Simple Summary
Cryptocaryon irritans is a primitive ciliate that parasitizes the skin of marine fish and can cause illness or even death in Fugu. In this study, we revealed an immune regulatory network of genes and non-coding RNAs co-modulating competing endogenous RNAs. Potential biomarkers of Fugu infection with Cryptocaryon irritans were analyzed by bioinformatics. Of these, the regulatory network showed that the LOC105418663-circ_0000361-fre-miR-204a-fzd3a ceRNA axis was potentially involved in modulating the immune response of Fugu against Cryptocaryon irritans infection. Our research results have laid a theoretical foundation for a deeper understanding of the immune mechanism of Fugu and proposed new treatment and prevention measures for resisting Cryptocaryon irritans infection.
Abstract
Cryptocaryon irritans (C. irritans) is a proto-ciliate parasite that infects marine fishes, including the cultured species Takifugu rubripes (T. rubripes), causing disease and potential mortality. In host organisms, infection by parasites triggers an immune response that is modulated by regulatory elements including proteins and non-coding RNAs. In this study, the whole transcriptome RNA sequencing of T. rubripes gill tissue before and after infection with C. irritans was performed to reveal the competitive endogenous RNA (ceRNA) regulatory network. Histomorphology revealed gill segment swelling and parasitic invasion in the infected group. The analysis identified 18 differentially expressed miRNAs (DEMs), 214 lncRNAs (DELs), 2501 genes (DEGs), and 7 circRNAs (DECs) in the infected group. Gene Ontology (GO) enrichment analysis revealed that these genes were notably enriched in the Wnt signaling pathway and mTOR signaling pathway. The co-expression networks (lncRNA/circRNA-miRNA-mRNA) were constructed based on correlation analysis of the differentially expressed RNAs. Further analysis suggested that the LOC105418663-circ_0000361-fru-miR-204a-fzd3a ceRNA axis was potentially involved in the regulation of immune responses against C. irritans infection. Finally, the expression levels of DEG, DEL, and DEM were validated. This study reveals the regulatory mechanism of a candidate ceRNA network, providing insights into the potential mechanism of T. rubripes’ infection with C. irritans.
1. Introduction
Takifugu rubripes (T. rubripes) is a well-studied model species of teleost fishes. This coastal fish holds economic significance and can be found in regions including China, Korea, Japan, and other areas [1,2,3]. T. rubipres is one of the most economically valuable species [4]. With breakthroughs in breeding technology and the maturation of culture techniques, the cultivation scale of T. rubripes has expanded, and production has stabilized in recent years [5]. However, during aquaculture production, fish are susceptible to a variety of infectious diseases that cause significant economic losses [6,7]. Of these, parasitic diseases are the primary diseases in T. rubripes aquaculture, posing serious threats to the health of the fish.
Cryptocaryon irritans (C. irritans) is a type of ciliate protozoan that primarily infects the gills, skin, and fins of its hosts, leading to serious and often deadly infections [1]. As aquaculture continues to expand globally, the threat posed by C. irritans in marine fish farming cannot be ignored [1,2]. C. irritans can infect almost all marine teleosts with broad host specificity, and high-density mariculture is more likely to lead to large outbreaks of infection and high lethality [2]. After being infected by C. irritans, the host produces innate and specific immune responses [8,9], inducing related inflammatory pathways, such as toll-like receptors (TLR) [10]. Currently, research on the control of C. irritans is centered on disease prevention, with fewer studies related to disease treatment. Therefore, comprehending the immune response of T. rubripes to C. irritans infection is crucial for preventing and controlling the disease.
RNA-seq analysis has become a popular method for studying immune mechanisms in recent years. By analyzing expression profiles, researchers can identify immune-related genes that play a critical role in the immune response. This approach allows for a comprehensive understanding of the complex interactions between different genes and their impacts on the immune system [11,12,13]. For example, the cytosolic interleukin IL-34 was significantly up-regulated in the gills of golden pompano (Trachinotus ovatus) after stimulation with C. irritans infection [14]. There is growing evidence that insights into the dynamics of gene expression regulation can be gained through transcriptional products, including mRNAs and non-coding RNAs (ncRNAs). ncRNA includes microRNA (miRNA), long non-coding RNA (lncRNAs), and circular RNA (circRNA) [15,16]. In contrast, competitive endogenous RNAs (ceRNAs) involve more gene species than the miRNA regulatory network, and form a larger inter-regulatory network of RNA molecules [17,18]. It has been shown that in teleost, the lncRNA MARL can competitively bind miRNA-122 and weaken the targeted inhibition of MAVS by miRNA-122, elucidating the molecular response mechanism of the ceRNA machinery regulating the RIG-I antiviral signaling pathway [13]. In addition to influencing signaling pathways, ceRNAs also participate in the regulation of disease occurrence and development [19,20]. Therefore, association analysis of ceRNAs using transcriptome sequencing is an effective tool for exploring potential regulatory network mechanisms.
In this study, we infected T. rubripes with C. irritans and analyzed gill tissues for expression differences at the coding RNA and non-coding RNA levels, identifying differentially expressed miRNAs (DEMs), lncRNAs (DELs), circRNAs (DECs), and genes (DEGs). Functional enrichment analysis was then performed to predict key genes involved in immune-related pathways. In addition, we established ceRNA networks including lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA, and performed functional analyses to explore the regulatory network of the immune response in the organism following C. irritans infection. The research highlights how the immune system of T. rubripes responds to infections and provides insights into its defense strategies. Additionally, it sheds light on the immunological control mechanisms of C. irritans. This information is critical for developing new methods of managing infections caused by C. irritans, potentially leading to more effective treatments and preventive measures in aquatic environments.
2. Materials and Methods
2.1. Ethics Statement
The research procedures for all fish were carried out following the Guidelines for Experimental Animals set out by the Ministry of Science and Technology. The Institutional Animal Care and Use Committee at Dalian Ocean University (located in Dalian, China) reviewed and approved the experimental protocols. Anesthesia was induced using MS-222, and efforts were made to minimize pain and discomfort throughout the study.
2.2. Experimental Animals
Six healthy individuals of T. rubripes (mean length: 9.5 ± 0.9 cm; mean weight: 100 ± 5 g; age: 5 months) were sourced from Dalian Tianzheng Industrial Co., Ltd. (Dalian, China). These experimental fish were randomly distributed into two tanks, each containing three fish, forming three biological replicates: infection group (IG) and control group (CG). Before the experiment, the fish underwent an acclimation period of approximately two weeks in cycling-filtered tanks filled with fresh seawater. Tank conditions were maintained at a volume of 50 L, a temperature of 22.0 ± 0.5 °C, an average salinity of 31, a dissolved oxygen content exceeding 7.0 mg/L, and a pH of 7.5 ± 0.5. The fish density was set at 6 kg/m3, 12L:12D. Feeding occurred twice daily at 8:30 and 18:30 h.
2.3. Experimental Design
The infection protocol was conducted as previously described [21]. The IG was immersed and exposed to C. irritans at a concentration of 1 × 104/fish for three hours. Concurrently, the CG was exposed to the equivalent volume of phosphate-buffered saline (PBS) for an identical period. Subsequently, both groups were transferred to separate clean tanks. Throughout the seven-day experimental period, neither group exhibited any mortality. Anesthesia was induced in all six fish using MS-222, following which gill tissue samples were collected on the seventh day. The aseptic transfer of samples was conducted into sterile containers, followed by flash-freezing in liquid nitrogen, and then storage at a temperature of −80 °C until additional analysis.
2.4. Library Construction and Illumina Sequencing
Total RNA was extracted by Trizol, RNA concentration and purity were determined, and, after passing the quality check, the total RNA was sent to Novogene (Beijing, China) to complete sequencing. miRNA-seq, lncRNA-seq, circRNA-seq, and mRNA-seq are described in the Supplementary Materials File S2, which gives details of the methodological steps and differential expression analysis. A total of six libraries were constructed. In miRNA library construction, total RNA was used as the starting sample. Small RNA was constructed for Illumina SE50 sequencing. Bowtie 2.1.1 [22] software was used for small RNA mapping reads. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were implemented using miRanda 2.2.1 [23] and RNAhybrid 2.1.2 [24] for target gene prediction of miRNAs, as well as miRNA quantification and differential expression analysis (Supplementary Table S5).
lncRNA, circRNA, and mRNA libraries were constructed for Illumina PE150 sequencing (Supplementary Table S4). Hisat2 2.1.0 [25] software was used for comparative analysis, and Cuffmerge software 2.0.2 was used to merge transcripts obtained from the splicing of each sample. The circRNAs were identified by find_circ and CIRI 1.1.0 software, and the raw sequences were checked for error rate and comparison rate, and then compared, spliced, screened, quantified, and analyzed for the significance of differences, predicted target gene, and functional enrichment. DEMs, DELs, and DEGs were determined using edgeR 4.0 software and based on padj < 0.05 and |log2(foldchange)| > 1.
2.5. Construction of ceRNA Networks
The screened differentially expressed miRNAs, lncRNAs, circRNAs, and mRNAs were analyzed. The target genes of DEMs enriched in the immune pathway were predicted. DEL and DEC, which have targeted relationships and differential expression with target genes and DEM, were predicted separately. The correlation coefficient was calculated simultaneously. Previous studies have shown that lncRNAs and circRNAs act as miRNA sponges to competitively bind to miRNAs, affecting the regulation of the target genes by the miRNAs, and thus modulating the expression of the target genes. The ceRNA network was visualized using Cytoscape (Cytoscape 3.4.0, http://www.cytoscape.org/download.php, accessed on 15 May 2024) and Figdraw (https://www.figdraw.com/static/index.html#/, accessed on 17 June 2024).
2.6. Quantitative Real-Time PCR Verification
To further validate the RNA-seq data, 5 DEMs, 5 DELs, and 5 DEGs were randomly identified for qRT-PCR. The total RNA was extracted by TRIzol. RNA was reverse transcribed using a cDNA synthesis kit (Vazyme, China). miRNA first strand cDNA synthesis (Vazyme, China) was performed. Validation of miRNA, lncRNA, and mRNA was conducted with LightCycler 96 using qRT-PCR. U6 served as an internal reference control for miRNA normalization, while β-actin was utilized for mRNA and lncRNA. The primer design used Primer 5 and the primer sequences can be found in Supplementary Table S1. Each qRT-PCR assay was independently performed three times, and the 2−△△Ct method was applied for quantification of mRNAs, miRNAs, and lncRNAs.
2.7. Statistical Analysis
SPSS 19.0 software (IBM) was used to develop statistics and perform analysis of all data. A Student’s t-test was used for the comparison of differences between the two groups. padj < 0.05 was statistically significant.
3. Results
3.1. Pathological Tissue
After the infection of the C. irritans in the T. rubripes, an optical microscope was used to observe the parasitic trophozoites on the gill filaments, which were round or oval in shape (Figure 1A). HE staining was used to further observe the microstructure of the gills in the CG and IG. The findings indicated that the CG exhibited a distinct gill structure, characterized by slender filaments devoid of mucous cells on the gill surface, and an absence of inflammation (Figure 1B). Conversely, the IG displayed gill filaments covered in numerous mucous cells, which had led to swelling and inflammation. The accumulation of mucus obstructed the capillaries within the gills, indicative of hypoxic conditions. At the same time, parasitic trophozoites were observed in the paraffin section of the gill filaments in the IG (Figure 1C).
