Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
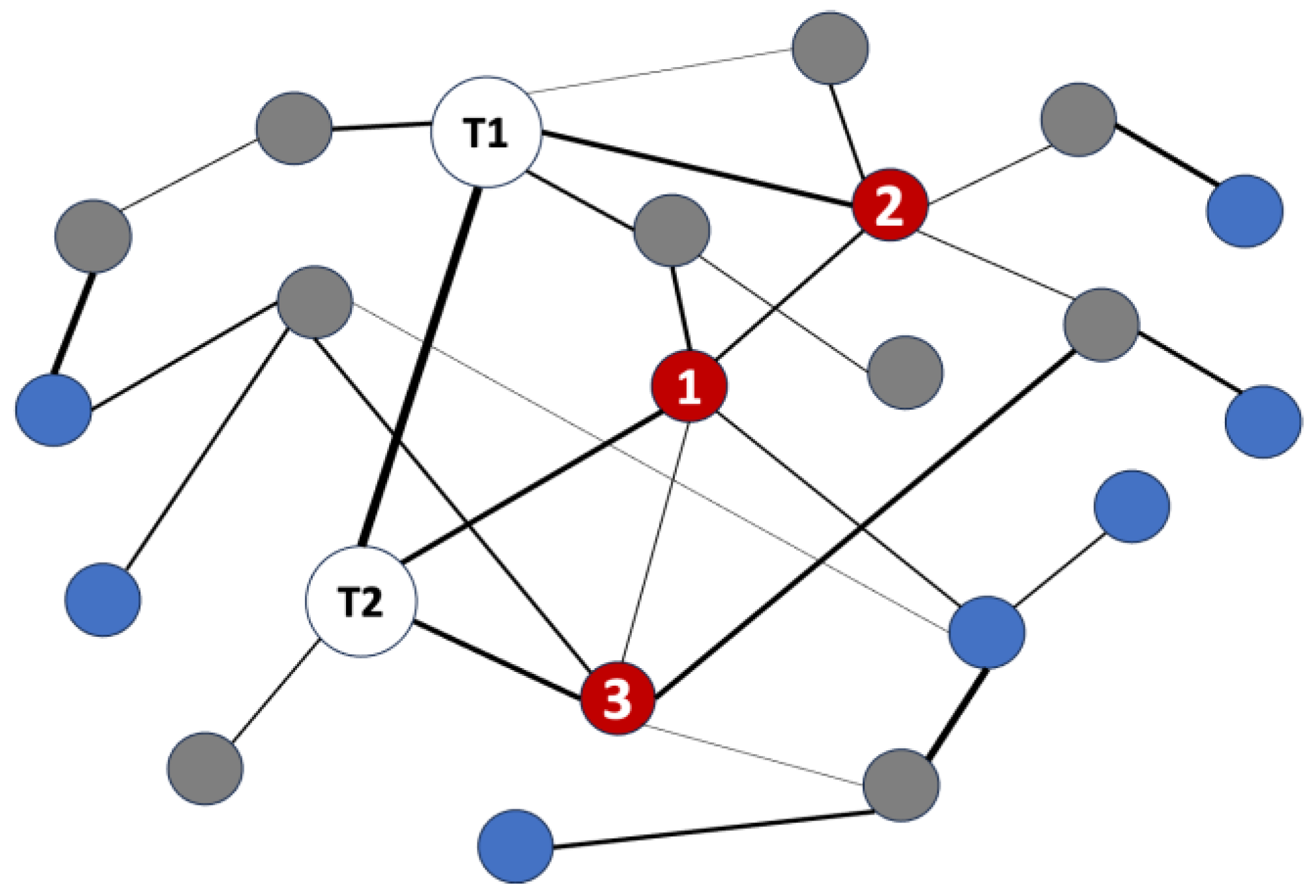
2.1. Overview of SemNet 2.0 Software for Literature-Based Discovery
2.2. Hub Network Analysis for Deeper, Cross-Domain Text Mining
2.3. Analysis Architecture and Simulation Parameters
2.4. Justification for Target Node and Hub Node Selection
2.5. Validation of Results and Determination of Themes
3. Results
3.1. Analysis Metadata
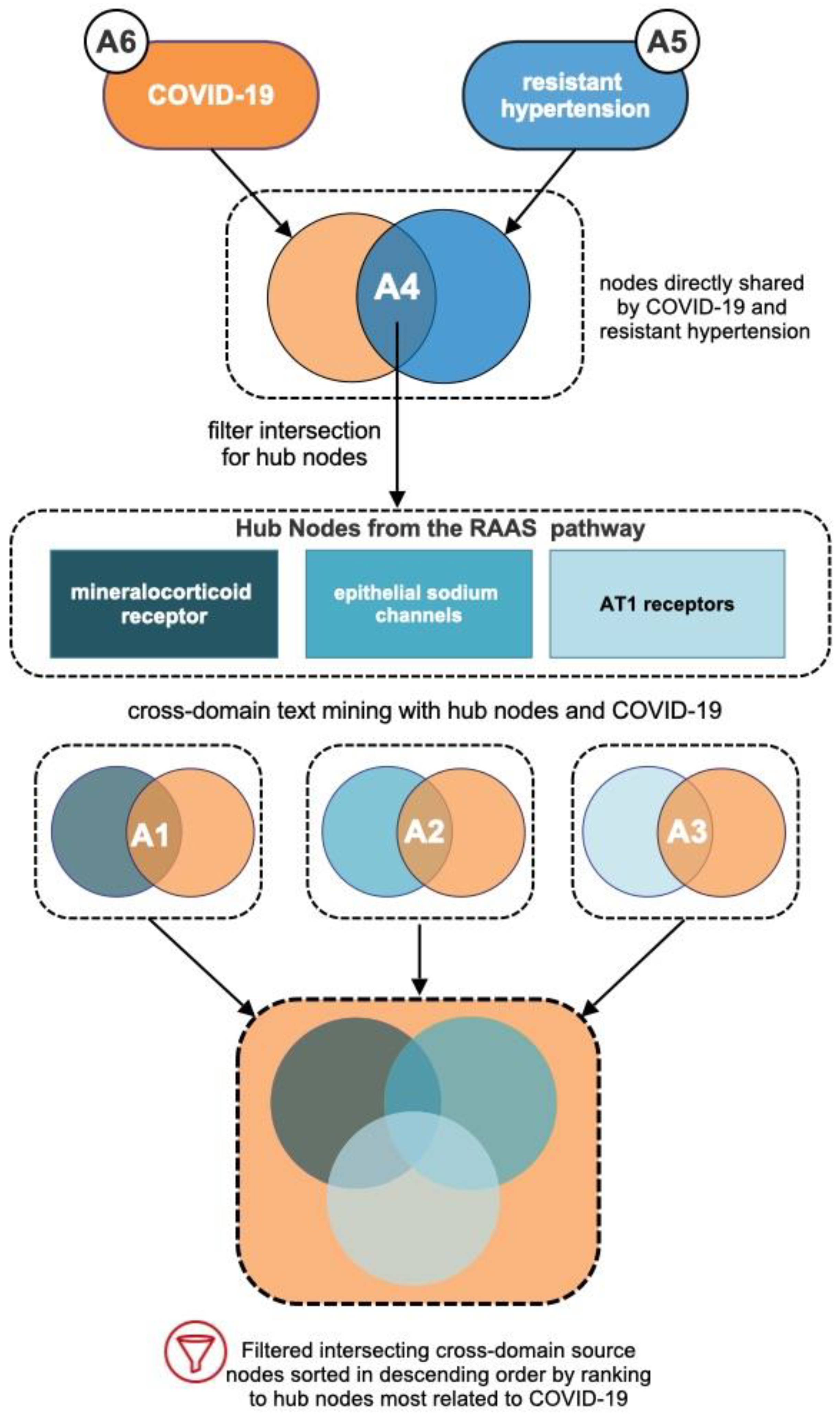
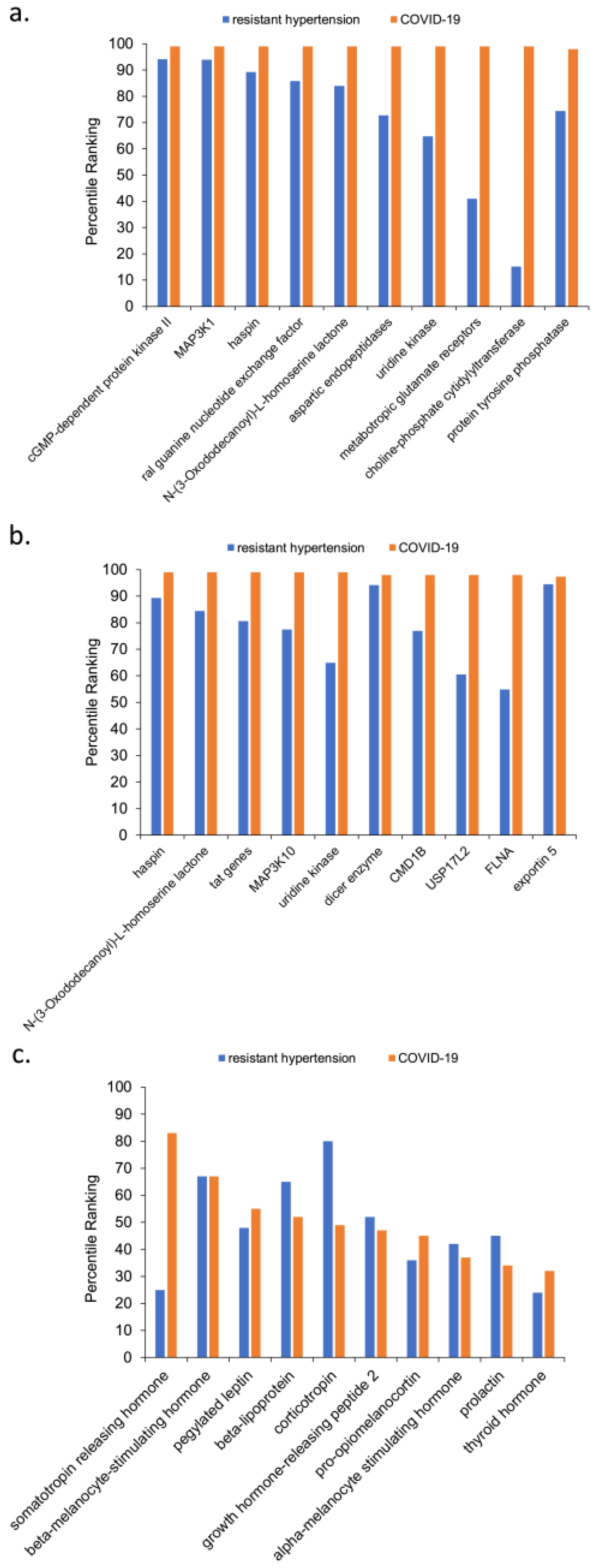
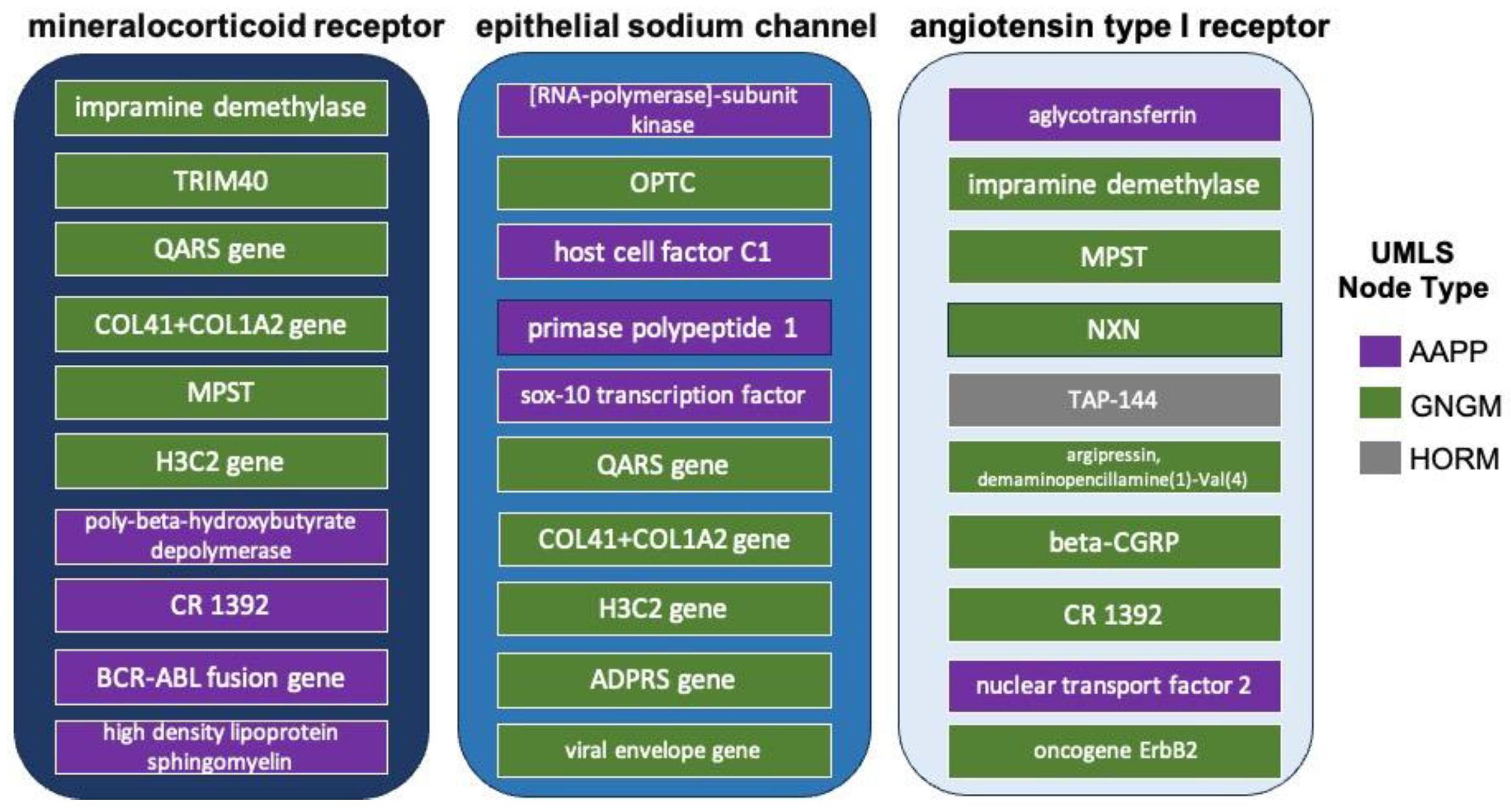
3.2. Analysis of Intersecting Nodes Related to COVID-19 and Resistant Hypertension
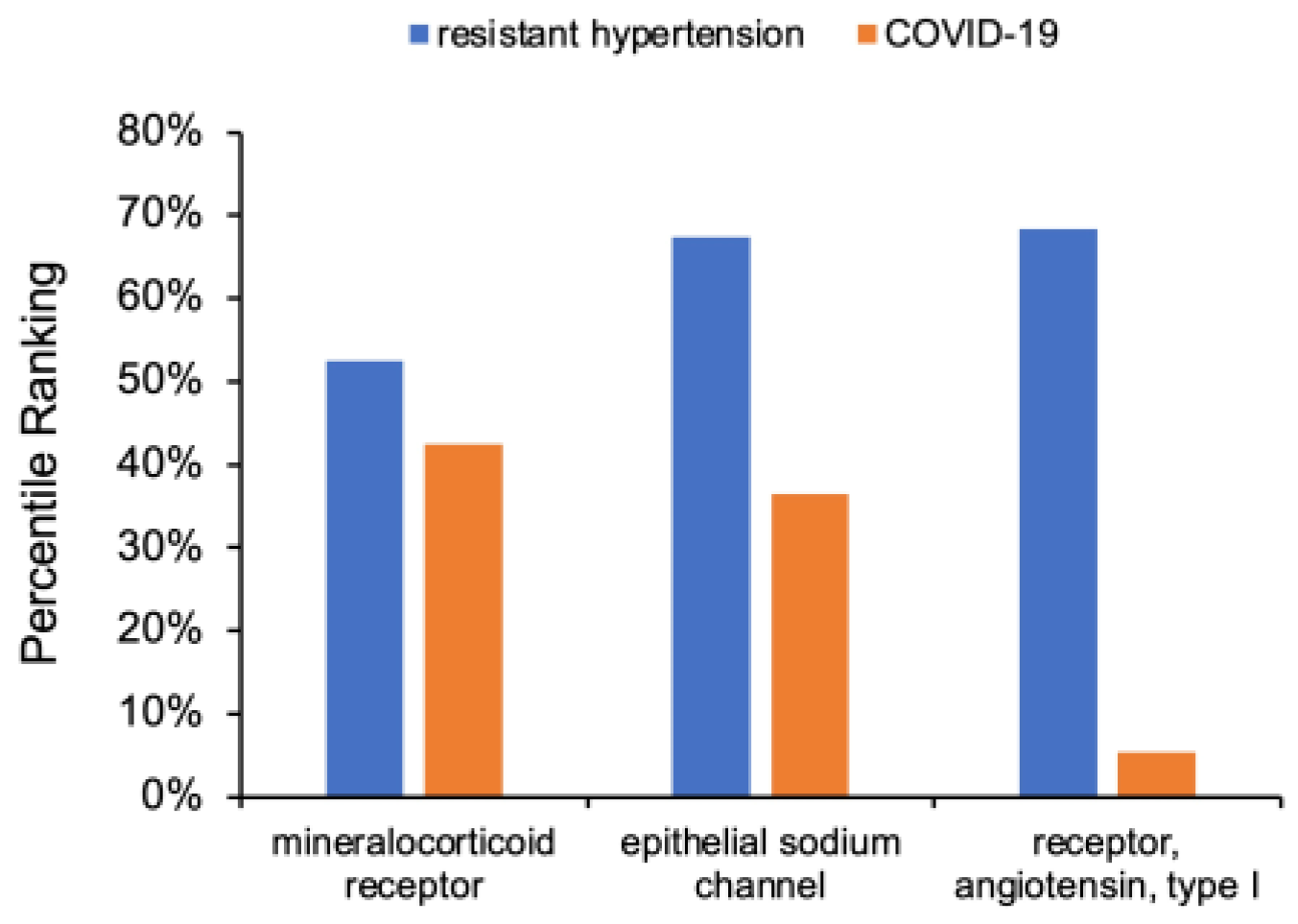
3.3. Determination of RAAS Receptor Hub Nodes Related to COVID-19
3.4. Cross-Domain Analysis with RAAS Receptor Hub Nodes and COVID-19
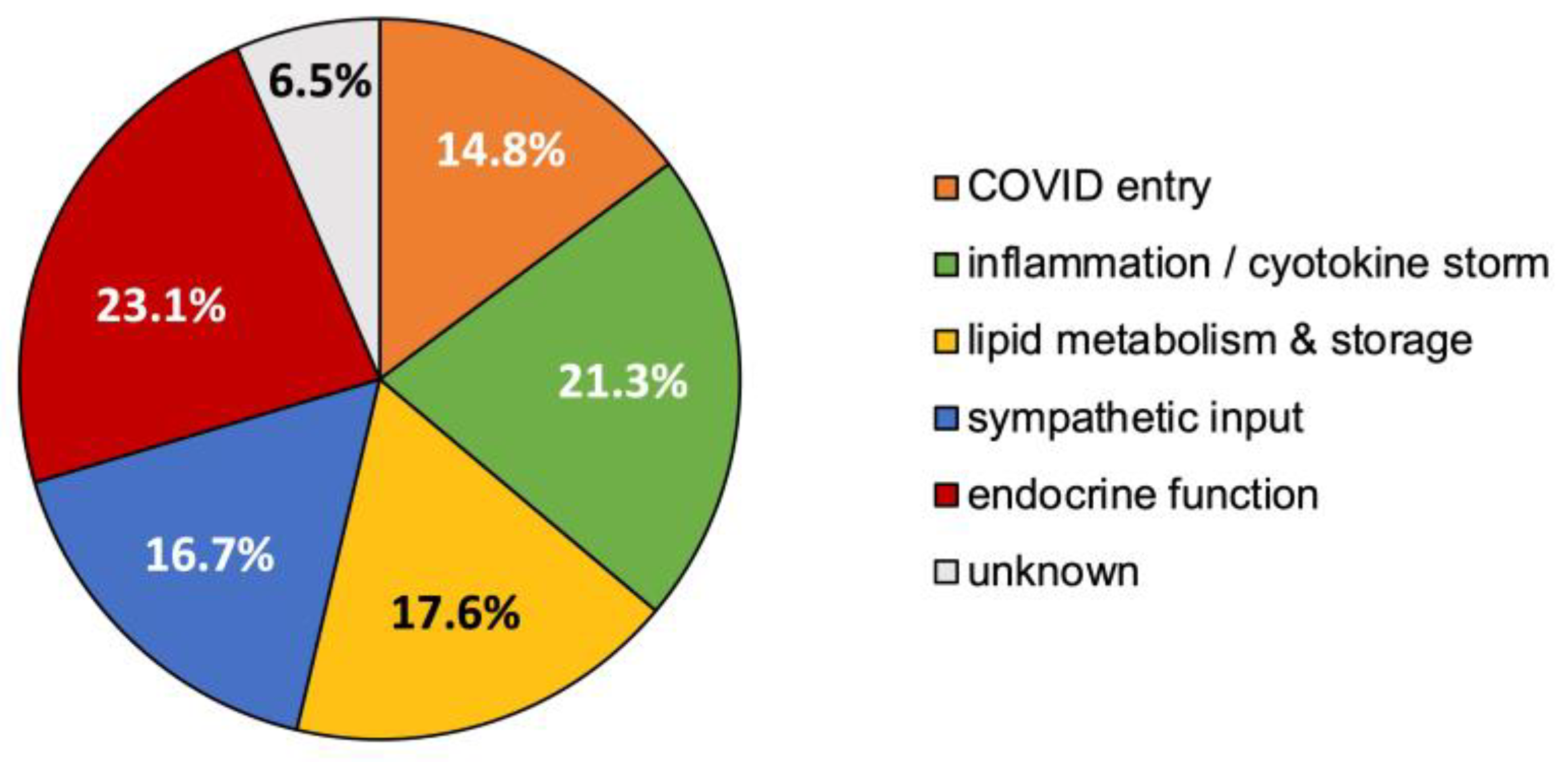
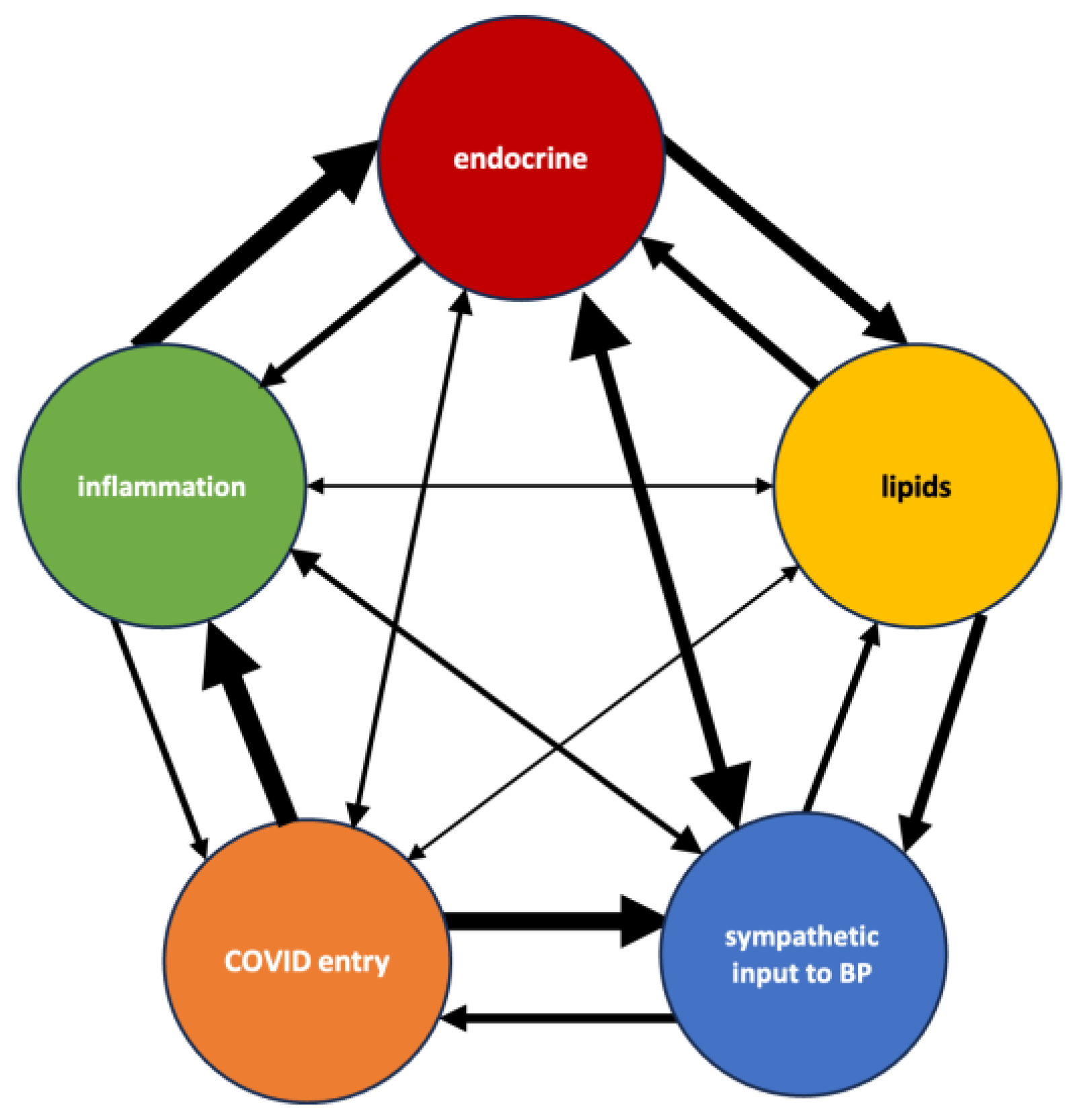
3.5. Synthesized Biological Themes Comprised by High-Ranking Concepts
- Concept relationships that modulate endocrine function, predominantly via the hypothalamic–pituitary–adrenal axis, 23.1%;
- Concept relationships mapping to inflammation and/or the cytokine storm, 21.3%;
- Concept relationships associated with lipid storage, metabolism, or atherosclerosis, 17.6%;
- Concept relationships that alter sympathetic drive for blood pressure regulation, 16.7%;
- Concept relationships that explained the entry or altered uptake of COVID-19, 14.8%;
- Concept relationships that did not clearly map to one of the above specified themes were labeled as “unknown”, 6.5%.
4. Discussion
4.1. Explicit Literature Relationships between Resistant Hypertension and COVID-19
