Simple Summary
Glaucoma is a common eye condition linked to genes and aging. We studied genetic factors related to primary open-angle glaucoma (POAG) in adult Arabs from Saudi Arabia. We focused on two specific variations in apolipoprotein gene (APOE), namely rs429358 and rs7412, to examine whether these variations are more common in people with POAG. We compared the DNA from 179 people with POAG and 251 without. Our results showed that these genetic changes were not significantly linked to POAG. We also checked different combinations of these variations but did not observe a strong connection with POAG risk. The gene variations also did not affect eye pressure or other eye indicators, such as cup/disc, ratio linked to POAG. Overall, in our Saudi group, these specific gene variations do not seem to be major factors causing POAG. However, more studies with larger groups are needed to confirm this.
Abstract
Adult-onset glaucoma, an age-related neurodegenerative disease, is very prevalent among the elderly Arabs of Saudi origin. This study investigated the association between apolipoprotein E (APOE) gene variants (rs429358 and rs7412) and primary open-angle glaucoma (POAG) in Arabs of Saudi origin. A case-control genetic association study involving 179 POAG patients and 251 controls utilized Sanger sequencing to genotype APOE gene variants. The allele frequencies and genotype distributions for rs429358 and rs7412 did not show significant associations with POAG. The haplotype analysis revealed apoε3 (87.6% and 87.4%) as the most prevalent, followed by ε4 (2.8% and 3.6%) and ε2 (9.6% and 8.9%) in the controls and POAG patients, respectively. Although the ε2/ε3 genotype and ε2-carriers displayed a more than two-fold increased risk, statistical significance was not reached. Notably, these polymorphisms did not affect clinical markers, such as intraocular pressure and cup/disc ratio. The logistic regression analysis demonstrated no significant influence of age, sex, rs429358, or rs7412 polymorphisms on POAG. In conclusion, within the Saudi cohort, APOE variants (rs429358 and rs7412) do not appear to be associated with POAG and are not substantial risk factors for its development. However, additional population-based studies are required to validate these findings.
Keywords:
APOE; genetics; glaucoma; intraocular pressure; POAG; polymorphisms; rs429358; rs7412; Saudi 1. Introduction
Adult-onset glaucoma is an age-related neurodegenerative disease and is very prevalent among the elderly Arabs of Saudi origin [1]. Primary open-angle glaucoma (POAG), a common form of glaucoma, is a complex, multifactorial ocular disorder characterized by elevated intraocular pressure (IOP), progressive retinal ganglion cell (RGC) death, optic nerve damage, and visual field loss [2]. It represents a significant global health concern, with an estimated 79.6 million individuals projected to be affected by 2020, a number expected to increase to 111.8 million by 2040 [3]. While the precise etiology of POAG remains elusive, genetic factors have been demonstrated to play a pivotal role in its pathogenesis, with numerous genome-wide and candidate-gene association studies highlighting the importance of genetic predisposition in disease susceptibility in various ethnicities [4,5]. Nonetheless, the underlying contribution of genes and genetic polymorphisms in such complex polygenic disease in POAG patients of Saudi Arabian descent is still unclear.
Apolipoprotein E (APOE) is a well-studied gene located on chromosome 19q13.32, known for its critical involvement in lipid metabolism and transport [6]. It exists in three major isoforms, denoted as ε2, ε3, and ε4, which are the result of two common single-nucleotide polymorphisms (SNPs): rs429358 (Cys112Arg) and rs7412 (Arg158Cys) [6]. These polymorphisms give rise to six possible genotypes, each contributing to variations in APOE protein function and expression levels [7]. Emerging evidence suggests that APOE polymorphisms may be implicated in the development and progression of various neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease, and multiple sclerosis [8,9]. However, the potential association between APOE polymorphisms and POAG has garnered increasing interest in recent years, as both conditions share common underlying pathological mechanisms, including oxidative stress, inflammation, and vascular dysfunction [10,11,12,13].
The APOE protein is expressed in the retina and optic nerve head tissues [14]. The ε4/ε4 genotype has been reported to be associated with increased risk of POAG in Asians [15,16,17], but not in populations from the United Kingdom [18] or Sweden [19]. According to certain recent reports, E4 has a protective effect on the development of POAG [20] or a favorable correlation with glaucoma risk [21]. However, others have discovered a conflicting relationship or a negative correlation between APOE E4 and glaucoma [22]. Therefore, although studies have suggested a potential link between APOE variants and POAG, the evidence is conflicting, with varying degrees of outcomes in different population [23,24].