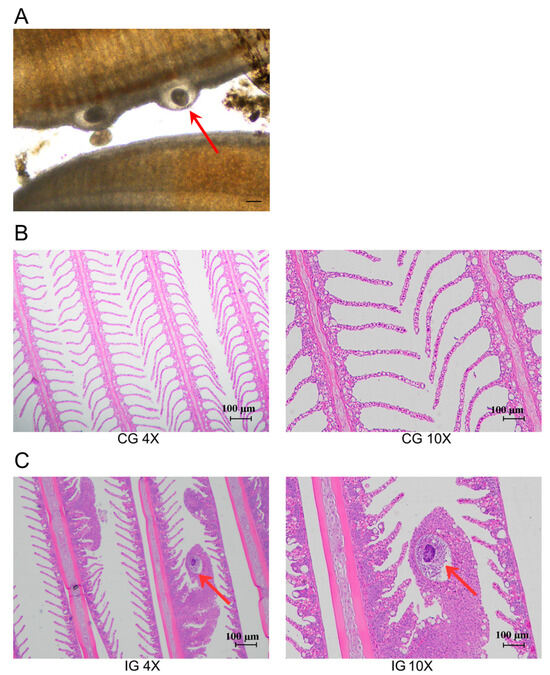
Figure 1.
T. rubripes infected with C. irritans. (A) Gill parasitic trophozoite (B) HE staining of gills in the CG (C) HE staining of gills in the IG. The red arrow points to the trophont of C. irritans. The parasitic stage on the T. rubripes is called the trophont and is round or pear-shaped, capable of rotational movement within the superficial epithelial layer.
3.2. RNA Sequencing and cDNA Library Construction
In this study, we aimed to obtain a global view of the gill tissue transcriptome (including mRNAs, miRNAs, lncRNAs, and circRNAs) of the T. rubripes, to explore which mRNA and ncRNA were involved in T. rubripes resisting C. irritans infection (Figure 2). Six RNA-seq libraries were constructed. For miRNA sequencing, 93,436,063 reads and 91,896,386 clean reads were obtained from six libraries. For each library with a data volume exceeding 0.76G, the percentage of clean reads in the raw reads was more than 97%. The Q30 values were above 97%. The error rate was 0.01%. After small RNA length screening, 86,636,517 total sRNA readings were produced with more than 0.28 GB. The clean reads for each sample were individually aligned to the specified reference genome, with matching rates between 94.55% and 97.89%. The miRNA underwent length screening, repeat sequence alignment, and exon/intron alignment. The analysis of miRNA lengths revealed a characteristic distribution, predominantly centered at 22 nucleotides (Supplementary Table S2). For sequencing of mRNA, lncRNA, and circRNA, six libraries yielded a cumulative 94.99 GB of clean data, exceeding 15.55 GB per sample, with Q30 base percentages surpassing 90.85%. Each sample’s clean reads were separately mapped to the designated reference genome, yielding alignment rates from 87.21% to 90.52% (Supplementary Table S3). In order to perform feature analysis of lncRNA, transcript length, exon count, and ORF length were analyzed. Based on this, the screened lncRNAs were divided into 29.5% LincRNAs, 28.3% antisense lncRNAs, and 42.2% sense overlapping. In comparing length, ORF length, and exon numbers, both annotated and novel lncRNAs are generally shorter or smaller than mRNA. The ORF length of messenger ribonucleic acid and ribonucleic acid is mostly within 1000, and the lncRNA density reaches 0.0100, while messenger ribonucleic acid is less than 0.0025 (Figure S1).
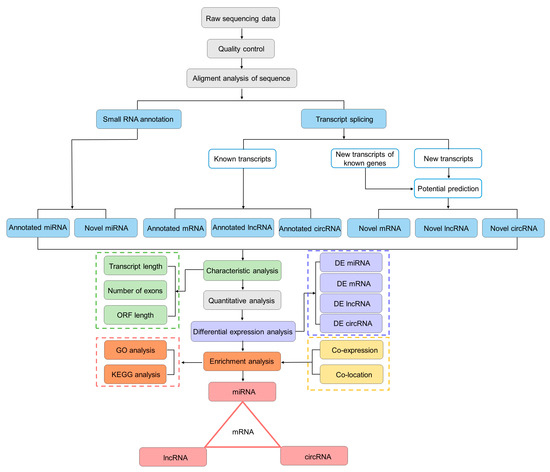
Figure 2.
The flowchart of whole-transcriptome association analysis.
3.3. Expression Profiles and Enrichment Analysis of DEMs
TPM density distribution can comprehensively examine the gene expression patterns of samples (Figure S2A). We conducted a correlation check on gene expression levels between samples (Figure S2B). The Venn shows that 259 DEMs were expressed in both CG and IG, 10 DEMs were specifically expressed in the CG, and 9 DEMs were specifically expressed in the IG (Figure 3A). According to padj < 0.05 and |log2(foldchange)| > 1, 18 DEMs were identified, including 10 known and 8 novel miRNAs. The volcano plot shows the screening of 10 up-regulated DEMs and 8 down-regulated DEMs (Figure 3B). Cluster analysis was performed on differentially expressed genes to obtain a heatmap, which was divided into two clusters (Figure 3C). In the CG, the first type was highly expressed, whereas it exhibited low expression in the IG, with a total of eight differentially expressed miRNAs, such as fru-miR-455, fru-miR-140, fru-miR-338, fru-miR-204a, and fru-miR-26. The second type showed low expression in the control group and high expression in the IG, with a total of ten DEMs, such as fru-miR-30b, fru-miR-221, fru-miR-192, fru-miR-194, and fru-miR-222.
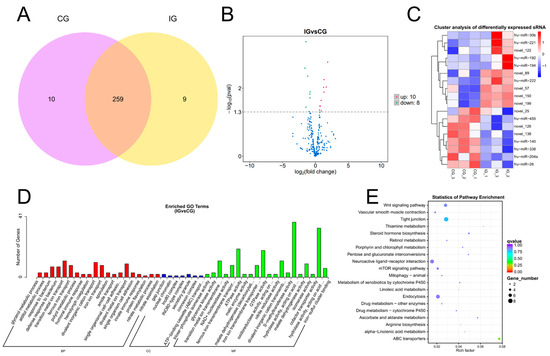
Figure 3.
Detection and functional analysis of DEMs associated with T. rubripes exposure to C. irritans infection. (A) Venn diagram of DEMs. (B) Volcano map of DEM. No significance is shown in blue, up-regulated DEMs are shown in red, and down-regulated DEMs are shown in green. (C) Heat map of DEMs. Identification of DEMs between the CG and IG based on the criteria of padj < 0.05 and |log2FC| > 1. Up-regulated DEMs are depicted in red, while down-regulated DEMs are depicted in blue. (D) GO analysis of DEM target genes. (E) KEGG pathway analysis of DEM target genes. Statistical significance threshold set at padj < 0.05.
We selected mRNA targets predicted by miRanda and RNAhydrid. A cumulative total of 9623 target genes were obtained. We performed GO and KEGG pathway enrichment analysis of the genes targeted by differentially expressed miRNAs within each experimental group. GO showed that target genes of DEMs were enriched in humoral immune response, innate immune response, and hydrolase activity (Figure 3D). The primary KEGG pathways enriched for the differential target genes in the CG and the IG were the Wnt signaling pathway and the mTOR signaling pathway. These pathways play a crucial role in immune function (Figure 3E).
3.4. Expression Profiles and Enrichment Analysis of DELs
The expression of lncRNAs in each group was stable and the sequencing data were of good quality (Supplementary Figure S3, Supplementary Table S4). Venn analysis revealed that there were a total of 4762 DELs that were common between the IG and CG. Additionally, 604 DELs were expressed in the CG, while 478 DELs were identified in the IG (Figure 4A). A total of 214 significant DELs were identified according to the threshold padj < 0.05. As shown in Figure 4B, the IG showed 103 up-regulated genes and 111 down-regulated genes compared to the CG. Hierarchical clustering analysis as shown in Figure 4C shows that each group had a unique set of lncRNAs that were significantly up-regulated in expression only within each group, and within the other groups were significantly down-regulated in expression. The difference in lncRNA expression within groups was not significant and the trend was consistent. There was a significant difference between groups; the trend of up-regulation and down-regulation was obvious. The results indicated that stimulation of the C. irritans infection affected the lncRNA expression pattern in the gills of T. rubripes.
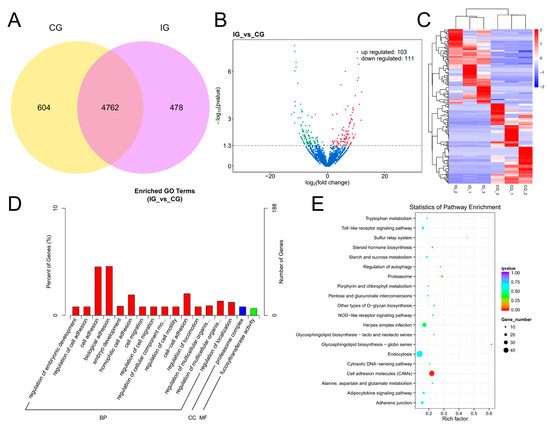
Figure 4.
Detection and functional analysis of DELs associated with T. rubripes exposure to C. irritans infection. (A) Venn diagram of DELs. (B) Volcano map of DELs. No significance is shown in blue, up-regulated DELs are shown in red, and down-regulated DELs are shown in green. (C) Heat map of DELs. Identification of DELs between the CG and IG based on the criteria of padj < 0.05 and |log2FC| > 1. Up-regulated DELs are depicted in red, while down-regulated DELs are depicted in blue. (D) GO analysis of DEL target genes. (E) KEGG pathway analysis of DEL target genes.
Through analyzing the positional relationship and expression correlation between lncRNAs and protein-coding genes, we were able to predict the cis and trans target genes of DELs. This allowed us to gain insight into the potential regulatory roles these lncRNAs may have on the expression of nearby or distant genes. Additionally, we further investigated the functions of these target genes by analyzing their distribution and utilizing GO and KEGG. This comprehensive analysis provided valuable information on the biological processes and signaling pathways that could be impacted by the activity of DELs, shedding light on their potential roles in gene regulation. The GO analysis of co-location is shown in Figure 4D; the target genes of DELs in the IG were annotated to 2801 GO categories compared to the control group, of which 1588 annotations were for biological processes, 395 annotations for cellular components, and 818 to molecular functions. The DELs were predominantly enriched in cell migration, cell adhesion, and proteasome complex. KEGG analysis showed that compared with the CG, the DEL target genes were mainly enriched in TLR signaling pathway, NOD-like receptor (NLR) signaling pathway, p53 signaling pathway, and PPAR signaling pathway (Figure 4E). There were 17 DELs enriched in TLR signaling pathway, 11 DELs enriched in NLR signaling pathway, 36 DELs enriched in p53 signaling pathway, and 38 DELs enriched in PPAR signaling pathway.