- haspin—Haspin is a mitotic kinase for Histone H3, is regarded as a promising anti-tumor therapy; it is overexpressed in malignant tissues due to its requirement for cancer cell proliferation [35].
- ral guanine nucleotide exchange factor—Ral guanine nucleotide exchange factor was previously identified as an important signaling component that regulates transcriptional responses in myocardial cells [36]. It also promotes cardiomyocyte survival and inhibits cardiac fibrosis [37]. Interestingly, the Ral and Ras pathways have also been implicated in myeloid differentiation [38] and especially the BCR-ABL mutation that causes chronic myeloid leukemia [39].
- N-(3-Oxododecanoyl)-L-homoserine lactone—Prior work found that N-3-oxododecanoyl homoserine lactone exacerbates endothelial cell death by inducing receptor-interacting protein kinase 1-dependent apoptosis [40]. It also been reported to induce apoptosis in various types of tumor cells, primarily through a mitochondrial pathway [41]. Notably, N-(3-Oxododecanoyl)-L-homoserine lactone is categorized as a bitter taste receptor. Neutrophils, monocytes, and lymphocytes can express bitter taste receptors being involved in immune response [42]. For this reason, N-(3-Oxododecanoyl)-L-homoserine lactone was hypothesized as one possible therapy for COVID-19 [43].
- uridine kinase—The UK2 gene encodes a uridine kinase protein that catalyzes the phosphorylation of uridine and cytidine to uridine monophosphate (UMP) and cytidine monophosphate (CMP). Uridine is phosphorylated to nucleotides [44]. Most notably, the presence of uridine in RNA and DNA nucleotides allows the SARS-CoV-2 enzyme to cleave RNA because SARS-CoV-2 is a uridine-specific endoribonuclease. Its active site binds to the uridines that are phosphorylated to RNA and DNA nucleotides, allowing the enzyme to cleave RNA [45]. Uridine nucleotides also play an important role in achieving homeostasis in the vascular system. Augmented contractile responses to uridine nucleotides in the femoral arteries of spontaneously hypertensive rates were much higher than in non-hypertensive rats [46]. Uridine kinase has long been known to be involved in the heart physiology. Earlier studies also showed a positive correlation between thyroid hormone and presence of uridine kinase in cardiac cells [47]. More recent work showed that uridine has a hypoglycemic effect that protects against diabetes-mediated functional and structural damage to cardiac mitochondria and disruption of mitochondrial quality-control systems in the diabetic heart [48].
- metabotropic glutamate receptors—Chronic stimulation of group II metabotropic glutamate receptors in the medulla oblongata attenuates hypertension development in spontaneously hypertensive rats [49]. Group III metabotropic glutamate receptors regulate hypothalamic pre-sympathetic neurons through opposing presynaptic and postsynaptic actions in hypertension [50]. Metabotropic glutamate receptors are also important in the regulation of steroidogenesis in the human adrenal gland [51]. As for ties to COVID-19, SARS-CoV-2 uses metabotropic glutamate receptor subtype 2 as an internalization factor to infect cells [52].
- aspartic endopeptidases—This group of endopeptidases is closely tied to the RAAS pathway that induces hypertension and has also been utilized in HIV therapies [53]. Aspartic endopeptidases have also been investigated to reduce dexamethasone-induced hypertension and associated fibrosis in rat models [54]. Their role in the RAAS pathway has also been explicitly noted during and after COVID-19 infection [13].