The Saudi population presents a unique genetic landscape, characterized by distinct allele frequencies and genetic variations, which may contribute to the variability in disease susceptibility and presentation. Despite this, limited studies have investigated the role of APOE variations in Saudi patients with POAG [25]. However, more research is needed to clarify the role of this gene in POAG susceptibility in this population. Therefore, the present study aims to examine the association between the rs429358 and rs7412 variants of APOE and POAG in a Saudi cohort.
2. Materials and Methods
2.1. Ethical Considerations
This retrospective case–control study was conducted in accordance with the principles outlined in the Declaration of Helsinki. Ethical approval was obtained from the Institutional Review Board/Ethics Committee prior to the commencement of the study (protocol number #09–657). Informed written consent was obtained from all participants after providing them with a detailed explanation of the study objectives, procedures, and potential risks involved.
2.2. Study Participants
A total of 179 patients clinically diagnosed with POAG and 251 age- and ethnically-matched controls were recruited for this study. All participants were of Saudi Arabian descent and provided informed consent prior to inclusion. The diagnosis of POAG was based on established clinical criteria, including characteristic optic-nerve-head changes, visual field defects, and elevated IOP. The IOP assessment employed Goldmann applanation tonometry, while the cup-to-disc ratio was ascertained through the comparison of the vertical diameter of the optic cup, representing the central depression in the optic nerve head, with the vertical diameter of the entire optic disc. Exclusion criteria for both cases and controls encompassed the presence of secondary glaucomas, congenital anomalies, previous ocular surgeries, and other significant ocular or systemic comorbidities [26].
2.3. DNA Preparation
Genomic DNA was extracted from peripheral EDTA blood samples using the QIAamp DNA Mini Kit protocol, as suggested by the manufacturer (cat. no. 51306, Qiagen, Hilden, Germany). The DNA aliquots were stored at –80 °C until further use.
2.4. Genotyping of APOE Variants rs429358 (T>C) and rs7412 (C>T)
The rs429358 (T>C) and rs7412 (C>T) polymorphisms of the APOE gene were genotyped using Sanger sequencing. The region encompassing the polymorphisms were PCR-amplified using primers and conditions described in Table 1. The PCR reaction consisted of standard reagents: 1X PCR buffer, 250 µM dNTP mix, 100 pmoles of each specific primer, 1.5 U Taq polymerase, and 20 ng of DNA, along with 1X Q-solution (cat. no. 203205, Qiagen). The PCR products were purified using a QIAquick PCR Purification Kit (cat. no. 28106, Qiagen) and subjected to sequencing using M13 primers in both forward and reverse directions using the BigDye Terminator V3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocol. Samples were electrophoresed on the ABI 3730 XL sequencer (Applied Biosystems). The sequencing data were read using CLC Sequence Viewer 6.0 (Qiagen, Hilden, Germany) to identify the nucleotide variations and APOE genotypes in comparison to APOE reference sequence (NG_007084.2).

Table 1.
PCR primers and cycling condition used for APOE genotyping.
2.5. Statistical Analysis
Demographic data, including age and sex distribution, were compared between cases and controls using Mann–Whitney U test (for continuous variables) and chi-squared tests (for categorical variables). Deviations from Hardy–Weinberg equilibrium for APOE genotypes in control subjects were assessed using chi-squared tests.
The association between APOE variants (rs429358 and rs7412) and POAG risk was evaluated using logistic regression models, adjusting for potential confounding variables, such as age and sex. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. The potential association of polymorphisms with clinical factors such as IOP and cup/disc ratio was determined by non-parametric method in dominant model. Furthermore, a binary logistic regression model was applied to assess the combined effect of age, sex, rs429358, and rs7412 on the likelihood of developing POAG.
All statistical analyses were performed using SPSS version 25 (IBM Inc., Chicago, IL, USA) and SNPStats web tool (https://www.snpstats.net/start.htm (accessed on 11 December 2023)), and p-values < 0.05 were considered statistically significant. Power analysis was performed using the PS program (version 3.1.2).
3. Results
3.1. Demographic Characteristics
In this study, a total of 430 samples were analyzed, comprising 179 controls and 251 POAG patients. The mean ages of the two groups were 61.1 (±10.1) years for the POAG patients and 59.7 (±7.0) years for the controls, with no statistically significant difference in age. Furthermore, there were 101 (56%) males and 78 (44%) females in the patient group, compared to 136 (54%) males and 115 (46%) females among the controls. The gender distribution also showed no significant variations between the two groups (Figure 1).
