 |
| C | 975 | 975.7 | 1.022 | 976.381 | 14.344 | 5.745 | 0.992 | 120.852 | 165.762 | 0.999 |
| D | 326 | 326.231 | 0.588 | 327.103 | 6.366 | 4.559 | 0.971 | 33.899 | 44.860 | 0.999 |
| E | 1088 | 1103.294 | 6.617 | 1099.633 | 16.391 | 4.302 | 0.845 | 6.471 | 188.359 | 0.999 |
| Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Alkalihalobacillus | 4 |
| Bacillus | 37 | ||||
| Peribacillus | 1 | ||||
| Cytobacillus | 19 | ||||
| Domibacillus | 3 | ||||
| Fictibacillus | 9 | ||||
| Mesobacillus | 8 | ||||
| Niallia | 2 | ||||
| Neobacillus | 2 | ||||
| Oceanobacillus | 2 | ||||
| Ornithinibacillus | 1 | ||||
| Priestia | 1 | ||||
| Robertmurraya | 2 | ||||
| Rossellomorea | 1 | ||||
| Sutcliffiella | 4 | ||||
| Litchfieldia | 5 | ||||
| Metabacillus | 22 | ||||
| Terribacillus | 1 | ||||
| Radiobacillus | 1 | ||||
| Falsibacillus | 1 | ||||
| Paenibacillaceae | Paenibacillus | 12 | |||
| Brevibacillus | 2 | ||||
| Xylanibacillus | 1 | ||||
| Ammoniphilus | 1 | ||||
| Exiguobacteriaceae | Exiguobacterium | 1 | |||
| Staphylococcaceae | Staphylococcus | 1 | |||
| Planococcaceae | Ureibacillus | 4 | |||
| Planococcus | 11 | ||||
| Bacteroidetes | Cytophagia | Cytophagales | Hymenobacteraceae | Adhaeribacter | 2 |
| Pontibacter | 9 | ||||
| Rufibacter | 1 | ||||
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingomonas | 1 |
| Erythrobacteraceae | Altericroceibacterium | 1 | |||
| Altererythrobacter | 1 | ||||
| Croceibacterium | 5 | ||||
| Rhizobiales | Brucellaceae | Brucella | 5 | ||
| Chelatococcaceae | Chelatococcus | 2 | |||
| Devosiaceae | Devosia | 4 | |||
| Methylobacteriaceae | Microvirga | 4 | |||
| Phyllobacteriaceae | Nitratireductor | 1 | |||
| Chelativorans | 1 | ||||
| Aliihoeflea | 1 | ||||
| Devosiaceae | Pelagibacterium | 3 | |||
| Rhizobiaceae | Rhizobium | 2 | |||
| Agrobacterium | 2 | ||||
| Salinarimonadaceae | Salinarimonas | 2 | |||
| Rhodobacterales | Rhodobacteraceae | Paracoccus | 7 | ||
| Plastorhodobacter | 4 | ||||
| Falsirhodobacter | 2 | ||||
| Halodurantibacterium | 1 | ||||
| Rhodospirillales | Azospirillaceae | Indioceanicola | 1 | ||
| Acetobacteraceae | Roseomonas | 2 | |||
| Caulobacterales | Caulobacteraceae | Brevundimonas | 3 | ||
| Betaproteobacteria | Burkholderiales | Comamonadaceae | Aquabacterium | 4 | |
| Oxalobacteraceae | Massilia | 11 | |||
| Noviherbaspirillum | 1 | ||||
| Actinobacteria | Gammaproteobacteria | Lysobacterales | Alcaligenaceae | Pigmentiphaga | 2 |
| Lysobacteraceae | Coralloluteibacterium | 2 | |||
| Alteromonadales | Alteromonadceae | Pararheinheimera | 1 | ||
| Actinomycetia | Oceanospirillales | Halomonadaceae | Lysobacter | 1 | |
| Halomonas | 2 | ||||
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | 4 | ||
| Enterobacterales | Enterobacteriaceae | Enterobacter | 1 | ||
| Micrococcales | Micrococcaceae | Escherichia | 5 | ||
| Arthrobacter | 15 | ||||
| Geodermatophilales | Geodermatophilaceae | Blastococcus | 4 | ||
| Mycobacteriales | Dietziaceae | Dietzia | 1 | ||
| Kineosporiales | Kineosporiaceae | Kineococcus | 2 | ||
| Micromonosporales | Micromonosporaceae | Micromonospora | 2 | ||
| Micrococcales | Micrococcaceae | Kocuria | 5 | ||
| Streptosporangiales | Microbacteriaceae | Microbacterium | 5 | ||
| Nocardiopsaceae | Nocardiopsis | 16 | |||
| Micrococcales | Micrococcaceae | Pseudarthrobacter | 4 | ||
| Pseudonocardiales | Pseudonocardiaceae | Saccharothrix | 5 | ||
| Pseudonocardia | 2 | ||||
| Saccharomonospora | 1 | ||||
| Amycolatopsis | 1 | ||||
| Streptomycetales | Streptomycetaceae | Streptomyces | 50 | ||
| Cellulomonadales | Thermomonosporaceae | Nonomuraea | 1 | ||
| Actinomadura | 1 | ||||
| Promicromonosporaceae | Cellulosimicrobium | 2 | |||
| Isoptericola | 1 | ||||
| Deinococcus-Thermus | Deinococci | Deinococcales | Deinococcaceae | Deinococcus | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, F.; Wu, S.; Luo, X.; Bai, L.; Xia, Z. Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods. Biology 2024, 13, 797. https://doi.org/10.3390/biology13100797
Wen F, Wu S, Luo X, Bai L, Xia Z. Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods. Biology. 2024; 13(10):797. https://doi.org/10.3390/biology13100797
Chicago/Turabian StyleWen, Feng, Siyuan Wu, Xiaoxia Luo, Linquan Bai, and Zhanfeng Xia. 2024. "Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods" Biology 13, no. 10: 797. https://doi.org/10.3390/biology13100797
APA StyleWen, F., Wu, S., Luo, X., Bai, L., & Xia, Z. (2024). Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods. Biology, 13(10), 797. https://doi.org/10.3390/biology13100797