- MAP3K1—MAP3K1 stands for MAP kinase kinase kinase 1. MAP kinase signaling plays a prominent role in the RAAS pathway. Prior work has shown that angiotensin II up-regulates angiotensin I-converting enzyme (ACE), but down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway [55]. Anti-cytokine therapies targeting JAK-STAT, such as Ruxolitinib, were shown to have a lesser incidence of cardiovascular adverse events compared to steroids given for COVID-19 [56].
- protein tyrosine phosphatase—One type of protein tyrosine phosphatase, CD45, was found to be altered in the leukocytes of COVID-19 patients [57]. Protein tyrosine phosphatase has been cited as having a role in both essential hypertension [58] and pulmonary hypertension [59]. Not surprisingly, tyrosine kinase inhibitors for CML therapy also cause similar adverse events [22]. Similarly, the related JAK pathway has overlap with both COVID-19 and CML [60]. Imatinib, a first-line therapy for CML, was tried as a potential COVID-19 therapy based on two hypothesized roles [61]: (1) potential intralysosomal entrapment of imatinib may increase endosomal pH and effectively decrease SARS-CoV-2/cell fusion, (2) the kinase inhibitory activity of imatinib may interfere with budding/release or replication of SARS-CoV-2.
- choline phosphate cytidylyltransferase—Choline phosphate cytidylyltransferase responsible for regulating phosphatidylcholine content in membranes. Fetal lung fatty acid synthase and choline phosphate cytidylyltransferase activities are increased by glucocorticoids [64].
- MAP3K10—MAP3K10 stands for MAP kinase kinase kinase 10. MAP3K10 activity has been cited in many cancers, namely pancreatic cancer. However, it has also been implicated in the atherosclerotic inflammatory process [65].
- tat genes—The tat (transactivator of transcription) gene transcribes the tat protein, a required transactivator for expression of full-length viral genes, which influences expression of cellular inflammatory genes [66]. Tat has also been implicated in pathways that to modulate angiotensin II-induced medial hypertrophy [67].
- FLNA—FLNA defects can be lethal as it leads to skeletal defects and defects which cause severe cardiac malformations [70].
- USP17L2—Ubiquitin-Specific Peptidase 17-Like Family Member 2 is most known for its role in cancers [71]. USP17 substrates populate two pathways that drive cell cycle progression: one that promotes and one that inhibits. This dual path could explain its both pro-cancer and anti-tumor effects.
- dicer enzyme—Dicer enzyme is a microRNA that has been associated with pregnancy-induced hypertension [68] as well as the brain renin–angiotensin II-induced hypertension and cardiac hypertrophy [72]. More recently, one isoform of Dicer, named antiviral Dicer (aviD), was found to protect tissue stem cells from RNA viruses—including Zika virus and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—by dicing viral double-stranded RNA to orchestrate antiviral RNAi [73].
- somatotropin-releasing hormone—Somatropin is responsible for modulating the release of growth hormone. There is a known delicate balance between endocrine and autonomic function that impact blood pressure. Somatropin hormone, thyrotropin, gonadotropin-releasing hormone, and corticotropin-releasing hormone and others impact the RAAS pathway via angiotensin II to modulate blood pressure [74]. Individuals with isolated congenital GH deficiency due to a GHRH receptor gene mutation appear to cope better with SARS-CoV-2 infection than controls [75]. Likewise, somatotropin-releasing hormone has been shown to be highly correlated to COVID-19. Observations show growth-stimulating hormone may pose as a predictor for severity of post-COVID-19 symptoms while also appearing somewhat relevant to resistant hypertension [76]. Prior research illustrated the anti-inflammatory properties of growth-stimulating hormone receptor antagonists to resistant hypertension [77].
- melanocyte-stimulating hormone—Melanocyte-stimulating hormones (MSHs) are involved in energy metabolism and in inflammation. While melanocyte-stimulating hormones are prevalently acknowledged to have anti-inflammatory and anti-hypertensive properties, their impact and role as a potential adjuvant treatment for COVID-19 remains under scrutiny [78]. Additionally, alpha- and gamma-MSH acutely elevate blood pressure and heart rate through central stimulation of sympathetic nervous outflow [79]. One study implicated neuropeptide Y and alpha-melanocyte-stimulating hormone in hypothalamic regulation of sympathetic nervous system activity [80].
- beta-lipotropin—Beta-lipotropin was initially reported to stimulate melanocytes to produce melanin. Later, it was found to perform lipid-mobilizing functions such as lipolysis and steroidogenesis [83].
- corticotropin—Also known as adrenocorticotropin hormone, corticotropin is produced by the anterior pituitary gland and is an important part of the hypothalamic–pituitary–adrenal axis. As such, it has direct roles in the stress response that increases blood pressure. Autopsy studies on patients who died from the SARS outbreak in 2003 had shown degeneration and necrosis of the adrenal cortical cells [84]. Additionally, SARS-CoV-2 (e.g., COVID-19) was identified in the adrenal glands, hinting towards a direct cytopathic effect of the virus and altered cortisol dynamics [84].
- growth-hormone-releasing peptide-2—Also known as GHRP2, it is a synthetic agonist of ghrelin, a gut peptide that binds to the growth hormone secretagogue receptor. Ghrelin increases growth hormone secretion and appetite initiation [85].
- pro-opiomineralocorticoid—This hormone is synthesized in the anterior pituitary and is part of the central mineralocorticoid system. It has been mentioned as part of the hypothalamic–pituitary autoimmunity seen in COVID-19 patients [86]. Mineralocorticoid receptors are thought to relieve the endothelial and systemic inflammatory mechanisms of respiratory viruses [87].
- prolactin—Prolactin is best known for its role in enabling milk production. However, it also has a role in immunity. For this reason, it was hypothesized that controlled augmentation of prolactin could provide protective benefits for patients infected with COVID-19 [88]. Anecdotal evidence was presented in seven patients when prolactin was initially recommended as a possible repurposed therapy [88].
- thyroid hormones—Thyroid hormones were a recurring top-ranked source node. The thyroid gland also express angiotensin-converting enzyme 2 (ACE2), the main protein that functions as a receptor to which SARS-CoV-2 binds to enter host cells. Immune system cells are targets for thyroid hormones and T3 and T4 modulate specific immune responses, including cell-mediated immunity, natural killer cell activity, the antiviral action of interferon and proliferation of T- and B-lymphocytes [89]. Thyroid pathology is a known event during and immediately after COVID-19 infection. There has been numerous established cases of COVID-19 induced pathology [89] ranging from thyrotoxicosis to suppressed thyroid function, which are largely attributed to the “cytokine storm” [90]. However, one study found that the thyroid hormones T3 and T4 are decreased during active COVID-19 infection compared to baseline [91].
4.2. Intersection of RAAS Hub Nodes and Relationship to COVID-19 and Resistant Hypertension
- poly-beta-hydroxybutyrate depolymerase—It is most known for its role in bacterial microbe metabolism. It is produced by penicillium expansum. Notably, the enzyme requires essential disulphide bonds (cystine residues) and tyrosine to maintain the native enzyme structure [92]. Moreover, patients with severe COVID-19 were found to have elevated levels of branched-chain amino acids and beta-hydroxybutyrate [93].
- CR 1392—It is most known for its role on pancreatic exocrine function and insulin release [94,95]. Glucose dysregulation among patients the COVID-19 is well known [96]. The inflammation of COVID-19 is believed to contribute to increased blood glucose levels. COVID-19 patients with new or worsened glucose dysregulation requiring additional or new insulin administration were associated with poorer outcomes in a study of 456 patients [96].
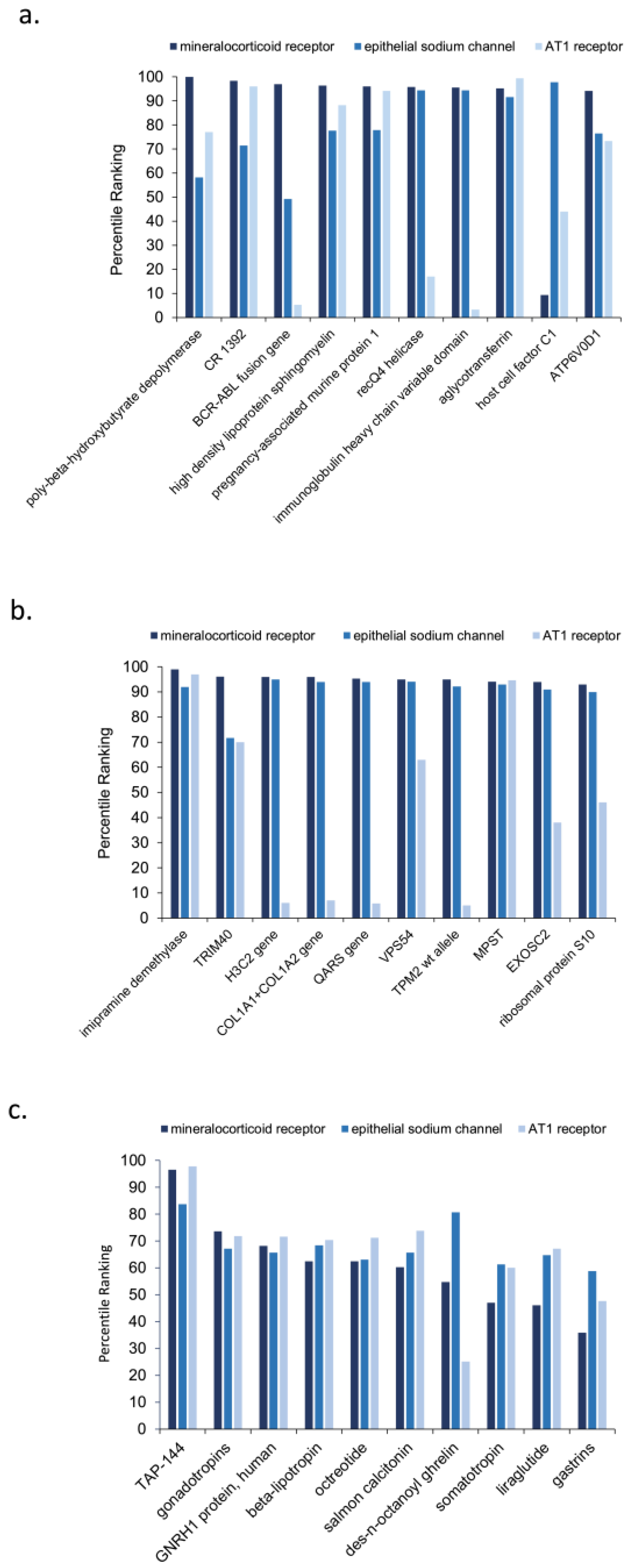
- BCR-ABL fusion gene—Within the list of highest percentile-ranked amino acids and proteins in Figure 6a, the BCR-ABL fusion gene possesses a percentile ranking for mineralocorticoid receptors that is nearly 50% higher than the next greatest percentile ranking for the source nodes of interest. While most clinicians associate the presence of BCR-ABL fusion gene with chronic myeloid leukemia (CML), about 10% of healthy control patients express at least some measurable copies in their peripheral blood [97]. Prior work relating COVID-19 to CML concisely summarizes the role of the gene in upregulating fusion transcripts and proteins, ultimately intensifying the presence and activity of tyrosine kinase within the body [98]. Recent extensive investigation of tyrosine kinase has revealed its direct association in vasculopathy as well as its antihypertensive properties from its interactions with endothelial receptors [99]. Such findings could explain the well-predicted increase in cardiovascular adverse events, including hypertension, in CML patients on tyrosine kinase inhibitor therapies [22]. Since protein kinase in integral in the signaling process of mineralocorticoid receptors both on and within the nucleus of endothelial cells [100], it can be hypothesized that COVID-19 provokes resistant hypertensive tendencies partially by manipulating the expression of BCR-ABL in the mineralocorticoid receptor pathway, decreasing the activity of tyrosine kinase [100].