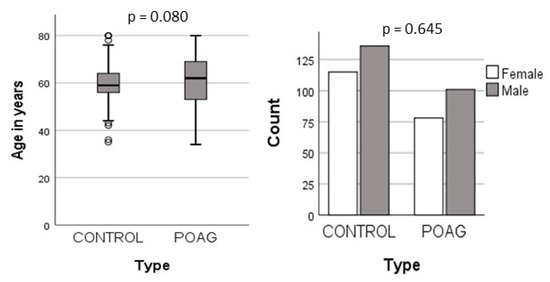
Figure 1.
Demographic distribution in POAG and controls.
3.2. Allele and Genotype Associations
The minor allele frequencies (MAFs) for rs429358 and rs7412 were determined in both the controls and the POAG patients. Both polymorphisms were found to be in Hardy–Weinberg equilibrium. The MAFs for rs429358 were 0.10 and 0.09 in the controls and the POAGs, respectively, while for rs7412, they were 0.03 and 0.04 in the controls and the POAGs, respectively. No significant associations were observed between the allele frequencies and the occurrence of POAG (Table 2).

Table 2.
Minor allele frequency distribution of APOE polymorphisms.
The genotype associations were assessed under different genetic models, including co-dominant, dominant, and recessive models. However, none of these models revealed a significant association between the APOE polymorphisms and the POAG patients. Although the frequency of the rs7412 C/T heterozygous genotype was higher in the POAGs (7.3%) than in the controls (4.8%), exhibiting a 1.55-fold increased risk of POAG. However, this difference did not reach statistical significance (Table 3).

Table 3.
Genotype analysis of rs429358 and rs7412 variants in APOE gene in POAG.
3.3. Association of APOE Genotypes with POAG
In this study, we also examined the association of the two APOE variants according to the three APOE alleles (ε3, ε2, and ε4) and six different genotypes (ε3/ε3, ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε4, and ε4/ε4). The APOE allele and genotype calling was based on the presence of the nucleotide sequences shown in Table 4, and their association analysis is shown in Table 5. The APOE ε3 was the most prevalent allele, followed by ε4 and ε2, in both the POAG patients and the controls. However, no significant associations were observed between the APOE alleles and the occurrence of POAG. The most common genotype in both the cases and the controls was ε3/ε3, and the overall distribution of different APOE genotypes did not show statistical significance (Pearson chi-squared = 4.61, df = 5, p = 0.465). The homozygous ε2/ε2 and ε4/ε4 genotypes were absent in the POAGs. The frequency of the heterozygous ε2/ε3 genotype was 1.6% in the controls, compared to 4.5% in the POAGs, showing a 2.8-fold increased risk of POAG. A further analysis of the APOE genotypes according to carrier status (Table 3) showed that the frequency of ε2 carriers (ε2/ε2 and ε2/ε3 genotypes) was also high in the POAGs (4.6%), compared to the 2.0% in the controls, exhibiting more than two-fold increased risk of POAG. However, none of these differences reached statistical significance (Table 3). The representative chromatograms of APOE sequencing of the rs429358 (T>C) and rs7412 (C>T) variants and the identified APOE genotypes is shown in Figure 2.

Table 4.
APOE allele and genotype calling based on the presence of nucleotide sequences at rs429358 and rs7412 variants.

Table 5.
Association analysis of APOE variants according to APOE alleles and genotypes in POAG.
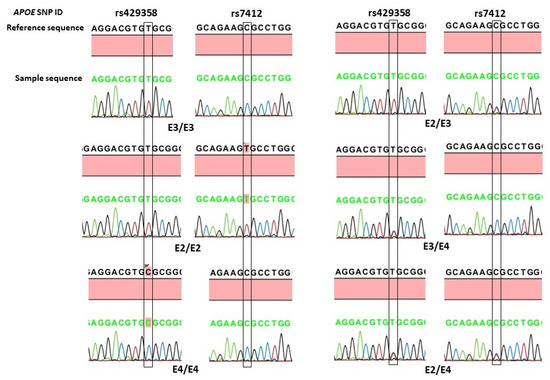
Figure 2.
Representative DNA-sequence chromatograms of APOE genotypes based on rs429358 (T>C) and rs7412 (C>T) variants. The position of the nucleotide change is boxed.