3.5. Expression Profiles and Enrichment Analysis of DECs
CircRNAs were identified using find_circ with CIRI, and the percentage of circRNA sources was 13% intron, 20% intergenic, and the rest were exon. circRNA expression was present in all groups. An analysis using Venn diagrams revealed that 451 DELs were found between the IG vs. CG, 495 DELs in the CG, and 237 DELs in the IG (Figure 5A). Based on padj < 0.05 and log2(foldchange)| > 1, DECs were screened as in Figure 5B, and the IG showed six up-regulated expressions and one down-regulated expression compared to the CG. Cluster analysis showed that DECs were divided into two groups, novel_circ_0001560 was highly expressed in the CG and expressed to a low degree in the IG. The other group was expressed to a low degree in the CG and highly expressed in the IG (Figure 5C). The target genes of DECs were annotated into 168 GO categories, of which 107 were annotated for biological processes, 20 for cellular components, and 41 for molecular functions (Figure 5D). The DECs were predominantly enriched for cellular protein metabolic process, cGMP biosynthetic process, and phosphorus–oxygen lyase activity. KEGG enrichment analysis connects biological signaling pathways (Figure 5E). Differentially expressed circRNA target genes are linked to metabolic pathways, various cellular processes, and immune response. In the IG compared to the CG, 11 target genes were enriched in the Calcium signaling pathway, three target genes were enriched in the PPAR signaling pathway, and eight target genes were enriched in the Apelin signaling pathway.
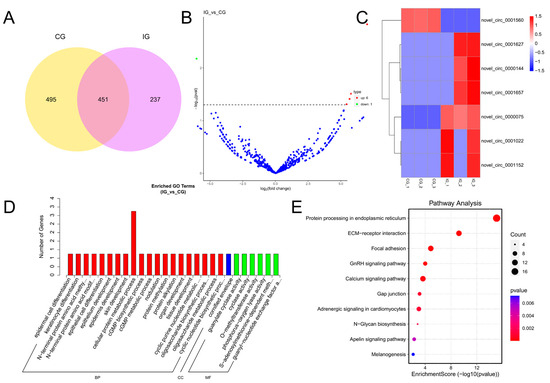
Figure 5.
Detection and functional analysis of DECs associated with T. rubripes exposure to C. irritans infection. (A) Venn diagram of DECs. (B) Volcano map of DECs. No significance is shown in blue, up-regulated DECs are shown in red, and down-regulated DECs are shown in green. (C) Heat map of DECs. Identification of DECs between the CG and IG based on the criteria of padj < 0.05 and |log2FC| > 1. Up-regulated DECs are depicted in red, while down-regulated DECs are depicted in blue. (D) GO analysis of DEC target genes. (E) KEGG pathway analysis of DEC target genes.
3.6. Expression Profiles and Enrichment Analysis of DEGs
For the screening of differentially expressed genes, as shown in Figure 6A, 20,563 DELs were identified between the IG and CG. Among these, 481 DELs were found in the CG, while 389 DELs were uniquely expressed in the IG. There were 2501 differentially expressed mRNAs in the IG compared to the CG, with 1100 up-regulated and 1401 down-regulated expressions (Figure 6B). As shown in Figure 6C, the DEGs that were down-regulated in the CG showed a trend of up-regulation in the IG. Furthermore, compared to the CG, the IG DEG is mainly annotated into the GO category for the ribonucleoprotein complex biogenesis, protein N-linked glycosylation, and NADH dehydrogenase activity (Figure 6D). The KEGG pathway analysis is shown in Figure 6E, and the 238 DEGs were enriched in the metabolic pathway, while 19 DEGs were highly expressed in the PPAR signaling pathway.
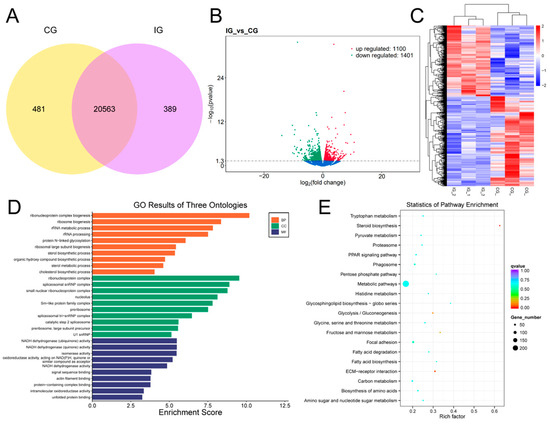
Figure 6.
Detection and functional analysis of DEGs associated with T. rubripes exposure to C. irritans infection. (A) Venn diagram of DEGs. (B) Volcano map of DEGs. No significance is shown in blue, up-regulated DEGs are shown in red, and down-regulated DEGs are shown in green. (C) Heat map of DEGs. Identification of DEGs between the CG and IG based on the criteria of padj < 0.05 and |log2FC| > 1. Up-regulated DEGs are depicted in red, while down-regulated DEGs are depicted in blue. (D) GO enrichment analysis of DEG target genes. (E) KEGG pathway analysis of DEG target genes.
3.7. CeRNA Network Construction
The differentially expressed miRNAs, lncRNAs, circRNAs and mRNAs were first screened, followed by the prediction of target genes of DEMs enriched in the immune pathway. In addition, DEL and DEC, which have target relationships with target genes and DEMs and are differentially expressed, were also predicted and correlation coefficients were calculated, respectively. Based on the correlation coefficients (Supplementary Materials File S1), using expressed gene pairs with the same miRNA binding sites, a key miRNA-lncRNA-mRNA-ceRNA regulatory network was constructed based on the ceRNA theory by the above DEM, DEL and DEG, revealing that non-coding RNAs play important roles in the immune response of T. rubripes to C. irritans infection. According to the prediction, six crucial mRNAs were expected to interact with three miRNAs, which would then interact with 64 lncRNAs (Figure 7A). As a result, an immune-related ceRNA network involving lncRNA-miRNA-mRNA interactions was formed, with five core miRNAs, competing with lncRNAs for mRNA interactions. Of these, fru-miR-194, fru-miR-204a, and fru-miR-338 interacted with lamtor1, rictor, fzd3a, chd9, sfrp1a, and nlk2 targets. fru-miR-204a-rictor up-regulated three lncRNA (LOC115251324, LOC115250966, col25a1) and down-regulated two lncRNA (LOC115250120, LOC105418663). fru-miR-204a-fzd3a up-regulated seven lncRNA (cd276, plch2, LOC115248313, LOC105417236, LOC115252978, LOC115252569, LOC105418562), and down-regulated LOC105418663. rictor competed with fzd3a for LOC105418663. fru-miR-204a-chd9 interacted with 14 down-regulated lncRNAs and six up-regulated lncRNAs. fru-miR-338-sfrp1a up-regulated 2 lncRNAs (LOC115248470, LOC105416523) and down-regulated 19 lncRNAs. Nine lncRNAs participated in the fru-miR-338-nlk2 interaction, with two up-regulated and seven down-regulated. The up-regulated fru-miR-194 was regulated by LOC115252404, LOC101077560, and LOC105417507 and affected the target gene lamtor1. fru-miR-338 was regulated by up-regulated LOC101075208 and LOC105419406, and targeted the nlk2 gene. In addition, fru-miR-194-lamtor1 was enriched in the mTOR signaling pathway, while fru-miR-338-nlk2 was enriched in the Wnt signaling pathway.
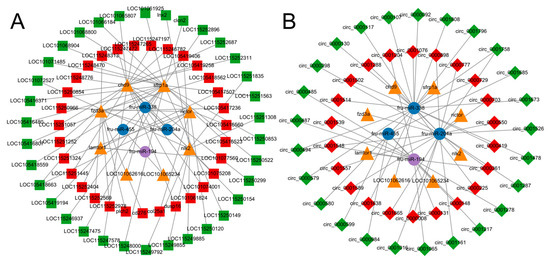
Figure 7.
Identification of the competing endogenous RNA network. (A) lncRNA-miRNA-mRNA network. (B) circRNA-miRNA-mRNA network. The blue circle indicates down-regulated miRNAs, the purple circle indicates up-regulated miRNAs, the yellow triangle indicates mRNAs, the red square indicates up-regulated lncRNAs, the green square indicates down-regulated lncRNAs, the red diamond indicates up-regulated cricRNAs, and the green diamond indicates down-regulated circRNAs.
Integrating the circRNA-miRNA and the miRNA-mRNA regulatory networks, a total of four key miRNAs can interact with 47 circRNAs, which can subsequently interact with eight mRNAs. Thus, an immune-associated miRNA-circRNA-mRNA-ceRNA network contains eight miRNA-centred interactions with circRNAs competing for mRNAs (Figure 7B). Unlike the lncRNA-miRNA-mRNA network, fru-miR-455-LOC101065234 appeared only in the circRNA-miRNA-mRNA network and four circRNAs were involved in regulation, including three down-regulated circRNAs (circ_0000984, circ_0000599, circ_0001685) and up-regulated circ_0000148. Four up-regulated circRNAs and five down-regulated circRNAs compete with lamtor for binding to fru-miR-194. fru-miR-204a is regulated by nine up-regulated circRNAs and five down-regulated circRNAs and targets rictor, fzd3a, and chd9. There are 12 down-regulated circRNAs and eight up-regulated circRNAs down-regulated fru-miR-338 and competed for sfrp1a, nlk2, and LOC101062616. The fru-miR-204a targeting chd9 was regulated by circ_0000361, circ_0000494, and circ_0001758 and was enriched in the Wnt signaling pathway. circ_0000361 up-regulated and simultaneously regulated fru-miR-194 expression, and down-regulated circ_0000494 and circ_0001758 regulated fru-miR-338 expression. This suggests that non-coding RNAs play an important role in the immune process of the T. rubripes against stimulation of C. irritans infection.