- high-density lipoprotein sphingomyelin—It is part of high-density lipoprotein (HDL). HDL levels are an inverse risk factor for cardiovascular diseases, and sphingomyelin is the second most abundant phospholipid component and the major sphingolipid in HDL [101]. In one large-scale study, patients with severe COVID-19 were reported to have low levels of total cholesterol, HDL-cholesterol, and LDL-cholesterol, but elevated levels of triglycerides [93].
- immunoglobulin heavy chain variable domain—It is important for binding the antigen and the chain variable constant domains necessary for successful B cell maturation. The heavy chain variables have been suggested as a point of entry for useful applications for prophylaxis and therapy of COVID-19 alone or in combination [102].
- pregnancy-associated murine protein 1—This particular protein is from the mouse domain. However, there could be some relevant ties to clinical COVID-19. Prior work demonstrated a distinct difference in immune modulation between the non-pregnant and pregnant states in COVID-19 patients, which may provide some insight into the pathogenesis of COVID-19 and perhaps explain the more severe outcomes observed in pregnant women [103].
- recQ4 helicase—RecQ4 helicase is a family of helicase enzymes that has been shown to be important in genome maintenance and stability [104]; they catalyze the reaction of ATP and water to drive the unwinding of paired DNA. Relatively little is known about recQ4 other than its important role in genetic stability. Other helicases besides recQ4 have been suggested as potential therapies for COVID-19 based on their antiviral activity [105].
- HCF1—HCF1 is involved in control of the cell cycle and has regulatory roles in a multitude of processes related to transcription. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components [106]. Recent works point to HCF1 as being a putative longevity determinant [107]. Additionally, prior work with herpes simplex virus (HSV) illustrated that multiple chromatin modulation components associated with HCF-1 lead to the initiation of immediate early gene expression in HSV [108].
- ATP6V0D1—ATP6V0D1 encodes a component of vacuolar ATPase (V-ATPase) and is involved in lysosome homeostasis. Clinically, a high expression of ATP6V0D1 was correlated with prolonged survival of patients with pancreatic ductal adenocarcinoma [109]. It has been shown to correlate with thyroid hormones [110] and proinsulin processing [111]. Collectively, these data suggested that the S protein from COVID-19 increased V-ATPase in SARS-CoV-2 infection, which provided a microenvironment easier for the cleavage of S protein by making the activation of inflammatory cells in the respiratory epithelium easier [112].
- imipramine demethylase—Imipramine is a tricyclic antidepressant that undergoes demethylation as part of metabolism. Imipramine increases dopamine in the striatum [113]. Imipramine demethylation is associated with the CYP2D6 and CYP2C19 genotype. While there is no straightforward reason for its predicted association with COVID-19, it is known that dopamine increases hypertension [114]. A meta-analysis examining the association of anti-depressants with COVID-19 incidence and severity found that most either had no impact or had have slight protective effects [115].
- TRIM40—Tripartite motif (TRIM)-containing proteins are E3 ubiquitin ligases that possess crucial regulatory functions in innate immunity [116]. In particular, they attenuate antiviral immune response [117]. TRIM40 also has associations with IgA nephropathology, which causes kidney disease; it is thought to suppress IgA1-induced GMC proliferation by inhibiting the activation of NLRP3 inflammasome [116]. TRIM40′s associations with kidney disease and regulatory functions in immunity both align with a role in resistant hypertension and COVID-19, respectively.
- QARS gene—Encoding glutaminyl-tRNA synthetase QARS has been implicated in progressive microcephaly, severe seizures in infancy, atrophy of the cerebral cortex and cerebellar vermis, and mild atrophy of the cerebellar hemispheres [118]. Its repeated selection by the algorithm as a potential important source node for the relationship between COVID-19 and resistant hypertension remains unclear.
- COL1A1+COL1A2 gene—are collagen I and II genes that encode primarily for connective tissue. COL1A1+COL1A2 gene mutations are most implicated in osteogenesis impefecta (OI), which is also known as brittle bone disease. Additionally, patients with COL1A1+COL1A2 gene mutations have shown various forms of cardiovascular pathology, particularly regurgitation of the heart valves, attributed to poor collagen [119]. Interestingly, a small study examining pediatric OI patients found that they had no difference in outcome after COVID-19 infection [120]. Moreover, two therapies used for OI, mesenchymal stem cells and decidua stromal cells were used to treat SARS-CoV-2 coronavirus-induced disease [121] as an anti-inflammatory treatment to reduce COVID-induced cytokine storm.
- MPST—The biological function of MPST remains unclear. It may be involved in cyanide detoxification, biosynthesis of thiosulfate, production of the signaling molecule hydrogen sulfide, or the degradation of cysteine. Sulfur metabolism in the liver has been strongly correlated with hypertension in animal models [122]. Sulfur dioxide also increases pulmonary hypertension [123]. Interesting, air pollutants like sulfur dioxide were found to be associated with increased incidence of COVID-19 [124]. Additionally, liver dysfunction marked by elevation of lipoproteins X and Z has been found in patients with severe COVID-19 [93].
- H3C2 gene—It encodes for histone 3, which is fundamental in development. H3C2 has been associated with several pediatric gliomas [125]. Notably, haspin, which encodes for histone, was found to be among the top-ranking nodes for both AAPP and GNGM node types in analysis A4, which examined the intersecting source nodes for resistant hypertension and COVID-19.
- VPS54—VPS54 is most known for its role in motor pathology, such as ALS phenotypes, including the wobbler mouse [126]. However, the vacuole protein sorting gene VPS54 was also recently shown to be required for extracellular virus (EV) formation in monkeypox infection [127]. It is possible VPS54 may play a similar role in COVID-19.
- EXOSC2—Low expression of EXOSC2 was found to protect against clinical COVID-19 and impedes SARS-CoV-2 replication [131]. Likewise, aggregating COVID-19 GWAS statistics revealed an association between increased expression of EXOSC2 and higher risk of clinical COVID-19 [131]. EXOSC2 is a component of the RNA exosome that LC-MS/MS analysis demonstrated an interaction between the SARS-CoV-2 RNA polymerase and the majority of human RNA exosome components [131]. Additionally, EXOSC2 has been linked to sudden cardiac death [132] due to cardiac conduction abnormalities and arrhythmia from QTc prolongation. As such, EXOSC2 has clear ties to both COVID-19 and cardiovascular disease.
- ribosomal protein S10—The role of ribosomal protein S10 is not clear with COVID-19. However, there is an association with lupus autoantibodies [133]. COVID-19 and subsequent cytokine storm was associated with both new onset lupus [134] as well as lupus flares in existing patients with systemic lupus erythematosus (SLE) [135]. Additionally, hydroxychloroquine, a common drug for lupus, was found in computer simulations to illustrate a potential benefit [20]. While hydroxychloroquine was initially tried as an adjuvant therapy for COVID-19, its efficacy for COVID-19 was ultimately found to be controversial [135]. Hydroxychloroquine for COVID-19 was largely stopped over concerns of sudden cardiac death from arrhythmia [136]. Interesting, an increase in new onset SLE has been noted after COVDI-19 infection [134].
- TAP-144—TAP-144 is also known as leuprorelin or leuprolide, which is a gonadotropin-releasing hormone analogue family of medications. Side effects may include high blood sugar and problems with the pituitary gland. Interestingly, previous artificial intelligence work identified leuprolide as a possible repurposed drug for COVID-19 based on its structural biology [137]. TAP-144 has a high binding affinity to COVID-19 and was thought to modify the functional ability of the spike protein. Leuprolide was validated by molecular docking against the spike protein complex with ACE receptor [137].
- des-n-octanoyl ghrelin—Ghrelin is involved in appetite stimulation and growth hormone release. Des-n-octanoyl ghrelin has some distinct functions from ghrelin. Namely, the lack of acylation prevents binding to the ghrelin receptor and growth hormone release. Interestingly, patients with cardiovascular events were found to have lower levels of des-acyl ghrelin at baseline than those without cardiovascular events [138].
- beta-lipotropin—Both beta-lipotropin and beta-endorphin are present in cardiac tissue. The amounts and ratio of beta-endorphin and beta-lipotropin in the heart appear to be modulated by testosterone propionate [139]. Circulating beta-endorphin and beta-lipotropin concentrations increase after the administration of acetylcholine or serotonin agonist drugs [140]. Patients with heart disease, namely congestive heart failure, were found to have lower amounts of beta-lipotropin than control patients [141].