3.4. Regression Analysis
The effects of risk factors including age, gender, and APOE gene variants on POAG risk were assessed through binary logistic regression analysis. The results demonstrated no significant impact of age (p = 0.098), sex (p = 0.622), rs429358 (p = 0.939), or rs7412 (p = 0.605) on the risk of developing POAG. The effect of the variants was examined in both co-dominant and dominant models (Table 6).

Table 6.
Binary logistic regression analysis.
3.5. Association with Clinical Variables
The study further examined the potential association between APOE polymorphisms and clinical parameters of POAG, including IOP and cup/disc ratio. However, no significant effects of either SNP on these clinical variables were observed (Figure 3).
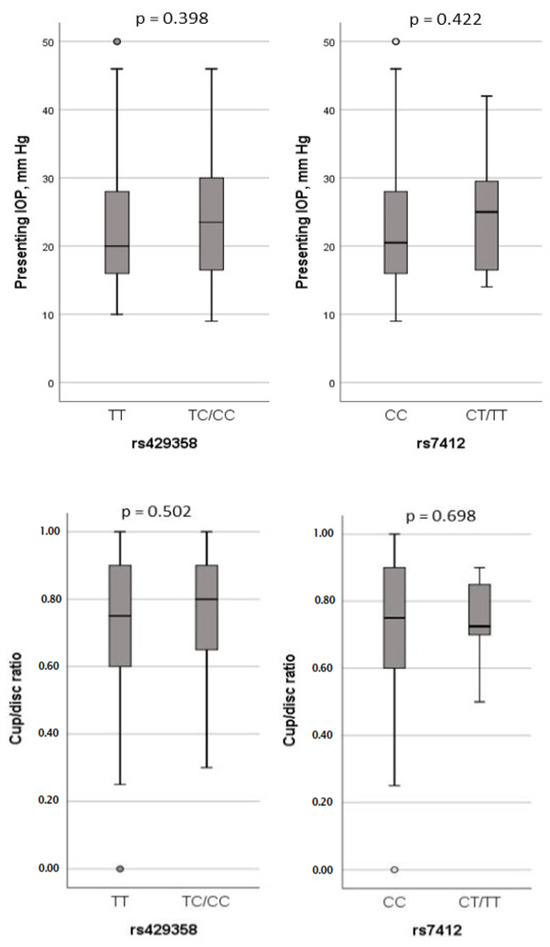
Figure 3.
Association analysis of IOP and cup/disc ratio with rs429358 and rs7412 genotypes in dominant model.
4. Discussion
In our study, we explored the potential association between two prevalent polymorphisms of the APOE gene, rs429358 and rs7412, and POAG. We conducted an analysis to assess the independent association of these polymorphisms and their haplotypes in relation to APOE alleles/genotypes.
Specifically, for the rs429358 polymorphism, where T is the reference allele and C is the alternate allele, we observed C allele frequencies of 0.1 in the controls and 0.09 in the POAG cases within our cohort. This frequency was higher than in Europeans (0.07) and Asians (0.037), but lower than in Africans (0.13) and African Americans (0.13), according to NCBI ALFA allele frequency data. Notably, our study did not reveal any allelic or genotype associations of rs429358 polymorphism with POAG. Additionally, this polymorphism did not have an impact on IOP or cup/disc ratio.
For the rs7412 polymorphism, where C is the reference allele and T is the alternate allele, the T allele frequency was 0.03 in the controls and 0.04 in the POAG cases. Compared to the ALFA allele frequency data, this frequency was higher than in Latin Americans (0.026) and South Asians (0.021), but much lower than in Europeans (0.083), Africans (0.10), African Americans (0.10), and East Asians (0.07). As with rs429358, this polymorphism did not demonstrate a significant effect on POAG or related clinical factors, such as IOP and cup/disc ratio.
Regarding the haplotypes of these polymorphisms (rs429358 and rs7412), referred to as APOE alleles and genotypes, numerous studies have been conducted. In most populations, ε3 is reported as the most common allele, followed by ε4 and ε2 [27]. Our study aligned with this pattern, with the prevalence of APOE alleles found to be in the following order of ε3 > ε4 > ε2. Nevertheless, our study did not detect any significant associations between APOE alleles/genotypes and POAG. Furthermore, the examination of the association of APOE genotypes with POAG also yielded no significant findings. It is worth noting that the ε2/ε3 genotype appeared to increase the risk of POAG by over two-fold, although this effect did not reach statistical significance. This trend was consistently observed when considering ε2-carrier status. Similarly, no significant impact of APOE ε2, ε3, or ε4-containing genotypes on IOP and cup/disc ratio was observed in the dominant models. Moreover, the binary logistic regression analysis showed no significant influence of age, sex, rs429358, or rs7412 polymorphisms on POAG outcomes. In summary, our study demonstrated no significant associations between APOE alleles/genotypes and POAG, or its associated clinical markers, such as IOP and cup/disc ratio, within our Saudi cohort.