3.8. lncRNA-circRNA-miRNA-mRNA Construction
Combined analysis of lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA revealed an interaction network consisting of 64 lncRNAs, 47 circRNAs, 4 miRNAs, and 8 mRNAs (Figure 8). At the same time, in order to further clarify the anti-C. irritans immune mechanism of T. rubripes, the pathway relationship enriched by the screened immune genes is shown in Figure 9. These were mainly enriched in the Wnt signaling pathway and mTOR signaling pathways. Among them, chd9, nlk2, sfrp1a, and rictor gene expression was down-regulated and fzd3a and lamtor1 gene expression was up-regulated. Other genes were screened and linked to immune genes. fru-miR-194-lamtor1 interacted with three up-regulated lncRNAs such as LOC115252404, LOC101077560, LOC105417507, and nine circRNAs such as circ_0000487, circ_0000485, circ_0001539, which constitute a ceRNA regulatory network, in which five circRNAs were down-regulated and four circRNAs were up-regulated. fru-miR-204a, with 16 circRNAs and 32 lncRNAs, regulated the expression of rictor, fzd3a, and chd9. Among them, there were 24 up-regulated lncRNAs and 9 up-regulated circRNAs, 8 down-regulated lncRNAs and 7 down-regulated circRNAs, and fru-miR-204a expression was down-regulated. In addition, fru-miR-204a competed with fru-miR-194 for binding to circ_0000361, and competed with fru-miR-338 for binding to circ_0000494 and circ_0001758. fru-miR-338 targeted 3 DEGs (sfrp1a, nlk2, and LOC101062616), and regulated the expression of 20 circRNAs and 30 lncRNAs. Among them, there were 4 up-regulated lncRNAs and 8 up-regulated circRNAs, 26 down-regulated lncRNAs and 12 down-regulated circRNAs, and fru-miR-338 expression was down-regulated. However, mRNA LOC101062616 did not match significantly differentially expressed lncRNAs. fru-miR-204a targeted fzd3a and chd9 was enriched in the Wnt signaling pathway, while fru-miR-204a targeted fzd3a and rictor was enriched in the mTOR signaling pathway. fru-miR-204a and fzd3a were simultaneously involved in two important immune pathways, so we hypothesized that the interactions between fru-miR-204a and fzd3a play an important role in the immune response against C. irritans in T. rubripes. Since LOC105418663 was involved in both rictor and fzd3a expression, and circ_0000361 regulated both fru-miR-204a and fru-miR-194, a lncRNA-circRNA-miRNA-mRNA ceRNA network composed of LOC105418663-circ_0000361-fru-miR-204a-fzd3a was formed. The only up-regulated key fru-miR-194 targeted lamtor1 was enriched in the mTOR signaling pathway.
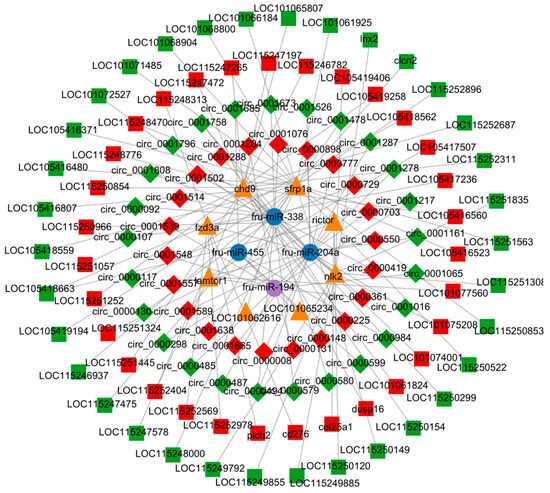
Figure 8.
Identification of the lncRNA-circRNA-miRNA-mRNA network.
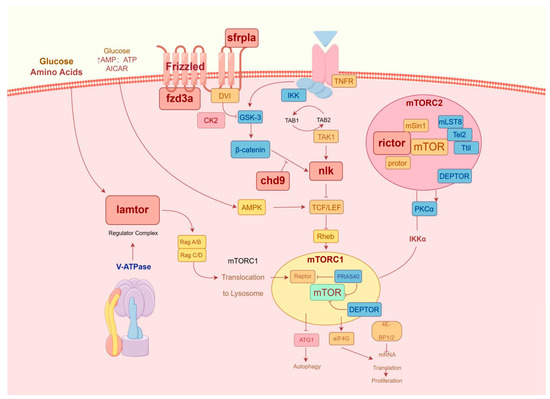
Figure 9.
Immune-related pathways of gene interaction.
3.9. Validation of ncRNAs
To further validate the accuracy of the RNA-seq data, four DEMs, five DELs, and five DEGs in the ceRNA were randomly identified. Correlation analysis was performed (Supplementary Figure S4). The findings showed that the RNA molecules in qRT-PCR followed the same trend as those in RNA-seq, and the results were stable (Figure 10A–C).
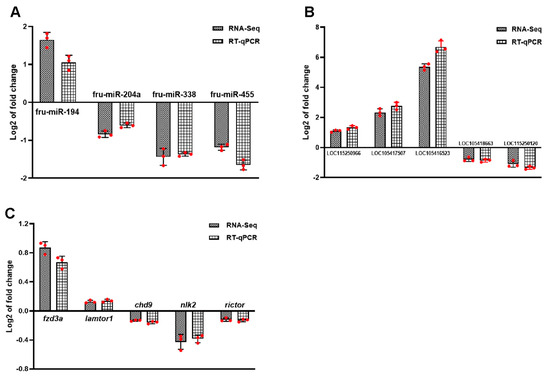
Figure 10.
Identification of ncRNAs by qRT-PCR. (A) DEMs. (B) DELs. (C) DEGs. Log2 FC was expressed as mean ± SD. n = 3. The statistical significance of all genes reached padj < 0.05.
4. Discussion
Currently, parasites are the main problem affecting fish farming. Parasites can even use immune evasion to invade the host, such as avoiding immune system surveillance, molecular mimicry, immunosuppression, immune regulation, and manipulation of the host’s endocrine and nervous systems [26,27]. The host can have different physiological responses to parasites at different stages of development. The immune response of Salmo trutta to developing parasites is that most immune-related genes are up-regulated, while in Oncorhynchus mykiss at the later stage of parasite development, the immune response is weakened and metabolic processes are increased [28,29]. Related studies on fish gill tissues before and after infection with Ichthyophthirius multifiliis found that some of the up-regulated differentially expressed genes were enriched in pathways related to inducing immune responses [30]. However, there are few relevant studies on C. irritans.
MicroRNA (miRNA) is a type of short, single-stranded, ncRNA molecule that can regulate gene expression by binding to the 3′UTR of target genes, leading to protein translation obstruction or mRNA degradation [31]. Previous studies have shown that a large number of fish miRNAs are involved in immune responses associated with pathogen infections, for example, infectious pancreatic necrosis virus (IPNV)-induced Atlantic salmon (Salmo salar) [32], tilapia lake virus (TiLV)-induced tilapia (Oreochromis spp.) [33], and Japanese flounder (Paralichthys olivaceus) induced by Vibrio anguillarum [34]. These miRNAs are mainly enriched in the TLR signaling pathway, the NLR signaling pathway, the Wnt signaling pathway, and the mTOR signaling pathway. They are involved in regulating innate and acquired immune responses by regulating immune cell activity, inflammatory response, antimicrobial protein expression, and immune-related signaling pathways. Additionally, fish primarily rely on their innate immune system to resist pathogen invasion, with the gill mucosa being a critical component. As the first line of defense, gills are often the initial site of pathogen attack during infection. Therefore, elucidating the immune response mechanism against infection is crucial to investigate the essential signaling pathways and immune genes in the gill mucosa.
MiR-194 is a newly identified molecule that plays a significant role in various biological processes. It is extensively involved in the regulation of innate immunity, which is the first line of defense against pathogens. Additionally, miR-194 is critical in controlling apoptosis, the process of programmed cell death, essential for maintaining cellular homeostasis and eliminating damaged or unneeded cells. Furthermore, miR-194 has a substantial impact on inflammation, a complex biological response to harmful stimuli such as infections or injuries. Lastly, miR-194 is deeply integrated into the mechanisms of autophagy. Previous research has indicated that miR-194 has dual characteristics in a variety of diseases. In the inflammatory response, miR-194 has been identified as a promising anti-inflammatory agent [35]. In this study, we found that miR-194 was significantly up-regulated in the case of C. irritans infection, which was consistent with results from studies on inflammation in Danio rerio, Cyprinus carpio, and Mus musculus [36]. Two weeks post-recovery from gossypol poisoning, the expression of miR-194 in C. carpio was significantly reduced. This suggests that miR-194, as a new inflammatory response regulator, actively participates in the immune response. Meanwhile, miR-194 was significantly up-regulated in zebrafish infected with V. harveyi, and overexpression of miR-194 affected LPS-induced increase in immune-related genes [37]. It can suppress the activation of genes associated with inflammation caused by lipopolysaccharide, such as IL-1 and TNF-α [38,39,40]. MiR-194 plays a role in the NF-κB pathway by interacting with TRAF6 [39], TRIM23, and C21ORF91, and several other target genes [41]. In addition, there is evidence that circ_0001278 and circ_0001204 are involved in the regulation of cancer progression, and circ_0001204 can be used as a new biomarker for tuberculosis diagnosis [42,43,44]. The results of this study suggest that circ_0001278 and circ_0001204 may interact with miR-194 and regulate the downstream target gene lamtor1. The target gene lamtor1 is closely related to cell growth, autophagy and immune regulation [45,46]. Wu et al. found that lamtor1 could reduce MHC-II expression, thereby decreasing the anti-tumor response of T cells [47], suggesting that lamtor1 may have an oncogenic role. Knockdown of lamtor1 expression was reported to reduce the activity of non-small cell lung cancer (NSCLC) cells. This is consistent with our results, which showed that fru-miR-194 was up-regulated and targeted lamtor1 was down-regulated to resist stimulation of C. irritans infection. On the other hand, lamtor1 functions as a modulator of the mTORC1 protein. A decrease in lamtor1 expression results in the suppression of mTORC1 activity within retinal pigment epithelial cells [48]. This change has also been observed in NSCLC cells, where the attenuation of lamtor1 led to reduced levels of phosphorylation of mTOR expression. Lamtor1 facilitates the activation of mTORC1, influencing the advancement of colorectal cancer (CRC) induced by inflammation [49]. Moreover, the activation of mTORC1 relies heavily on the ubiquitination of lamtor1, making it a viable target for therapeutic interventions in various diseases.
miR-455 is an emerging molecule that regulates the immune response. miR-455-3p mainly targets genes related to apoptosis and inflammation. Reversing inflammation development and apoptosis induced by circ_102049 knockdown may be achieved through the down-regulation of miR-455-3p [50]. Interestingly, miR-455 expression in gill tissues was reduced after infection with C. irritans. We speculate that miR-455 is similarly involved in modulating anti-inflammatory responses in T. rubripes. miR-455 was down-regulated during Trionyx sinensis hemorrhagic syndrome virus infection, which was also consistent with our results. Research has demonstrated that miR-455 is crucial for Nile tilapia’s response to cold stress and also influences the rhythmic expression of skeletal muscle in Carassius auratus. However, another study showed that overexpression of miR-455-3p led to attenuation of excessive stress-induced cardiac hypertrophy. The results indicate that miR-455 may have a dual function in the inflammatory pathway. In this study, miR-455 targeted a key gene that was associated with four circRNAs, with circ_0000148 being up-regulated and the others down-regulated. Of these, circ_0000984 can up-regulate the expression of CDK6 gene by competitively binding to miR-106b, thereby playing a role in the growth and metastasis of tumor cells [51]. CircRNA is non-coding closed-loop RNA produced by pre-mRNA reverse splicing [52]. Existing literature has shown that circRNA can regulate inflammatory responses by affecting the level of downstream targets [53]. Additionally, because of its covalent closed-loop structure, circRNA is more stable than linear RNA and can be directly released from cells through extracellular vesicles. Therefore, circRNA, as a candidate for therapeutic vaccines, has potentially important functions in gene expression regulation. Based on the results of transcriptome analyses, we believe that the mechanism of miR-455 in the inflammatory response in fish needs to be further investigated.