- liraglutide—Liraglutide is anti-diabetic medication utilized to treat hyperglycemia due to type 2 diabetes. New onset diabetes is another disease found to be a sequala of COVID-19 [10,11]. Higher blood sugar tends to happen during and after COVID-19 infection [96,146] and is a comorbidity with other reported endocrine dysfunction brough on or exacerbated by COVID-19 [91].
- octreotide—Octreotide has been successfully utilized to treat portal hypertension in the liver associated with cirrhosis or other liver pathology [147]. As noted earlier, markers of hepatic dysfunction are also prominent in severe COVID-19 illness [93]. One study reported a patient with acromegaly and pre-diabetes with severe respiratory distress from COVID-19 that responded quite positively to the administration of octreotide to improve COVID-19 outcome [148]. Additionally, another structural biology simulation predicted octreotide as a viable repurposed drug for COVID-19 [149]. Acromegaly has been hypothesized by other studies to be a point of emphasis in COVID-19 etiology and treatment [150].
- somatotropin—is a recombinant form of human growth hormone. In a clinical study of 456 patients, growth hormone and IGF-1 deficiency were found in COVID-19 cases with lung involvement, regardless of age or gender; COVID-19 infection progressed worse in GH and IGF-1 deficiency [151]. Another study examining COVID-19 inflammation in cellular model found that growth hormone and estradiol improved inflammation, but testosterone had the opposite effect [152]. However, a potential complicating factor of growth hormone treatment for COVID-19 is that acromegaly, a state of endogenous GH excess, results in myocardial hypertrophy and decreased cardiac performance with increased cardiovascular mortality [153].
- gastrins—Hypertension is related to impaired metabolic homeostasis and can be regarded as a metabolic disorder [154]. Interestingly, an in silico modeling study examining molecular dynamics suggested that pentagastrin, a synthetic polypeptide that has effects like gastrin when given parenterally, could be a viable drug for COVID-19 [155]. Gastrin-releasing peptide has long been held for its possible role as a target for inflammatory disease [156], including cardiovascular disease, gastrointestinal disease, pulmonary disease, and of course, its roles in endocrine disorders, namely glucose metabolism.
- thyroid hormone—Thyroid hormone was a recurring highly ranked source node identified during both direct simulation and cross-domain analysis. It has been hypothesized that hyperinflammation, as reflected by the secretion of cytokines, might induce thyroid dysfunction among patients with COVID-19. Thyroid hormone involvement in the acute phase of symptomatic COVID-19 and its possible associations with cytokine levels and mortality risk have been explored [157]. Findings suggest that fluctuations of TSH levels in patients with COVID-19 may be influenced by circulating IL-8, IL-10, IL-15, IP-10, and GM-CSF as previously described in autoimmune thyroid diseases [157]. Thyroid sequala are well-documented in COVID-19 [84,89,91,157]. There are also documented correlations with thyroid hormone and salmon and human calcitonin peptides and uridine kinase [47], of which salmon calcitonin peptide was also a highly ranked AAPPs in the present study [144].
4.3. Explaining Synthesized Biological Themes Comprised by High-Ranking Concepts
4.4. Study Limitations
4.4.1. Technical Limitations
4.4.2. Applied Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, D.; Comish, P.; Kang, R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020, 16, e1008536. [Google Scholar] [CrossRef] [PubMed]
- Clerkin, K.J.; Fried, J.A.; Raikhelkar, J.; Sayer, G.; Griffin, J.M.; Masoumi, A.; Jain, S.S.; Burkhoff, D.; Kumaraiah, D.; Rabbani, L.; et al. COVID-19 and Cardiovascular Disease. Circulation 2020, 141, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, F.; Wang, L. COVID-19 and cardiovascular diseases. J. Mol. Cell Biol. 2021, 13, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Tadic, M.; Cuspidi, C. Resistant hypertension and COVID-19: Tip of the iceberg? J. Hum. Hypertens. 2022, 36, 693–694. [Google Scholar] [CrossRef]
- Dumitru, I.M.; Vlad, N.D.; Rugina, S.; Onofrei, N.; Gherca, S.; Raduna, M.; Trana, A.; Dumitrascu, M.; Popovici, E.; Bajdechi, M.; et al. SARS-CoV-2 Infection and Emery-Dreifuss Syndrome in a Young Patient with a Family History of Dilated Cardiomyopathy. Genes 2021, 12, 1070. [Google Scholar] [CrossRef]
- Isik, F.; Cap, M.; Akyuz, A.; Bilge, O.; Aslan, B.; Inci, U.; Kaya, I.; Tastan, E.; Oksul, M.; Cap, N.K.; et al. The effect of resistant hypertension on in-hospital mortality in patients hospitalized with COVID-19. J. Hum. Hypertens. 2022, 36, 846–851. [Google Scholar] [CrossRef]
- Dudenbostel, T.; Siddiqui, M.; Oparil, S.; Calhoun, D.A. Refractory Hypertension: A Novel Phenotype of Antihypertensive Treatment Failure. Hypertension 2016, 67, 1085–1092. [Google Scholar] [CrossRef]
- Delalic, D.; Jug, J.; Prkacin, I. Arterial Hypertension Following COVID-19: A Retrospective Study of Patients in a Central European Tertiary Care Center. Acta Clin. Croat. 2022, 61, 23–27. [Google Scholar] [CrossRef]
- Zhang, V.; Fisher, M.; Hou, W.; Zhang, L.; Duong, T.Q. Incidence of New-Onset Hypertension Post-COVID-19: Comparison with Influenza. Hypertension 2023, 80, 2135–2148. [Google Scholar] [CrossRef]
- Chen, G.; Li, X.; Gong, Z.; Xia, H.; Wang, Y.; Wang, X.; Huang, Y.; Barajas-Martinez, H.; Hu, D. Hypertension as a sequela in patients of SARS-CoV-2 infection. PLoS ONE 2021, 16, e0250815. [Google Scholar] [CrossRef]
- Wrona, M.; Skrypnik, D. New-Onset Diabetes Mellitus, Hypertension, Dyslipidaemia as Sequelae of COVID-19 Infection-Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 13280. [Google Scholar] [CrossRef]
- Ong, W.Y.; Satish, R.L.; Herr, D.R. ACE2, Circumventricular Organs and the Hypothalamus, and COVID-19. Neuromol. Med. 2022, 24, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Tsampasian, V.; Corballis, N.; Vassiliou, V.S. Renin-Angiotensin-Aldosterone Inhibitors and COVID-19 Infection. Curr. Hypertens. Rep. 2022, 24, 425–433. [Google Scholar] [CrossRef]
- Fountain, J.H.; Kaur, J.; Lappin, S.L. Physiology, Renin Angiotensin System; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Grant, S.N.; Lester, H.A. Regulation of epithelial sodium channel activity by SARS-CoV-1 and SARS-CoV-2 proteins. Biophys. J. 2021, 120, 2805–2813. [Google Scholar] [CrossRef]
- Swanson, D.R. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect. Biol. Med. 1986, 30, 7–18. [Google Scholar] [CrossRef]
- Al-Hussaini, I.; Nakajima An, D.; Lee, A.J.B.; Bi, S.; Mitchell, C.S. CCS Explorer: Relevance Prediction, Extractive Summarization, and Named Entity Recognition from Clinical Cohort Studies. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 17–20 December 2022. [Google Scholar]
- Kirkpatrick, A.; Onyeze, C.; Kartchner, D.; Allegri, S.; An, D.N.; McCoy, K.; Davalbhakta, E.; Mitchell, C.S. Optimizations for Computing Relatedness in Biomedical Heterogeneous Information Networks: SemNet 2.0. Big Data Cogn. Comput. 2022, 6, 27. [Google Scholar] [CrossRef]
- Sedler, A.R.; Mitchell, C.S. SemNet: Using Local Features to Navigate the Biomedical Concept Graph. Front. Bioeng Biotechnol. 2019, 7, 156. [Google Scholar] [CrossRef]
- McCoy, K.; Gudapati, S.; He, L.; Horlander, E.; Kartchner, D.; Kulkarni, S.; Mehra, N.; Prakash, J.; Thenot, H.; Vanga, S.V.; et al. Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19. Pharmaceutics 2021, 13, 794. [Google Scholar] [CrossRef]
- Pires, C. A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019. J. Pers. Med. 2021, 11, 926. [Google Scholar] [CrossRef]
- Mehra, N.; Varmeziar, A.; Chen, X.; Kronick, O.; Fisher, R.; Kota, V.; Mitchell, C.S. Cross-Domain Text Mining to Predict Adverse Events from Tyrosine Kinase Inhibitors for Chronic Myeloid Leukemia. Cancers 2022, 14, 4686. [Google Scholar] [CrossRef]
- Tandra, G.; Yoone, A.; Mathew, R.; Wang, M.; Hales, C.M.; Mitchell, C.S. Literature-Based Discovery Predicts Antihistamines Are a Promising Repurposed Adjuvant Therapy for Parkinson & rsquo’s Disease. Int. J. Mol. Sci. 2023, 24, 12339. [Google Scholar] [PubMed]