The APOE protein is a prominent apolipoprotein predominantly expressed in the central nervous system, synthesized by retinal Müller cells, neurons, and macrophages [14,28]. It plays a pivotal role in neural growth and repair processes. Associations between Apo E4 and neurodegenerative disorders, such as AD, and eye-related disorders, like AMD, have been documented [9,29]. Furthermore, POAG, characterized by a significant hereditary component, is also considered a neurodegenerative disorder. Since early reports of the APOE ε4 allele’s association with increased risk of normal tension glaucoma in the Tasmanian population [30], several investigators have examined this link in adult-onset POAG. However, the results regarding the association between APOE alleles/genotypes and POAG have been contradictory.
In a German study, it was noted that the presence of the ε2 allele influenced IOP levels in normal controls [31]. In Japanese patients with open-angle glaucoma (OAG), the ε3 allele was associated with an increased risk of OAG, while the ε2 allele was linked to a significant reduction in OAG risk, and the ε4 allele was associated with lower IOP levels [32]. In contrast to our findings, a smaller study conducted by Al-Dabbagh et al. in Riyadh, which included 60 Saudi-origin POAG patients, reported a significant association between the ε4 allele and POAG [25]. A large-scale study utilizing data from the NEIGHBOR Massachusetts Eye and Ear Infirmary dataset reported a protective effect of the ε4 allele in both high-tension and normal-tension POAG patients in this population [20]. A protective effect of the ε4 allele was also observed in the Canadian Longitudinal Study on Aging, in which it was found to be protective against glaucoma in individuals without systemic hypertension [21]. Conversely, in Brazilian POAG patients, individuals carrying the ε2 allele were reported to be at an increased risk of developing POAG [33]. In contrast, Mullany et al. reported that the ε4 allele was associated with an increased rate of neuroretinal thinning in eyes with normal-tension glaucoma [22]. Our study did not replicate the findings of these studies.
While numerous studies have emphasized the involvement of APOE in POAG, a subset of investigations has yielded contrasting results regarding the genetic association between APOE and POAG across diverse ethnic groups. Our own study is in agreement with these divergent findings. Notable examples include studies conducted on European [18,19,34], Chinese [15,35], Japanese [36], and Turkish [23] populations, as well as other meta-analysis studies [24,37]. The distribution of APOE allele frequencies, as observed in these distinct studies and our current research, is summarized in Table 7. Notably, the frequencies of the ε2, ε3, and ε4 alleles exhibit variability among different ethnicities. Across all populations, ε3 consistently emerges as the prevailing allele, followed by ε4 and ε2, in descending order of prevalence.

Table 7.
APOE allele frequencies reported in POAG patients with different ethnicities.
Several factors may contribute to the disparities in APOE findings in POAG. These encompass the potential impact of clinical diversity within the study population, variations in functional properties of different APOE isoforms in different cell types [7], and potential interactions with other genetic elements involved in lipid metabolism or neuroinflammation [28,38]. It is also crucial to recognize that genetic associations are inherently complex and can diverge across populations due to age, sex, race, and differing genetic backgrounds and environmental influences [39]. Moreover, discrepancies in sample size or study power can also contribute to conflicting results, even within the same population, as reported by Mullany et al. in the UK population [34], by Al-dabbagh et al. in patients of Saudi origin [25], and our study. In our study, considering the observed allele frequency, the estimated power to detect an odds ratio of 2.0 at a significance level of 0.05 was 0.6 per allele for ε2 and 0.9 per allele for ε4. However, to substantiate our findings, a significantly larger sample size would be necessary, especially when aiming to detect an odds ratio of 1.5, which is commonly observed in genetic association studies with more subtle effects.
POAG is a complex polygenic and multifactorial disease. Given the strong evidence for the role of APOE in AD [9], the role of APOE in glaucoma is still unclear and the exact mechanisms by which APOE might contribute to the risk of the development and progression of glaucoma or may be protective needs further investigation. Although our study reports no associations between the two common variants, rs429358 and rs7412, and POAG, the role of other polymorphism(s), including the promoter polymorphism, which influences APOE expression, and those in linkage disequilibrium, cannot be ruled out and needs to be investigated.