In related studies of teleost, miR-338 has been shown to participate in regulating the immune response. Studies have shown that miR-338 may have protective or harmful effects in different diseases. For example, miR-338 can inhibit angiogenesis by targeting vascular endothelial growth factor (VEGF) [54]. miR-338 inhibits the transition of gastric cancer cells by inactivating the Wnt signaling pathway [55]. Another study showed that miR-338 has an inhibitory effect on RCC [56,57]. Moreover, the overexpression of miR-338 reduced LPS-induced damage to 16HBE cells [58]. miR-338 targeted at least three key immune-related genes after Oncorhynchus mykiss was infected with hematopoietic necrosis virus (IHNV). In this study, fru-miR-338 also targeted three key genes, including nlk2, sfrp1a, and LOC101062616. Moreover, the expression of fru-miR-338 decreased after infection with C. irritans, suggesting that it may participate in the immune response against C. irritans and alleviate fish infection. Additionally, studies have shown that miRNA can affect the stability of lncRNA by interacting with lncRNA [59,60]. Notably, we predicted and verified the binding relationship between fru-miR-338 and lncRNA, which further suggested that fru-miR-338 and lncRNA interacted in the anti C. irritans response. CircRNAs can regulate inflammatory response by affecting the level of downstream targets [53]. At present, circRNAs in aquatic organisms against C. irritans or other parasitic diseases, such as circMIB2, novel_circ_002325, have been discovered and identified. Our results showed that circRNA was abnormally elevated in the infection group of T. rubripes, and fru-miR-338 targeted nlk2 through circ_0001502 to ultimately inhibit cell apoptosis, thereby affecting the Wnt signaling pathway. miR-338 can reduce the inflammatory damage to cells, and circ0038467 plays a role through the molecular sponge of miR-338 [61]. Circ_0026579 has a similar effect in targeting miR-338 [62]. In addition, among the 20 circRNAs that have a targeted relationship with fru-miR-338, circ_0000419 and circ_0000729 have been shown to play a certain promoting role in the proliferation, migration, and other developmental processes of cancer cells [63,64]. This is consistent with our research findings, as both circ_0000419 and circ_0000729 in the infected group showed up-regulation in the interaction between circ_0000419-fru-miR-338 and circ_0000729. This indicated that the expression of circ_0000419 and circ_0000729 was positively correlated with fish before and after infection. Additionally, studies have shown that hsa_circ_0000092 has a targeted relationship with microRNA-338-3p. The interaction between hsa_circ_0000092 and microRNA-338-3p can regulate HN1 to affect life processes such as cell proliferation, invasion and angiogenesis [65]. Similarly, our research findings suggest that circ_0000092 interacts with fru-miR-338 and forms axial relationships with three target genes. Among them, nlk2 and sfrp1a have been shown to be enriched in the Wnt signaling pathway and involved in innate immune response. In summary, the pathogenesis of C. irritans is complex and multifactorial, including pathophysiological factors such as immune dysfunction and inflammatory response, which ultimately lead to organ dysfunction [66]. Although the relationship between circRNA, fru-miR-338, and mRNA is not entirely clear, our results still provide a new theoretical basis for anti-C. irritans infection.
In this research, the low expression of fru-miR-338 targeted nlk2, sfrp1a and LOC101062616 mediated immune response, indicating that fru-miR-338 regulation can drive immune response progression. The key genes nlk2 and sfrp1a have been confirmed to be enriched in the Wnt signaling pathway, contributing to the innate immune response. NLK, a protein kinase known as Nemo-like kinase, plays a crucial role in both the Wnt signaling pathway and immune response [67,68,69,70,71,72]. It is reported that nlk is a regulator of primary neurogenesis in the neural plate, widely expressed in early development, and acts as a positive regulator of Wnt signaling [73]. nlk not only participates in the development of the nervous system in vertebrates but is also involved in various diseases [4,5,6,7,8]. nlk2 plays a crucial role in viral infections in mammals. Specifically, when HEK293T cells are infected with SEV, nlk2 suppresses the production of ISG56, IFN-β, and ISG15 [74,75]. nlk positively regulates Wnt signal transduction in mammalian neural progenitor cell (NPC)-like cell lines, negatively regulates HEK293 and HeLa cells, and inhibits β-Catenin-mediated transcription [67,76]. Additionally, nlk gene sequences have been reported in many aquatic organisms. For example, the nlk2 of grass carp (Ctenopharyngodon idella) responds to grass carp reovirus (GCRV) stimulation and is highly expressed in the gill and kidney, suggesting that nlk2 is involved in the immune response [77]. Generally, few studies focus on the immune function of nlk2 in teleost. However, it is worth noting that nlk2 is highly conserved from worms to humans [78], suggesting that nlk2 is the target gene of fru-miR-338 in this study. fru-miR-338 is down-regulated to regulate the expression of nlk2 and innate immune response, revealing a new role for nlk2 in activating the Wnt signaling pathway. In addition to nlk, the key gene sfrp can bind Wnt ligands, regulate Wnt signaling and signal through Wnt receptors. sfrp1 has been identified in medaka, zebrafish, and xenopus [79,80] as a Wnt antagonist related to frizzled, which is expressed in the central nervous system and intestine of zebrafish [81]. Functional analysis of Drosophilid showed that sfrp could act as both an inhibitor and promoter, affecting signal activation [82]. However, there are no reports about the expression of sfrp1 in T. rubripes.
miR-204a plays a crucial role in regulating cytokine levels within cellular structures. MiRNA generally regulates its targets through translation inhibition and mRNA instability [83,84]. In the immune system, miRNA is frequently associated with abnormal expression [85]. Currently, miR-204 has been confirmed to be dysregulated in several cancer cells. For example, miR-204 overexpression can trigger cell apoptosis [86]. The overexpression of miR-204 in macrophages can inhibit TLR4/JNK signal transduction, reduce pro-inflammatory cytokine release, and consequently inhibit macrophage inflammation [87]. Furthermore, studies have confirmed miR-204’s role in regulating the proliferation and metastasis of cancer cells [88,89]. Marco Galasso et al. found that, in vitro, miR-204 has a significant effect on melanin [90]. miR-204 is involved in several inflammatory suppression processes and has become a potential therapeutic target for some diseases. CircRNAs with targeting relationships with fru-miR-204a play an important role in regulating key genes. Studies have shown that circ_0001589 and circ_0001665 are significantly up-regulated in clinical tissue samples and cell lines, and they regulate tumor cell proliferation, invasion, and apoptosis [91,92]. The ceRNA network showed that circ_0001589 and circ_0001665 interacted with fru-miR-204a and regulated three target genes, respectively, and were highly expressed in infected tissues. In addition, the expression of some circRNAs was down-regulated, such as circ_0001287, circ_0001016, circ_0001526, and circ_0001161. Circ_0001287 has been shown to be down-regulated in NSCLC tissues, and its overexpression inhibits proliferation, migration, and invasion, and can be involved in the regulation of NSCLC cell proliferation and migration by indirectly modulating PTEN expression [93]. Similarly, circ_0001016 has been shown to promote the growth of prostate cancer cells by targeting miR-23a during cancer progression [94]. This evidence indicates that non-coding RNAs are important in the development of the disease. It has also been reported that the ceRNA network of Sebastes schlegelii after infection with Aeromonas salmonicida shows up-regulation of circ_0000298 expression and a positive correlation with infection time [95]. This is consistent with our research findings. Therefore, circ_0000298 may play an important role in disease immunity in fish. There are limited studies on the interaction between miR-204 and circRNAs together in the immune response of fish. Our research revealed a significant decrease in the expression of miR-204a in the gills of T. rubripes following infection with C. irritans, aligning with findings from prior investigations [96]. In addition, transcriptome data analysis showed that miR-204 simultaneously targeted three key genes, rictor, fzd3a, and chd9. rictor was enriched in the mTOR signaling pathway, chd9 was enriched in the Wnt signaling pathway, and fzd3a was enriched in both the mTOR and Wnt signaling pathways.
The conventional understanding is that Wnt signaling is primarily mediated through seven transmembrane proteins belonging to the frizzled (Fzd) family and the low-density lipoprotein receptor-related protein as co-receptors, which subsequently trigger the activation of the canonical Wnt signaling pathway [97,98]. Deletion of fzd3a may lead to disorder in the Wnt signaling pathway [99]. It has been reported that the xenopus’s fzd3 may inhibit typical Wnt signaling by recruiting transfer-related kinases [100]. This shows that the fzd3 signal has significant biological importance for the common neural circuit across species. On the other hand, knocking down fzd3 could inhibit the growth and metastasis of melanoma cells [101]. The ncRNAs (such as lncRNA and miRNA) can directly or indirectly target fzd3, regulate the Wnt signaling pathway, and affect the proliferation and metastasis of cancer cells [102,103,104]. However, the relationship between fzd3-Wnt signaling and the fish immune response has not been reported. In this study, eight lncRNAs associated with fzd3 were identified, which together with miR-204a constituted the lncRNA-miRNA-mRNA network. This is significant for further screening of functional lncRNAs. Chromodomain helicase DNA binding 9 (chd9) is a regulatory factor that facilitates chromatin structural reorganization [105]. The function of chd9 in human cancer is controversial, and it is considered to be an oncogene in renal cell carcinoma [106] or a tumor suppressor gene in colorectal cancer (CRC) [107]. The role of chd9 in the immune response to C. irritans infection in T. rubripes remains uncharacterized. Through bioinformatics analysis, chd9 has been found to have a binding site for fru-miR-204a. After infection with C. irritans, the expression of fru-miR-204a in the gill of T. rubripes was down-regulated, while the expression of chd9 was up-regulated. This is consistent with the role of chd9 in CRC. Meanwhile, circRNA can act as a sponge to absorb miRNA [87]. Reports indicate that circpdzd8 up-regulates chd9 and promotes cancer progression by sponging miR-197-5p [108]. In this study, fru-miR-204a regulates the Wnt signaling pathway by binding to chd9 and simultaneously targeting multiple circRNAs. In conclusion, the combination of chd9 and fru-miR-204a can serve as a potential therapeutic target, and inhibiting chd9 expression may be a key mechanism in combating C. irritans infection.