- Xiao, Y.; Zhang, B.; Cloyd, J.M.; Alaimo, L.; Xu, G.; Du, S.; Mao, Y.; Pawlik, T.M. Novel Drug Candidate Prediction for Intrahepatic Cholangiocarcinoma via Hub Gene Network Analysis and Connectivity Mapping. Cancers 2022, 14, 3284. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.; McInnes, B.T. Literature Based Discovery: Models, methods, and trends. J. Biomed. Inform. 2017, 74, 20–32. [Google Scholar] [CrossRef]
- Pimenta, E.; Calhoun, D.A. Treatment of resistant hypertension. J. Hypertens. 2010, 28, 2194–2195. [Google Scholar] [CrossRef]
- Judd, E.K.; Calhoun, D.A.; Warnock, D.G. Pathophysiology and treatment of resistant hypertension: The role of aldosterone and amiloride-sensitive sodium channels. Semin. Nephrol. 2014, 34, 532–539. [Google Scholar] [CrossRef][Green Version]
- Gorini, S.; Kim, S.K.; Infante, M.; Mammi, C.; La Vignera, S.; Fabbri, A.; Jaffe, I.Z.; Caprio, M. Role of Aldosterone and Mineralocorticoid Receptor in Cardiovascular Aging. Front. Endocrinol. 2019, 10, 584. [Google Scholar] [CrossRef]
- Gomez-Sanchez, C.E.; Gomez-Sanchez, E.P. The Mineralocorticoid Receptor and the Heart. Endocrinology 2021, 162, bqab131. [Google Scholar] [CrossRef]
- Kim, J.; Miyazaki, K.; Shah, P.; Kozai, L.; Kewcharoen, J. Association between Mineralocorticoid Receptor Antagonist and Mortality in SARS-CoV-2 Patients: A Systematic Review and Meta-Analysis. Healthcare 2022, 10, 645. [Google Scholar] [CrossRef]
- Shimosawa, T. Salt, the renin-angiotensin-aldosterone system and resistant hypertension. Hypertens. Res. 2013, 36, 657–660. [Google Scholar] [CrossRef]
- Zaika, O.; Mamenko, M.; Staruschenko, A.; Pochynyuk, O. Direct activation of ENaC by angiotensin II: Recent advances and new insights. Curr. Hypertens. Rep. 2013, 15, 17–24. [Google Scholar] [CrossRef]
- Kawai, T.; Forrester, S.J.; O’Brien, S.; Baggett, A.; Rizzo, V.; Eguchi, S. AT1 receptor signaling pathways in the cardiovascular system. Pharmacol. Res. 2017, 125, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Rothlin, R.P.; Duarte, M.; Pelorosso, F.G.; Nicolosi, L.; Salgado, M.V.; Vetulli, H.M.; Spitzer, E. Angiotensin Receptor Blockers for COVID-19: Pathophysiological and Pharmacological Considerations About Ongoing and Future Prospective Clinical Trials. Front. Pharmacol. 2021, 12, 603736. [Google Scholar] [CrossRef] [PubMed]
- Quadri, R.; Sertic, S.; Muzi-Falconi, M. Roles and regulation of Haspin kinase and its impact on carcinogenesis. Cell Signal 2022, 93, 110303. [Google Scholar] [CrossRef] [PubMed]
- Post, G.R.; Swiderski, C.; Waldrop, B.A.; Salty, L.; Glembotski, C.C.; Wolthuis, R.M.; Mochizuki, N. Guanine nucleotide exchange factor-like factor (Rlf) induces gene expression and potentiates alpha 1-adrenergic receptor-induced transcriptional responses in neonatal rat ventricular myocytes. J. Biol. Chem. 2002, 277, 15286–15292. [Google Scholar] [CrossRef] [PubMed]
- Scotland, R.L.; Allen, L.; Hennings, L.J.; Post, G.R.; Post, S.R. The ral exchange factor rgl2 promotes cardiomyocyte survival and inhibits cardiac fibrosis. PLoS ONE 2013, 8, e73599. [Google Scholar] [CrossRef] [PubMed]
- Omidvar, N.; Pearn, L.; Burnett, A.K.; Darley, R.L. Ral is both necessary and sufficient for the inhibition of myeloid differentiation mediated by Ras. Mol. Cell Biol. 2006, 26, 3966–3975. [Google Scholar] [CrossRef]
- Fredericks, J.; Ren, R. The role of RAS effectors in BCR/ABL induced chronic myelogenous leukemia. Front. Med. 2013, 7, 452–461. [Google Scholar] [CrossRef]
- Shin, J.; Ahn, S.H.; Kim, S.H.; Oh, D.J. N-3-oxododecanoyl homoserine lactone exacerbates endothelial cell death by inducing receptor-interacting protein kinase 1-dependent apoptosis. Am. J. Physiol. Cell Physiol. 2021, 321, C644–C653. [Google Scholar] [CrossRef]
- Zhao, G.; Neely, A.M.; Schwarzer, C.; Lu, H.; Whitt, A.G.; Stivers, N.S.; Burlison, J.A.; White, C.; Machen, T.E.; Li, C. N-(3-oxo-acyl) homoserine lactone inhibits tumor growth independent of Bcl-2 proteins. Oncotarget 2016, 7, 5924–5942. [Google Scholar] [CrossRef]
- Dhanaraj, P.; Muthiah, I.; Rozbu, M.R.; Nuzhat, S.; Paulraj, M.S. Computational Studies on T2Rs Agonist-Based Anti-COVID-19 Drug Design. Front. Mol. Biosci. 2021, 8, 637124. [Google Scholar] [CrossRef]
- Esam, Z. Protective potential of expectorants against COVID-19. Med. Hypotheses. 2020, 142, 109844. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.N.; Koizumi, K.; Fukushima, M.; Matsuda, A.; Inagaki, F. Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase. Structure 2004, 12, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wower, J.; Maltseva, N.; Chang, C.; Jedrzejczak, R.; Wilamowski, M.; Kang, S.; Nicolaescu, V.; Randall, G.; Michalska, K.; et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Takayanagi, K.; Katome, T.; Kojima, M.; Taguchi, K.; Kobayashi, T. Extracellular Uridine Nucleotides-Induced Contractions Were Increased in Femoral Arteries of Spontaneously Hypertensive Rats. Pharmacology 2021, 106, 435–445. [Google Scholar] [CrossRef]
- Gertz, B.J.; Haugaard, E.S.; Haugaard, N. Effects of thyroid hormone on UTP content and uridine kinase activity of rat heart and skeletal muscle. Am. J. Physiol. 1980, 238, E443–E449. [Google Scholar] [CrossRef]
- Belosludtseva, N.V.; Starinets, V.S.; Mikheeva, I.B.; Belosludtsev, M.N.; Dubinin, M.V.; Mironova, G.D.; Belosludtsev, K.N. Effect of Chronic Treatment with Uridine on Cardiac Mitochondrial Dysfunction in the C57BL/6 Mouse Model of High-Fat Diet-Streptozotocin-Induced Diabetes. Int. J. Mol. Sci. 2022, 23, 10633. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.C.; Sekizawa, S.I.; Tochinai, R.; Kuwahara, M. Chronic stimulation of group II metabotropic glutamate receptors in the medulla oblongata attenuates hypertension development in spontaneously hypertensive rats. PLoS ONE 2021, 16, e0251495. [Google Scholar] [CrossRef]
- Zhou, J.J.; Pachuau, J.; Li, D.P.; Chen, S.R.; Pan, H.L. Group III metabotropic glutamate receptors regulate hypothalamic presympathetic neurons through opposing presynaptic and postsynaptic actions in hypertension. Neuropharmacology 2020, 174, 108159. [Google Scholar] [CrossRef]
- Felizola, S.J.A.; Nakamura, Y.; Satoh, F.; Morimoto, R.; Kikuchi, K.; Nakamura, T.; Hozawa, A.; Wang, L.; Onodera, Y.; Ise, K.; et al. Glutamate receptors and the regulation of steroidogenesis in the human adrenal gland: The metabotropic pathway. Mol. Cell Endocrinol. 2014, 382, 170–177. [Google Scholar] [CrossRef]
- Wang, J.; Yang, G.; Wang, X.; Wen, Z.; Shuai, L.; Luo, J.; Wang, C.; Sun, Z.; Liu, R.; Ge, J.; et al. SARS-CoV-2 uses metabotropic glutamate receptor subtype 2 as an internalization factor to infect cells. Cell Discov. 2021, 7, 119. [Google Scholar] [CrossRef]
- Dash, C.; Kulkarni, A.; Dunn, B.; Rao, M. Aspartic peptidase inhibitors: Implications in drug development. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 89–119. [Google Scholar] [CrossRef] [PubMed]
- Savitha, M.N.; Suvilesh, K.N.; Siddesha, J.M.; Milan Gowda, M.D.; Choudhury, M.; Velmurugan, D.; Umashankar, M.; Vishwanath, B.S. Combinatorial inhibition of Angiotensin converting enzyme, Neutral endopeptidase and Aminopeptidase N by N-methylated peptides alleviates blood pressure and fibrosis in rat model of dexamethasone-induced hypertension. Peptides 2020, 123, 170180. [Google Scholar] [CrossRef] [PubMed]
- Koka, V.; Huang, X.R.; Chung, A.C.; Wang, W.; Truong, L.D.; Lan, H.Y. Angiotensin II up-regulates angiotensin I-converting enzyme (ACE), but down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway. Am. J. Pathol. 2008, 172, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Stanevich, O.V.; Fomina, D.S.; Bakulin, I.G.; Galeev, S.I.; Bakin, E.A.; Belash, V.A.; Kulikov, A.N.; Lebedeva, A.A.; Lioznov, D.A.; Polushin, Y.S.; et al. Ruxolitinib versus dexamethasone in hospitalized adults with COVID-19: Multicenter matched cohort study. BMC Infect. Dis. 2021, 21, 1277. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.G.T.; Limmer, A.; Sucker, C.; Fares, K.M.; Mohamed, S.A.; Othman, A.H.; Berger, M.M.; Brenner, T.; Hartmann, M. Differential Regulation of CD45 Expression on Granulocytes, Lymphocytes, and Monocytes in COVID-19. J. Clin. Med. 2022, 11, 4219. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hsiung, C.A.; Chuang, L.M.; Ho, L.T.; Ting, C.T.; Bustos, V.I.; Lee, T.M.; De Witte, A.; Chen, Y.D.; Olshen, R.; et al. Single nucleotide polymorphisms in protein tyrosine phosphatase 1beta (PTPN1) are associated with essential hypertension and obesity. Hum. Mol. Genet. 2004, 13, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Jiang, W.; Du, H.; Shao, J.; Lu, B.; Wang, J.; Zou, D. Protein tyrosine phosphatase 1B gene polymorphisms and essential hypertension: A case-control study in Chinese population. J. Endocrinol. Investig. 2010, 33, 483–488. [Google Scholar] [CrossRef]