5. Conclusions
Our findings indicate that there is no apparent link between these APOE variants and POAG within our cohort of individuals of Saudi Arabian descent. This implies that APOE might not be a significant risk factor for POAG in this particular ethnic group. However, further population-based studies are required to validate these results.
Author Contributions
A.A.K.: conceptualization, investigation, methodology, analysis, writing draft, review and editing, and supervision; T.S. and T.A.A.: investigation, methodology, data curation, manuscript review, and editing; T.K. and G.P.L.: analysis, data curation, data interpretation, resources, manuscript review, and editing; A.A.A., E.A.O. and F.A.A.: data curation, resources, manuscript review, and editing; S.A.A.-O.: concept, resources, funding acquisition, project administration, manuscript review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by King Saud University through the Vice Deanship of Scientific Research Chair and Glaucoma Research Chair in Ophthalmology (GRC-2023).
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board Committee of the College of Medicine, King Saud University (protocol number #09–657).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data supporting the conclusions of this article are all presented within the report.
Acknowledgments
We would like to thank the Vice Deanship of Scientific Research Chair, Glaucoma Research Chair in Ophthalmology at King Saud University. We thank Abdulrahman Al-Mosa for his clinical assistance during the study.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Khandekar, R.; Chauhan, D.; Yasir, Z.H.; Al-Zobidi, M.; Judaibi, R.; Edward, D.P. The prevalence and determinants of glaucoma among 40 years and older saudi residents in the riyadh governorate (except the capital)—A community based survey. Saudi J. Ophthalmol. 2019, 33, 332–337. [Google Scholar] [CrossRef]
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Gharahkhani, P.; Jorgenson, E.; Hysi, P.; Khawaja, A.P.; Pendergrass, S.; Han, X.; Ong, J.S.; Hewitt, A.W.; Segre, A.V.; Rouhana, J.M.; et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 2021, 12, 1258. [Google Scholar] [CrossRef]
- Han, X.; Gharahkhani, P.; Hamel, A.R.; Ong, J.S.; Renteria, M.E.; Mehta, P.; Dong, X.; Pasutto, F.; Hammond, C.; Young, T.L.; et al. Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci. Nat. Genet. 2023, 55, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Rall, S.C., Jr. Apolipoprotein e: Far more than a lipid transport protein. Annu. Rev. Genom. Hum. Genet. 2000, 1, 507–537. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, D.; Bertrand, P.; Dea, D.; Davignon, J.; Poirier, J. Apolipoprotein e and low-density lipoprotein binding and internalization in primary cultures of rat astrocytes: Isoform-specific alterations. J. Neurochem. 1996, 66, 2410–2418. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calle, R.; Konings, S.C.; Frontinan-Rubio, J.; Garcia-Revilla, J.; Camprubi-Ferrer, L.; Svensson, M.; Martinson, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. Apoe in the bullseye of neurodegenerative diseases: Impact of the apoe genotype in alzheimer’s disease pathology and brain diseases. Mol. Neurodegener. 2022, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. Apoe and alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- Chan, J.W.; Chan, N.C.Y.; Sadun, A.A. Glaucoma as neurodegeneration in the brain. Eye Brain 2021, 13, 21–28. [Google Scholar] [CrossRef]
- Kuang, G.; Halimitabrizi, M.; Edziah, A.A.; Salowe, R.; O’Brien, J.M. The potential for mitochondrial therapeutics in the treatment of primary open-angle glaucoma: A review. Front. Physiol. 2023, 14, 1184060. [Google Scholar] [CrossRef]