The mTOR pathway is essential in controlling the differentiation processes of both innate and adaptive immune cells [109,110]. The role of mTOR in immune cell activation and functional characteristics has been well-established [111]. Similarly, we observed that miR-204a was down-regulated and targeted rictor after infection with C. irritans. As the core component of mTORC2, rictor plays a role in amino acid sensing [112]. rictor affects intestinal inflammation in hybrid grouper [113]. Knockdown of rictor can induce apoptosis in breast cancer cells, inhibit cell migration and metastasis [114,115], and alleviate pancreatic tumor occurrence [115]. miR-16 and let-7 target rictor and regulate the induction of T cells [116]. Therefore, compared to the CG group, regulating the key gene rictor in the mTOR pathway by down-regulating miRNA expression is an important immune process in T. rubripes against infection. The mTOR pathway integrates multiple environmental signals [117]. Dysregulation of the mTOR pathway has been found in many diseases [118,119,120]. Therefore, targeting the mTOR represents a potential therapeutic strategy [121]. Concurrently, the immune mechanism in T. rubripes was induced by C. irritans. A total of 16 circRNAs and five lncRNAs combined to form two ceRNA networks, which regulate the mTOR pathway through miR-204a, thereby resisting infection. The research indicates that miR-204a is a crucial component in the immune response mechanism of marine fish to C. irritans. In this study, the CG and the IG were mainly enriched in the Wnt signaling pathway and mTOR signaling pathway, which have immune functions. We speculate that immune pathway signals are activated when resisting C. irritans infection. This discovery offers a key theoretical foundation for future advancements in vaccine development in related areas.
5. Conclusions
In this study, the expression profile of ncRNA after infection with C. irritans by T. rubripes was comprehensively analyzed. We identified 18 DEMs, 214 DELs, 7 DECs, and 2501 DEGs, and performed functional enrichment analysis. We comprehensively screened and analyzed ncRNAs related to immune pathways including the Wnt signaling pathway and mTOR signaling pathway, and constructed lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks. Based on this, we constructed an integrated network of lncRNA-circRNA-miRNA-mRNA and predicted the LOC105418663-circ_0000361-fru-miR-204a-fzd3a axis, further revealing ceRNA function and providing new insights into and support for the study of immune mechanisms related to ncRNAs in T. rubripes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology13100788/s1, File S1: Correlation Coefficients; File S2: Supplementary methods and materials; Table S1: Primers used in real-time PCR for validation, Table S2: Summary of microRNA sequencing data, Table S3: Summary of lncRNA sequencing data; Table S4: Reference genome mapped information of LncRNA; Table S5: Reference genome mapped information of miRNA; Figure S1: Statistics on the length distribution of total sRNA fragments and lncRNA Characteristics, Figure S2: Analysis of miRNA expression levels. Figure S3: FPKM expression distribution, Supplementary Figure S4: Correlation analysis between RNA-seq and qRT-PCR.
Author Contributions
Data curation, Y.X. and X.Y.; Formal analysis, Y.X. and X.Y.; Funding acquisition, Y.L.; Investigation, X.Y.; Methodology, Y.L. and Z.Y.; Project administration, Y.L.; Resources, Z.Y. and Y.Y.; Supervision, Y.L.; Validation, Y.Y.; Writing—original draft, Y.X.; Writing—review & editing, Y.L. and Y.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Innovation Support Program for High-level Talents of Dalian City [grant number 2021RQ089]. The APC was funded by the Innovation Support Program for High-level Talents of Dalian City [grant number 2021RQ089].
Institutional Review Board Statement
The study was conducted according to the Guidelines for Experimental Animals set out by the Ministry of Science and Technology and approved by the Institutional Animal Care and Use Committee at Dalian Ocean University (located in Dalian, China); Approval Code: DLOU2024006; Approval Date: 28 June 2024.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article and Supplementary Materials; further inquiries can be directed to the author.
Acknowledgments
We would like to express our gratitude to our colleagues who have contributed to this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, Y.; Jiang, B.; Mo, Z.; Li, A.; Dan, X. Cryptocaryon irritans (Brown, 1951) is a serious threat to aquaculture of marine fish. Rev. Aquac. 2021, 14, 218–236. [Google Scholar] [CrossRef]
- Hirazawa, N.; Oshima, S.I.; Hara, T.; Mitsuboshi, T.; Hata, K. Antiparasitic effect of medium-chain fatty acids against the ciliate Cryptocaryon irritans infestation in the red sea bream Pagrus major. Aquaculture 2001, 198, 219–228. [Google Scholar] [CrossRef]
- Chang, L.C.; Karin, M. Mammalian MAP kinase signaling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef]
- Liao, K.; Lou, X.; Yang, Z. Comparative analysis of the economic feasibility of Tiger puffer (Takifugu rubripes) aquaculture in China. Aquacult. Int. 2023, 32, 939–961. [Google Scholar] [CrossRef]
- Xia, Y.Q.; Hua, X.T.; Zhu, S.; Liu, Y.; Liu, P.F. Characterization and expression analysis of the complement component 8β from pufferfish (Takifugu rubripes). J. Aquac. Rep. 2021, 21, 100861. [Google Scholar] [CrossRef]
- Reyes-Cerpa, S.; Reyes-Lopez, F.; Toro-Ascuy, D.; Montero, R.; Maisey, K.; Acuna-Castillo, C.; Sunyer, J.O.; Parra, D.; Sandino, A.M.; Imarai, M. Induction of anti-inflammatory cytokine expression by IPNV in persistent infection. Fish Shellfish Immunol. 2014, 41, 172–182. [Google Scholar] [CrossRef]
- Rigos, G.; Padrós, F.; Golomazou, E.; Zarza, C. Antiparasitic approaches and strategies in European aquaculture, with emphasis on Mediterranean marine finfish farming: Present scenarios and future visions. Rev. Aquac. 2024, 16, 622–643. [Google Scholar] [CrossRef]
- Witzany, G.; Nowacki, M. Interactions Between Parasitic Ciliates and Their Hosts: Ichthyophthirius multifiliis and Cryptocaryon irritans as Examples. Biocommunication Ciliates 2016, 327–350. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Alvarez-Pellitero, P. Fish immunity and parasite infections: From innate immunity to immunoprophylactic prospects. Vet. Immunol. Immunopathol. 2008, 126, 171–198. [Google Scholar] [CrossRef]
- Ma, R.; Yu, Y.; Liu, X.; Lei, Y.; Zhou, S.; Xie, X.; Jin, S.; Qian, D.; Yin, F. Transcriptomic analysis of Nibea albiflora skin in response to infection by Cryptocaryon irritans. Fish Shellfish Immunol. 2020, 98, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Josepriya, T.A.; Chien, K.H.; Lin, H.Y.; Huang, H.N.; Wu, C.J.; Song, Y.L. Immobilization antigen vaccine adjuvanted by parasitic heat shock protein 70C confers high protection in fish against cryptocaryonosis. Fish Shellfish Immunol. 2015, 45, 517–527. [Google Scholar] [CrossRef]
- Chu, Q.; Xu, T.; Zheng, W.; Chang, R.; Zhang, L. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PLoS Pathog. 2020, 16, e1008670. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, Y.; Cao, Z.; Sun, Y.; Chen, Y.; Xiang, Y.; Wang, L.; Zhang, S.; Guo, W. Comparative analysis of the expression patterns of IL-1beta, IL-11, and IL-34 in golden pompano (Trachinotus ovatus) following different pathogens challenge. Fish Shellfish Immunol. 2019, 93, 863–870. [Google Scholar] [CrossRef]
- Wilhelm, B.T.; Marguerat, S.; Watt, S.; Schubert, F.; Wood, V.; Goodhead, I.; Penkett, C.J.; Rogers, J.; Bahler, J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 2008, 453, 1239–1243. [Google Scholar] [CrossRef]
- Chen, R.; Mao, Y.; Wang, J.; Liu, M.; Qiao, Y.; Zheng, L.; Su, Y.; Ke, Q.; Zheng, W. Molecular mechanisms of an antimicrobial peptide piscidin (Lc-pis) in a parasitic protozoan, Cryptocaryon irritans. BMC Genom. 2018, 19, 192. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Karreth, F.A.; Reschke, M.; Ruocco, A.; Ng, C.; Chapuy, B.; Leopold, V.; Sjoberg, M.; Keane, T.M.; Verma, A.; Ala, U.; et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 2015, 161, 319–332. [Google Scholar] [CrossRef]
- Tang, J.; Zhuo, H.; Zhang, X.; Jiang, R.; Ji, J.; Deng, L.; Qian, X.; Zhang, F.; Sun, B. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014, 5, e1549. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xie, Q.; Xu, H.; Li, J.; Li, Y. Circular RNAs and cancer. Cancer Lett. 2017, 396, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.M.; Li, A.X.; Lin, X.T.; Teng, N.; Zhu, X.Q. A standardized method to propagate Cryptocaryon irritans on a susceptible host pompano Trachinotus ovatus. Aquaculture 2006, 258, 127–133. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Faber, M.; Shaw, S.; Yoon, S.; de Paiva Alves, E.; Wang, B.; Qi, Z.; Okamura, B.; Hartikainen, H.; Secombes, C.J.; Holland, J.W. Comparative transcriptomics and host-specific parasite gene expression profiles inform on drivers of proliferative kidney disease. Sci. Rep. 2021, 11, 2149. [Google Scholar] [CrossRef]
- Bailey, C.; von Siebenthal, E.W.; Rehberger, K.; Segner, H. Transcriptomic analysis of the impacts of ethinylestradiol (EE2) and its consequences for proliferative kidney disease outcome in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 222, 31–48. [Google Scholar] [CrossRef]
- Sudhagar, A.; Ertl, R.; Kumar, G.; El-Matbouli, M. Transcriptome profiling of posterior kidney of brown trout, Salmo trutta, during proliferative kidney disease. Parasit. Vectors 2019, 12, 569. [Google Scholar] [CrossRef]