- Kuang, G.; Salowe, R.; O’Brien, J. Genetic factors implicated in the investigation of possible connections between alzheimer’s disease and primary open angle glaucoma. Genes 2023, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Civeira-Marin, M.; Cenarro, A.; Marco-Benedi, V.; Bea, A.M.; Mateo-Gallego, R.; Moreno-Franco, B.; Ordovas, J.M.; Laclaustra, M.; Civeira, F.; Lamiquiz-Moneo, I. Apoe genotypes modulate inflammation independently of their effect on lipid metabolism. Int. J. Mol. Sci. 2022, 23, 12947. [Google Scholar] [CrossRef] [PubMed]
- Amaratunga, A.; Abraham, C.R.; Edwards, R.B.; Sandell, J.H.; Schreiber, B.M.; Fine, R.E. Apolipoprotein e is synthesized in the retina by muller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J. Biol. Chem. 1996, 271, 5628–5632. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.J.; Wang, D.Y.; Fan, D.S.; Tam, P.O.; Lam, D.S.; Tham, C.C.; Lam, C.Y.; Lau, T.C.; Pang, C.P. Snps and interaction analyses of myocilin, optineurin, and apolipoprotein e in primary open angle glaucoma patients. Mol. Vis. 2005, 11, 625–631. [Google Scholar] [PubMed]
- Liao, R.; Ye, M.; Xu, X. An updated meta-analysis: Apolipoprotein e genotypes and risk of primary open-angle glaucoma. Mol. Vis. 2014, 20, 1025–1036. [Google Scholar]
- Wang, Y.; Zhou, Y.F.; Zhao, B.Y.; Gu, Z.Y.; Li, S.L. Apolipoprotein e gene epsilon4epsilon4 is associated with elevated risk of primary open angle glaucoma in asians: A meta-analysis. BMC Med. Genet. 2014, 15, 60. [Google Scholar] [CrossRef]
- Ressiniotis, T.; Griffiths, P.G.; Birch, M.; Keers, S.; Chinnery, P.F. The role of apolipoproteine gene polymorphisms in primary open-angle glaucoma. Arch. Ophthalmol. 2004, 122, 258–261. [Google Scholar] [CrossRef]
- Zetterberg, M.; Tasa, G.; Palmer, M.S.; Juronen, E.; Teesalu, P.; Blennow, K.; Zetterberg, H. Apolipoprotein e polymorphisms in patients with primary open-angle glaucoma. Am. J. Ophthalmol. 2007, 143, 1059–1060. [Google Scholar] [CrossRef]
- Margeta, M.A.; Letcher, S.M.; Igo, R.P., Jr.; Cooke Bailey, J.N.; Pasquale, L.R.; Haines, J.L.; Butovsky, O.; Wiggs, J.L. Association of apoe with primary open-angle glaucoma suggests a protective effect for apoe epsilon4. Investig. Ophthalmol. Vis. Sci. 2020, 61, 3. [Google Scholar] [CrossRef]
- Freeman, E.E.; Bastasic, J.; Grant, A.; Leung, G.; Li, G.; Buhrmann, R.; Roy-Gagnon, M.H. Inverse association of apoe epsilon4 and glaucoma modified by systemic hypertension: The canadian longitudinal study on aging. Investig. Ophthalmol. Vis. Sci. 2022, 63, 9. [Google Scholar] [CrossRef] [PubMed]
- Mullany, S.; Marshall, H.; Diaz-Torres, S.; Berry, E.C.; Schmidt, J.M.; Thomson, D.; Qassim, A.; To, M.S.; Dimasi, D.; Kuot, A.; et al. The apoe e4 allele is associated with faster rates of neuroretinal thinning in a prospective cohort study of suspect and early glaucoma. Ophthalmol. Sci. 2022, 2, 100159. [Google Scholar] [CrossRef] [PubMed]
- Saglar, E.; Yucel, D.; Bozkurt, B.; Ozgul, R.K.; Irkec, M.; Ogus, A. Association of polymorphisms in apoe, p53, and p21 with primary open-angle glaucoma in turkish patients. Mol. Vis. 2009, 15, 1270–1276. [Google Scholar] [PubMed]
- Wang, W.; Zhou, M.; Huang, W.; Chen, S.; Zhang, X. Lack of association of apolipoprotein e (apo e) epsilon2/epsilon3/epsilon4 polymorphisms with primary open-angle glaucoma: A meta-analysis from 1916 cases and 1756 controls. PLoS ONE 2013, 8, e72644. [Google Scholar]
- Al-Dabbagh, N.M.; Al-Dohayan, N.; Arfin, M.; Tariq, M. Apolipoprotein e polymorphisms and primary glaucoma in saudis. Mol. Vis. 2009, 15, 912–919. [Google Scholar]
- Kondkar, A.A.; Azad, T.A.; Almobarak, F.A.; Bahabri, I.M.; Kalantan, H.; Abu-Amero, K.K.; Al-Obeidan, S.A. Lack of association between variant rs7916697 in atoh7 and primary open angle glaucoma in a saudi cohort. Genet. Res. Int. 2018, 2018, 2148056. [Google Scholar] [CrossRef]
- Abondio, P.; Sazzini, M.; Garagnani, P.; Boattini, A.; Monti, D.; Franceschi, C.; Luiselli, D.; Giuliani, C. The genetic variability of apoe in different human populations and its implications for longevity. Genes 2019, 10, 222. [Google Scholar] [CrossRef]
- Anderson, D.H.; Ozaki, S.; Nealon, M.; Neitz, J.; Mullins, R.F.; Hageman, G.S.; Johnson, L.V. Local cellular sources of apolipoprotein e in the human retina and retinal pigmented epithelium: Implications for the process of drusen formation. Am. J. Ophthalmol. 2001, 131, 767–781. [Google Scholar] [CrossRef]
- Klaver, C.C.; Kliffen, M.; van Duijn, C.M.; Hofman, A.; Cruts, M.; Grobbee, D.E.; van Broeckhoven, C.; de Jong, P.T. Genetic association of apolipoprotein e with age-related macular degeneration. Am. J. Hum. Genet. 1998, 63, 200–206. [Google Scholar] [CrossRef]
- Vickers, J.C.; Craig, J.E.; Stankovich, J.; McCormack, G.H.; West, A.K.; Dickinson, J.L.; McCartney, P.J.; Coote, M.A.; Healey, D.L.; Mackey, D.A. The apolipoprotein epsilon4 gene is associated with elevated risk of normal tension glaucoma. Mol. Vis. 2002, 8, 389–393. [Google Scholar]
- Junemann, A.; Bleich, S.; Reulbach, U.; Henkel, K.; Wakili, N.; Beck, G.; Rautenstrauss, B.; Mardin, C.; Naumann, G.O.; Reis, A.; et al. Prospective case control study on genetic assocation of apolipoprotein epsilon2 with intraocular pressure. Br. J. Ophthalmol. 2004, 88, 581–582. [Google Scholar] [CrossRef]
- Mabuchi, F.; Tang, S.; Ando, D.; Yamakita, M.; Wang, J.; Kashiwagi, K.; Yamagata, Z.; Iijima, H.; Tsukahara, S. The apolipoprotein e gene polymorphism is associated with open angle glaucoma in the japanese population. Mol. Vis. 2005, 11, 609–612. [Google Scholar]
- Occhiutto, M.L.; de Melo, M.B.; Cabral de Vasconcellos, J.P.; Rodrigues, T.A.R.; Bajano, F.F.; Costa, F.F.; Costa, V.P. Association of apoe gene polymorphisms with primary open angle glaucoma in brazilian patients. Ophthalmic Genet. 2021, 42, 53–61. [Google Scholar] [CrossRef]
- Mullany, S.; Diaz-Torres, S.; Schmidt, J.M.; Thomson, D.; Qassim, A.; Marshall, H.N.; Knight, L.S.W.; Berry, E.C.; Kolovos, A.; Dimasi, D.; et al. No strong association between the apolipoprotein e e4 allele and glaucoma: A multicohort study. Ophthalmol. Sci. 2023, 3, 100287. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.Y.; Tam, P.O.; Chiang, S.W.; Ding, N.; Chen, L.J.; Yam, G.H.; Pang, C.P.; Wang, N.L. Multiple gene polymorphisms analysis revealed a different profile of genetic polymorphisms of primary open-angle glaucoma in northern chinese. Mol. Vis. 2009, 15, 89–98. [Google Scholar] [PubMed]
- Tamura, H.; Kawakami, H.; Kanamoto, T.; Kato, T.; Yokoyama, T.; Sasaki, K.; Izumi, Y.; Matsumoto, M.; Mishima, H.K. High frequency of open-angle glaucoma in japanese patients with alzheimer’s disease. J. Neurol. Sci. 2006, 246, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Chen, P.; Liu, Q. Role of the apoe epsilon2/epsilon3/epsilon4 polymorphism in the development of primary open-angle glaucoma: Evidence from a comprehensive meta-analysis. PLoS ONE 2013, 8, e82347. [Google Scholar] [CrossRef] [PubMed]
- Parhizkar, S.; Holtzman, D.M. Apoe mediated neuroinflammation and neurodegeneration in alzheimer’s disease. Semin. Immunol. 2022, 59, 101594. [Google Scholar] [CrossRef] [PubMed]
- Corbo, R.M.; Scacchi, R. Apolipoprotein e (apoe) allele distribution in the world. Is apoe*4 a ‘thrifty’ allele? Ann. Hum. Genet. 1999, 63, 301–310. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).