- Syahputra, K.; Kania, P.W.; Al-Jubury, A.; Jafaar, R.M.; Dirks, R.P.; Buchmann, K. Transcriptomic analysis of immunity in rainbow trout (Oncorhynchus mykiss) gills infected by Ichthyophthirius multifiliis. Fish Shellfish Immunol. 2019, 86, 486–496. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Woldemariam, N.T.; Agafonov, O.; Sindre, H.; Hoyheim, B.; Houston, R.D.; Robledo, D.; Bron, J.E.; Andreassen, R. miRNAs Predicted to Regulate Host Anti-viral Gene Pathways in IPNV-Challenged Atlantic Salmon Fry Are Affected by Viral Load, and Associated with the Major IPN Resistance QTL Genotypes in Late Infection. Front. Immunol. 2020, 11, 2113. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Li, Y.; Yin, J.; Ren, Y.; Shi, C.; Bergmann, S.M.; Zhu, X.; Zeng, W. Integrated analysis of mRNA-miRNA expression in Tilapia infected with Tilapia lake virus (TiLV) and identifies primarily immuneresponse genes. Fish Shellfish Immunol. 2020, 99, 208–226. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Sun, L. Micro-Transcriptome Analysis Reveals Immune-Related MicroRNA Regulatory Networks of Paralichthys olivaceus Induced by Vibrio anguillarum Infection. Int. J. Mol. Sci. 2020, 21, 4252. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, Y.; Liu, H.; Yang, A.K.; Di, J.M.; Tan, G.M.; Wang, H.F.; Qiu, J.G.; Zhang, W.J.; Jiang, Q.W.; et al. MiR-194 functions as a tumor suppressor in laryngeal squamous cell carcinoma by targeting Wee1. J. Hematol. Oncol. 2017, 10, 32. [Google Scholar] [CrossRef]
- Li, G.; Zhao, Y.; Wen, L.; Liu, Z.; Yan, F.; Gao, C. Identification and characterization of microRNAs in the spleen of common carp immune organ. J. Cell Biochem. 2014, 115, 1768–1778. [Google Scholar] [CrossRef]
- Wu, T.H.; Pan, C.Y.; Lin, M.C.; Hsieh, J.C.; Hui, C.F.; Chen, J.Y. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish. Physiol. Biochem. 2012, 38, 1299–1310. [Google Scholar] [CrossRef]
- Dinh, H.; Hong, Y.H.; Lillehoj, H.S. Modulation of microRNAs in two genetically disparate chicken lines showing different necrotic enteritis disease susceptibility. Vet. Immunol. Immunopathol. 2014, 159, 74–82. [Google Scholar] [CrossRef]
- Kong, L.; Sun, M.; Jiang, Z.; Li, L.; Lu, B. MicroRNA-194 Inhibits Lipopolysaccharide-Induced Inflammatory Response in Nucleus Pulposus Cells of the Intervertebral Disc by Targeting TNF Receptor-Associated Factor 6 (TRAF6). Med. Sci. Monit. 2018, 24, 3056–3067. [Google Scholar] [CrossRef]
- Wang, K.; Lai, C.; Gu, H.; Zhao, L.; Xia, M.; Yang, P.; Wang, X. miR-194 Inhibits Innate Antiviral Immunity by Targeting FGF2 in Influenza H1N1 Virus Infection. Front. Microbiol. 2017, 8, 2187. [Google Scholar] [CrossRef]
- Bao, C.; Li, Y.; Huan, L.; Zhang, Y.; Zhao, F.; Wang, Q.; Liang, L.; Ding, J.; Liu, L.; Chen, T.; et al. NF-kappaB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1alpha. Sci. Signal 2015, 8, ra75. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Yan, Y.; Wang, H.; Bian, J.; Deng, Z.; Su, X.; Yang, Z.; Song, J. Hsa_circ_0001278 Facilitates Colorectal Cancer Progression via Sponging miR-338-5p and Regulating AMOTL1 Expression. Comb. Chem. High Throughput Screen. 2023, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Su, R.; Yao, F.; Peng, Y.; Luo, Q.; Li, J. Circulating circular RNAs hsa_circ_0001204 and hsa_circ_0001747 act as diagnostic biomarkers for active tuberculosis detection. Int. J. Clin. Exp. Pathol. 2018, 11, 586–594. [Google Scholar] [PubMed]
- Ma, X.; Wang, F.; Zhen, L.; Cai, Q. Hsa_circ_0001204 modulates inflammatory response of macrophages infected by Mycobacterium tuberculosis via TLR4/NF-kappaB signalling pathway. Clin. Exp. Pharmacol. Physiol. 2023, 50, 132–139. [Google Scholar] [CrossRef]
- Kimura, T.; Kumanogoh, A.; Okada, M. Roles of Lamtor1 in Macrophages, CD4+ T-cells, and Regulatory T-cells. Crit. Rev. Immunol. 2018, 38, 403–414. [Google Scholar] [CrossRef]
- Sun, Y.; Guan, Z.; Sheng, Q.; Duan, W.; Zhao, H.; Zhou, J.; Deng, Q.; Pei, X. N-myristoyltransferase-1 deficiency blocks myristoylation of LAMTOR1 and inhibits bladder cancer progression. Cancer Lett. 2022, 529, 126–138. [Google Scholar] [CrossRef]
- Wu, B.; Wang, Q.; Li, B.; Jiang, M. LAMTOR1 degrades MHC-II via the endocytic in hepatocellular carcinoma. Carcinogenesis 2022, 43, 1059–1070. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, H.; Zhang, Y.F.; Zhou, Z.; Wu, S. MicroRNA-29 enhances autophagy and cleanses exogenous mutant alphaB-crystallin in retinal pigment epithelial cells. Exp. Cell Res. 2019, 374, 231–248. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, N.; Peng, X.; Chen, L.; Meng, T.; Jiang, C.; Jin, J.; Zhang, J.; Duan, Q.; Tian, H.; et al. TRAF4-Mediated LAMTOR1 Ubiquitination Promotes mTORC1 Activation and Inhibits the Inflammation-Induced Colorectal Cancer Progression. Adv. Sci. 2024, 11, e2301164. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Y.; Zhu, S.; Li, F.; Xu, J.; Zhang, L.; Shu, H. circRNA circ_102049 Implicates in Pancreatic Ductal Adenocarcinoma Progression through Activating CD80 by Targeting miR-455-3p. Mediators Inflamm. 2021, 2021, 8819990. [Google Scholar] [CrossRef]
- Xu, X.W.; Zheng, B.A.; Hu, Z.M.; Qian, Z.Y.; Huang, C.J.; Liu, X.Q.; Wu, W.D. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget 2017, 8, 91674–91683. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Li, M.; Hua, Q.; Shao, Y.; Zeng, H.; Liu, Y.; Diao, Q.; Zhang, H.; Qiu, M.; Zhu, J.; Li, X.; et al. Circular RNA circBbs9 promotes PM(2.5)-induced lung inflammation in mice via NLRP3 inflammasome activation. Environ. Int. 2020, 143, 105976. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, W.; Zeng, X.C.; Jiang, N.; Fu, B.S.; Guo, Y.; Yi, H.M.; Li, H.; Zhang, Q.; Chen, W.J.; et al. Down-regulation of microRNA-338-3p promoted angiogenesis in hepatocellular carcinoma. Biomed. Pharmacother. 2016, 84, 583–591. [Google Scholar] [CrossRef]
- Song, B.; Lin, H.X.; Dong, L.L.; Ma, J.J.; Jiang, Z.G. MicroRNA-338 inhibits proliferation, migration, and invasion of gastric cancer cells by the Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1290–1296. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Li, H.; Niu, X.; Liu, X.; Pei, D.; Guo, X.; Xu, X.; Li, Y. miR-338-3p inhibits the invasion of renal cell carcinoma by downregulation of ALK5. Oncotarget 2017, 8, 64106–64113. [Google Scholar] [CrossRef][Green Version]
- Tong, Z.; Meng, X.; Wang, J.; Wang, L. MicroRNA-338-3p targets SOX4 and inhibits cell proliferation and invasion of renal cell carcinoma. Exp. Ther. Med. 2017, 14, 5200–5206. [Google Scholar] [CrossRef]
- Yang, J.; Huang, Q.; Liao, P.; Zhang, P.; Sun, S.; Xu, Q. Mechanism of miR-338-3p in sepsis-induced acute lung injury via indirectly modulating ATF4. Transpl. Immunol. 2023, 76, 101681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, Y.; Liao, Q. Long noncoding RNA: A crosslink in biological regulatory network. Brief. Bioinform. 2018, 19, 930–945. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; He, Y.; Lin, L.; Qi, Z.; Ma, L.; Li, L.; Su, Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. 2016, 37, 1683–1691. [Google Scholar] [CrossRef]
- Liu, G.; Wan, Q.; Li, J.; Hu, X.; Gu, X.; Xu, S. Circ_0038467 regulates lipopolysaccharide-induced inflammatory injury in human bronchial epithelial cells through sponging miR-338-3p. Thorac. Cancer 2020, 11, 1297–1308. [Google Scholar] [CrossRef]
- Cheng, S.; Chen, C.; Wang, L. Knockdown of circ_0026579 ameliorates lipopolysaccharide (bacterial origin)-induced inflammatory injury in bronchial epithelium cells by targeting miR-338-3p/TBL1XR1 axis. Transpl. Immunol. 2022, 74, 101635. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Geng, Y.; Hu, C.; Wang, J.; Quan, Y.; Yu, T.; Zhou, X. Circ_0000419 acts as a miR-580 sponge to accelerate colorectal cancer progression via regulating DNMT3B. J. Biol. Regul. Homeost. Agents 2022, 36, 21–34. [Google Scholar]
- Li, S.; Sun, X.; Miao, S.; Lu, T.; Wang, Y.; Liu, J.; Jiao, W. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac. Cancer 2018, 9, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Wang, J.; Li, W.; Lu, Y.; Wu, X.; Long, X.; Luo, C.; Wei, H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J. Cell Mol. Med. 2024, 28, e15010. [Google Scholar] [CrossRef]
- Huang, M.; Cai, S.; Su, J. The Pathogenesis of Sepsis and Potential Therapeutic Targets. Int. J. Mol. Sci. 2019, 20, 5376. [Google Scholar] [CrossRef]
- Ishitani, T.; Ninomiya-Tsuji, J.; Nagai, S.; Nishita, M.; Meneghini, M.; Barker, N.; Waterman, M.; Bowerman, B.; Clevers, H.; Shibuya, H.; et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature 1999, 399, 798–802. [Google Scholar] [CrossRef]
- Ishitani, T.; Hirao, T.; Suzuki, M.; Isoda, M.; Ishitani, S.; Harigaya, K.; Kitagawa, M.; Matsumoto, K.; Itoh, M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat. Cell Biol. 2010, 12, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Kanei-Ishii, C.; Ninomiya-Tsuji, J.; Tanikawa, J.; Nomura, T.; Ishitani, T.; Kishida, S.; Kokura, K.; Kurahashi, T.; Ichikawa-Iwata, E.; Kim, Y.; et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes. Dev. 2004, 18, 816–829. [Google Scholar] [CrossRef]
- Ohkawara, B.; Shirakabe, K.; Hyodo-Miura, J.; Matsuo, R.; Ueno, N.; Matsumoto, K.; Shibuya, H. Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes. Dev. 2004, 18, 381–386. [Google Scholar] [CrossRef]
- Kojima, H.; Sasaki, T.; Ishitani, T.; Iemura, S.; Zhao, H.; Kaneko, S.; Kunimoto, H.; Natsume, T.; Matsumoto, K.; Nakajima, K. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 4524–4529. [Google Scholar] [CrossRef]
- Zeng, Y.A.; Rahnama, M.; Wang, S.; Sosu-Sedzorme, W.; Verheyen, E.M. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development 2007, 134, 2061–2071. [Google Scholar] [CrossRef] [PubMed]
- Ota, S.; Ishitani, S.; Shimizu, N.; Matsumoto, K.; Itoh, M.; Ishitani, T. NLK positively regulates Wnt/beta-catenin signalling by phosphorylating LEF1 in neural progenitor cells. EMBO J. 2012, 31, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Peng, C.; Wu, G.; Wang, Y.; Liu, R.; Yang, S.; He, S.; He, F.; Yuan, Q.; Huang, Y.; et al. Expression of NLK and its potential effect in ovarian cancer chemotherapy. Int. J. Gynecol. Cancer 2011, 21, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Emami, K.H.; Brown, L.G.; Pitts, T.E.; Sun, X.; Vessella, R.L.; Corey, E. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate 2009, 69, 1481–1492. [Google Scholar] [CrossRef]
- Li, S.Z.; Zhang, H.H.; Liang, J.B.; Song, Y.; Jin, B.X.; Xing, N.N.; Fan, G.C.; Du, R.L.; Zhang, X.D. Nemo-like kinase (NLK) negatively regulates NF-kappa B activity through disrupting the interaction of TAK1 with IKKbeta. Biochim. Biophys. Acta 2014, 1843, 1365–1372. [Google Scholar] [CrossRef][Green Version]
- Yu, T.; Zeng, Q.; Mao, H.; Liu, Y.; Zhang, H.; Wang, S.; Hu, C.; Xu, X. Grass carp (Ctenopharyngodon idella) NLK2 inhibits IFN I response through blocking MAVS-IRF3 axis. Fish Shellfish Immunol. 2022, 131, 206–217. [Google Scholar] [CrossRef]
- Kortenjann, M.; Wehrle, C.; Nehls, M.C.; Boehm, T. Only one nemo-like kinase gene homologue in invertebrate and mammalian genomes. Gene 2001, 278, 161–165. [Google Scholar] [CrossRef]
- Jones, S.E.; Jomary, C. Secreted Frizzled-related proteins: Searching for relationships and patterns. Bioessays 2002, 24, 811–820. [Google Scholar] [CrossRef]
- Esteve, P.; Lopez-Rios, J.; Bovolenta, P. SFRP1 is required for the proper establishment of the eye field in the medaka fish. Mech. Dev. 2004, 121, 687–701. [Google Scholar] [CrossRef]
- Pezeron, G.; Anselme, I.; Laplante, M.; Ellingsen, S.; Becker, T.S.; Rosa, F.M.; Charnay, P.; Schneider-Maunoury, S.; Mourrain, P.; Ghislain, J. Duplicate sfrp1 genes in zebrafish: sfrp1a is dynamically expressed in the developing central nervous system, gut and lateral line. Gene Expr. Patterns 2006, 6, 835–842. [Google Scholar] [CrossRef]
- Uren, A.; Reichsman, F.; Anest, V.; Taylor, W.G.; Muraiso, K.; Bottaro, D.P.; Cumberledge, S.; Rubin, J.S. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 2000, 275, 4374–4382. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes. Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Hu, B.; Sun, M.; Liu, J.; Hong, G.; Lin, Q. MicroRNA-204 suppressed proliferation and motility capacity of human hepatocellular carcinoma via directly targeting zinc finger E-box binding homeobox 2. Oncol. Lett. 2017, 13, 3823–3830. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Liu, X.; Yu, J.; Ban, T.; Zhang, Z.; Wang, P.; Huang, R.; Zheng, F.; Chang, Y.; Peng, W.; et al. Nuclear miR-204-3p mitigates metabolic dysfunction-associated steatotic liver disease in mice. J. Hepatol. 2024, 80, 834–845. [Google Scholar] [CrossRef]
- Wu, G.; Wang, J.; Chen, G.; Zhao, X. microRNA-204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting zinc-finger E-box-binding homeobox 1 (ZEB1). Am. J. Transl. Res. 2017, 9, 3599–3610. [Google Scholar]
- Canu, V.; Sacconi, A.; Lorenzon, L.; Biagioni, F.; Lo Sardo, F.; Diodoro, M.G.; Muti, P.; Garofalo, A.; Strano, S.; D’Errico, A.; et al. MiR-204 down-regulation elicited perturbation of a gene target signature common to human cholangiocarcinoma and gastric cancer. Oncotarget 2017, 8, 29540–29557. [Google Scholar] [CrossRef]
- Galasso, M.; Morrison, C.; Minotti, L.; Corra, F.; Zerbinati, C.; Agnoletto, C.; Baldassari, F.; Fassan, M.; Bartolazzi, A.; Vecchione, A.; et al. Loss of miR-204 expression is a key event in melanoma. Mol. Cancer 2018, 17, 71. [Google Scholar] [CrossRef]
- Ma, T.; Guo, J.; Han, J.; Li, L.; Ren, Y.; Huang, J.; Diao, G.; Zheng, X.; Zheng, Y. Circ_0001589/miR-1248/HMGB1 axis enhances EMT-mediated metastasis and cisplatin resistance in cervical cancer. Mol. Carcinog. 2023, 62, 1645–1658. [Google Scholar] [CrossRef]
- Wu, L.; Wang, Y.; Liu, Q.; Wu, J.; Zheng, H.; Lin, B.; Huang, S. Circ_0001665 Contributes to the Occurrence of Vestibular Schwannoma via Targeting miR-302a-3p/Adam9/EGFR Signaling Pathway. Neuroscience 2022, 490, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Li, Y.; Feng, X.Z.; Li, D.B. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered 2021, 12, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, P.; Yu, B.; Liu, J. The Circular RNA circXPO1 Promotes Tumor Growth via Sponging MicroRNA-23a in Prostate Carcinoma. Front. Oncol. 2021, 11, 712145. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, N.; Yu, H.; Zhang, X.; Cao, M.; Li, C. Construction and analysis of competing endogenous RNA ceRNA networks in the liver of black rockfish (Sebastes schlegelii) following Aeromonas salmonicida infection. Comp. Immunol. Rep. 2024, 6, 200124. [Google Scholar] [CrossRef]
- Wu, L.; Li, Y.; Fan, J.M.; Zhang, Z.M.; Ouyang, J.L.; Ni, T.T.; Wu, H.X.; Li, H. MicroRNA-204 targets signal transducer and activator of transcription 5 expression and inhibits proliferation of B-cell lymphoma cells. Mol. Med. Rep. 2015, 11, 4567–4572. [Google Scholar] [CrossRef]
- van Amerongen, R.; Nusse, R. Towards an integrated view of Wnt signaling in development. Development 2009, 136, 3205–3214. [Google Scholar] [CrossRef]
- Yanfeng, W.; Saint-Jeannet, J.P.; Klein, P.S. Wnt-frizzled signaling in the induction and differentiation of the neural crest. Bioessays 2003, 25, 317–325. [Google Scholar] [CrossRef]
- Slusarski, D.C.; Pelegri, F. Calcium signaling in vertebrate embryonic patterning and morphogenesis. Dev. Biol. 2007, 307, 1–13. [Google Scholar] [CrossRef]
- Kibardin, A.; Ossipova, O.; Sokol, S.Y. Metastasis-associated kinase modulates Wnt signaling to regulate brain patterning and morphogenesis. Development 2006, 133, 2845–2854. [Google Scholar] [CrossRef]
- Li, C.; Nguyen, V.; Clark, K.N.; Zahed, T.; Sharkas, S.; Filipp, F.V.; Boiko, A.D. Down-regulation of FZD3 receptor suppresses growth and metastasis of human melanoma independently of canonical WNT signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 4548–4557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, Y.; Wang, X.; Wang, J.; Xi, G. lncRNA SNHG10 Promotes the Proliferation and Invasion of Osteosarcoma via Wnt/beta-Catenin Signaling. Mol. Ther. Nucleic Acids 2020, 22, 957–970. [Google Scholar] [CrossRef]
- Kenneth, M.J.; Shishir, T.A.; Haque, F.K.M. In silico analysis reveals mir-98-5p as a potential inhibitor of tumor cell proliferation and metastasis in colorectal cancer by targeting the fzd3 receptor of the Wnt signaling pathway. J. Genet. Eng. Biotechnol. 2023, 21, 79. [Google Scholar] [CrossRef]
- Zhi, Y.; Du, P.; Li, Y.; Liu, H.; Jiang, T.; Zhao, X.; Li, X. SOX21-AS1 Augmented Cervical Cancer Growth by Triggering FZD3 to Activate the Wnt/beta-Catenin Signaling Pathway. Biochem. Genet. 2024. [Google Scholar] [CrossRef]
- Lamar, K.J.; Carvill, G.L. Chromatin Remodeling Proteins in Epilepsy: Lessons from CHD2-Associated Epilepsy. Front. Mol. Neurosci. 2018, 11, 208. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Ran, X.G.; Du, Y.Q.; Ren, F.; Tian, Y.; Wang, Y.; Chen, M.M. High CHD9 expression is associated with poor prognosis in clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 3697–3702. [Google Scholar] [PubMed]
- Xu, L.; Peng, H.; Huang, X.X.; Xia, Y.B.; Hu, K.F.; Zhang, Z.M. Decreased expression of chromodomain helicase DNA-binding protein 9 is a novel independent prognostic biomarker for colorectal cancer. Braz. J. Med. Biol. Res. 2018, 51, e7588. [Google Scholar] [CrossRef]
- Xia, T.; Pan, Z.; Zhang, J. CircPDZD8 promotes gastric cancer progression by regulating CHD9 via sponging miR-197-5p. Aging 2020, 12, 19352–19364. [Google Scholar] [CrossRef]
- Powell, J.D.; Pollizzi, K.N.; Heikamp, E.B.; Horton, M.R. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 2012, 30, 39–68. [Google Scholar] [CrossRef]
- Kim, L.C.; Cook, R.S.; Chen, J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2017, 36, 2191–2201. [Google Scholar] [CrossRef]
- Rostamzadeh, D.; Yousefi, M.; Haghshenas, M.R.; Ahmadi, M.; Dolati, S.; Babaloo, Z. mTOR Signaling pathway as a master regulator of memory CD8+ T-cells, Th17, and NK cells development and their functional properties. J. Cell Physiol. 2019, 234, 12353–12368. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, L.A.; Murray, P.J. Proliferating Helper T Cells Require Rictor/mTORC2 Complex to Integrate Signals from Limiting Environmental Amino Acids. J. Biol. Chem. 2016, 291, 25815–25822. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Liu, H.; Tan, B.; Dong, X.; Chi, S.; Yang, Q.; Zhang, S. MHC II-PI(3)K/Akt/mTOR Signaling Pathway Regulates Intestinal Immune Response Induced by Soy Glycinin in Hybrid Grouper: Protective Effects of Sodium Butyrate. Front. Immunol. 2020, 11, 615980. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, X.; Li, M.; Chen, P.; Zhang, B.; Guo, H.; Cao, W.; Wei, X.; Cao, X.; Hao, X.; et al. mTOR complex component Rictor interacts with PKCzeta and regulates cancer cell metastasis. Cancer Res. 2010, 70, 9360–9370. [Google Scholar] [CrossRef]
- Li, H.; Lin, J.; Wang, X.; Yao, G.; Wang, L.; Zheng, H.; Yang, C.; Jia, C.; Liu, A.; Bai, X. Targeting of mTORC2 prevents cell migration and promotes apoptosis in breast cancer. Breast Cancer Res. Treat. 2012, 134, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Marcais, A.; Blevins, R.; Graumann, J.; Feytout, A.; Dharmalingam, G.; Carroll, T.; Amado, I.F.; Bruno, L.; Lee, K.; Walzer, T.; et al. microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. J. Exp. Med. 2014, 211, 2281–2295. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Sabatini, D.M. mTOR and cancer: Insights into a complex relationship. Nat. Rev. Cancer 2006, 6, 729–734. [Google Scholar] [CrossRef]
- Dancey, J. mTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol. 2010, 7, 209–219. [Google Scholar] [CrossRef]
- Mossmann, D.; Park, S.; Hall, M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 2018, 18, 744–757. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).