- Satarker, S.; Tom, A.A.; Shaji, R.A.; Alosious, A.; Luvis, M.; Nampoothiri, M. JAK-STAT Pathway Inhibition and their Implications in COVID-19 Therapy. Postgrad. Med. 2021, 133, 489–507. [Google Scholar] [CrossRef]
- Emadi, A.; Chua, J.V.; Talwani, R.; Bentzen, S.M.; Baddley, J. Safety and Efficacy of Imatinib for Hospitalized Adults with COVID-19: A structured summary of a study protocol for a randomised controlled trial. Trials 2020, 21, 897. [Google Scholar] [CrossRef]
- Savoia, C.; Ebrahimian, T.; He, Y.; Gratton, J.P.; Schiffrin, E.L.; Touyz, R.M. Angiotensin II/AT2 receptor-induced vasodilation in stroke-prone spontaneously hypertensive rats involves nitric oxide and cGMP-dependent protein kinase. J. Hypertens. 2006, 24, 2417–2422. [Google Scholar] [CrossRef]
- Patrucco, E.; Domes, K.; Sbroggio, M.; Blaich, A.; Schlossmann, J.; Desch, M.; Rybalkin, S.D.; Beavo, J.A.; Lukowski, R.; Hofmann, F. Roles of cGMP-dependent protein kinase I (cGKI) and PDE5 in the regulation of Ang II-induced cardiac hypertrophy and fibrosis. Proc. Natl. Acad. Sci. USA 2014, 111, 12925–12929. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Smart, D.A.; Rooney, S.A. Glucocorticoid induction of fatty-acid synthase mediates the stimulatory effect of the hormone on choline-phosphate cytidylyltransferase activity in fetal rat lung. Biochim. Biophys. Acta 1990, 1044, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, T.; Yang, L.; Li, Z.; Wong, M.M.; Zheng, X.; Pan, X.; Zhang, L.; Yan, H. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS ONE 2012, 7, e46551. [Google Scholar] [CrossRef]
- Cota-Gomez, A.; Flores, A.C.; Ling, X.F.; Varella-Garcia, M.; Flores, S.C. HIV-1 Tat increases oxidant burden in the lungs of transgenic mice. Free Radic. Biol. Med. 2011, 51, 1697–1707. [Google Scholar] [CrossRef]
- Liu, J.; Ormsby, A.; Oja-Tebbe, N.; Pagano, P.J. Gene transfer of NAD(P)H oxidase inhibitor to the vascular adventitia attenuates medial smooth muscle hypertrophy. Circ. Res. 2004, 95, 587–594. [Google Scholar] [CrossRef]
- Huang, X.; An, Y.; Li, X.; Wang, D.; Tan, H.; Lei, J. Genetic variants in DICER1, DROSHA, RAN, and XPO5 genes and risk of pregnancy-induced hypertension. Pregnancy Hypertens. 2019, 16, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Martins, E.; Cardoso, J.S.; Abreu-Lima, C. Familial dilated cardiomyopathy. Rev. Port. Cardiol. 2002, 21, 1487–1503. [Google Scholar]
- Hart, A.W.; Morgan, J.E.; Schneider, J.; West, K.; McKie, L.; Bhattacharya, S.; Jackson, I.J.; Cross, S.H. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum. Mol. Genet. 2006, 15, 2457–2467. [Google Scholar] [CrossRef]
- Ducker, C.; Shaw, P.E. USP17-mediated de-ubiquitination and cancer: Clients cluster around the cell cycle. Int. J. Biochem. Cell Biol. 2021, 130, 105886. [Google Scholar] [CrossRef]
- Lafarga, M.; Hervas, J.P. Intranuclear inclusions in pericytes of the hypothalamus of the rat. Cell Tissue Res. 1978, 193, 315–322. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Buck, M.D.; Chakravarty, P.; Carvalho, J.; Frederico, B.; Cardoso, A.; Healy, L.; Ulferts, R.; Beale, R.; Reis e Sousa, C. An isoform of Dicer protects mammalian stem cells against multiple RNA viruses. Science 2021, 373, 231–236. [Google Scholar] [CrossRef]
- Ganong, W.F. Circumventricular organs: Definition and role in the regulation of endocrine and autonomic function. Clin. Exp. Pharmacol. Physiol. 2000, 27, 422–427. [Google Scholar] [CrossRef]
- Melo, M.A.; Borges, L.P.; Salvatori, R.; Souza, D.R.V.; Santos-Junior, H.T.; de Neto, J.M.R.; Campos, V.C.; Santos, A.A.; Oliveira, C.R.P.; da Invencao, G.B.; et al. Individuals with isolated congenital GH deficiency due to a GHRH receptor gene mutation appear to cope better with SARS-CoV-2 infection than controls. Endocrine 2021, 72, 349–355. [Google Scholar] [CrossRef]
- Elkarow, M.H.; Hamdy, A. A Suggested Role of Human Growth Hormone in Control of the COVID-19 Pandemic. Front. Endocrinol. 2020, 11, 569633. [Google Scholar] [CrossRef]
- Cen, L.P.; Ng, T.K.; Chu, W.K.; Pang, C.P. Growth hormone-releasing hormone receptor signaling in experimental ocular inflammation and neuroprotection. Neural. Regen. Res. 2022, 17, 2643–2648. [Google Scholar] [CrossRef]
- Singh, M.; Mukhopadhyay, K. Alpha-melanocyte stimulating hormone: An emerging anti-inflammatory antimicrobial peptide. Biomed. Res. Int. 2014, 2014, 874610. [Google Scholar] [CrossRef]
- Humphreys, M.H. Cardiovascular and renal actions of melanocyte-stimulating hormone peptides. Curr. Opin. Nephrol. Hypertens. 2007, 16, 32–38. [Google Scholar] [CrossRef]
- Baltatzi, M.; Hatzitolios, A.; Tziomalos, K.; Iliadis, F.; Zamboulis, C. Neuropeptide Y and alpha-melanocyte-stimulating hormone: Interaction in obesity and possible role in the development of hypertension. Int. J. Clin. Pract. 2008, 62, 1432–1440. [Google Scholar] [CrossRef]
- Mela, V.; Diaz, F.; Gertler, A.; Solomon, G.; Argente, J.; Viveros, M.P.; Chowen, J.A. Neonatal treatment with a pegylated leptin antagonist has a sexually dimorphic effect on hypothalamic trophic factors and neuropeptide levels. J. Neuroendocr. 2012, 24, 756–765. [Google Scholar] [CrossRef]
- Obradovic, M.; Sudar-Milovanovic, E.; Soskic, S.; Essack, M.; Arya, S.; Stewart, A.J.; Gojobori, T.; Isenovic, E.R. Leptin and Obesity: Role and Clinical Implication. Front. Endocrinol. 2021, 12, 585887. [Google Scholar] [CrossRef]
- Li, C.H.; Chung, D. Isolation and structure of an untriakontapeptide with opiate activity from camel pituitary glands. Proc. Natl. Acad. Sci. USA 1976, 73, 1145–1148. [Google Scholar] [CrossRef]
- Pal, R. COVID-19, hypothalamo-pituitary-adrenal axis and clinical implications. Endocrine 2020, 68, 251–252. [Google Scholar] [CrossRef]
- Laferrere, B.; Abraham, C.; Russell, C.D.; Bowers, C.Y. Growth hormone releasing peptide-2 (GHRP-2), like ghrelin, increases food intake in healthy men. J. Clin. Endocrinol. Metab. 2005, 90, 611–614. [Google Scholar] [CrossRef]
- Gonen, M.S.; De Bellis, A.; Durcan, E.; Bellastella, G.; Cirillo, P.; Scappaticcio, L.; Longo, M.; Bircan, B.E.; Sahin, S.; Sulu, C.; et al. Assessment of Neuroendocrine Changes and Hypothalamo-Pituitary Autoimmunity in Patients with COVID-19. Horm. Metab. Res. 2022, 54, 153–161. [Google Scholar] [CrossRef]
- Lonati, C.; Gatti, S.; Catania, A. Activation of Melanocortin Receptors as a Potential Strategy to Reduce Local and Systemic Reactions Induced by Respiratory Viruses. Front. Endocrinol. 2020, 11, 569241. [Google Scholar] [CrossRef]
- Sen, A. Repurposing prolactin as a promising immunomodulator for the treatment of COVID-19: Are common Antiemetics the wonder drug to fight coronavirus? Med. Hypotheses. 2020, 144, 110208. [Google Scholar] [CrossRef]
- Rossetti, C.L.; Cazarin, J.; Hecht, F.; Beltrao, F.E.L.; Ferreira, A.C.F.; Fortunato, R.S.; Ramos, H.E.; de Carvalho, D.P. COVID-19 and thyroid function: What do we know so far? Front. Endocrinol. 2022, 13, 1041676. [Google Scholar] [CrossRef]
- Croce, L.; Gangemi, D.; Ancona, G.; Liboa, F.; Bendotti, G.; Minelli, L.; Chiovato, L. The cytokine storm and thyroid hormone changes in COVID-19. J. Endocrinol. Investig. 2021, 44, 891–904. [Google Scholar] [CrossRef]
- Khoo, B.; Tan, T.; Clarke, S.A.; Mills, E.G.; Patel, B.; Modi, M.; Phylactou, M.; Eng, P.C.; Thurston, L.; Alexander, E.C.; et al. Thyroid Function Before, During, and After COVID-19. J. Clin. Endocrinol. Metab. 2021, 106, e803–e811. [Google Scholar] [CrossRef]
- Gowda, U.S.V.; Shivakumar, S. Poly(-beta-hydroxybutyrate) (PHB) depolymerase PHAZ (Pen) from Penicillium expansum: Purification, characterization and kinetic studies. 3 Biotech 2015, 5, 901–909. [Google Scholar] [CrossRef]
- Ballout, R.A.; Kong, H.; Sampson, M.; Otvos, J.D.; Cox, A.L.; Agbor-Enoh, S.; Remaley, A.T. The NIH Lipo-COVID Study: A Pilot NMR Investigation of Lipoprotein Subfractions and Other Metabolites in Patients with Severe COVID-19. Biomedicines 2021, 9, 1090. [Google Scholar] [CrossRef] [PubMed]
- Okabayashi, Y.; Otsuki, M.; Nakamura, T.; Fujii, M.; Tani, S.; Fujisawa, T.; Koide, M.; Hasegawa, H.; Baba, S. Proglumide analogues CR 1409 and CR 1392 inhibit cholecystokinin-stimulated insulin release more potently than exocrine secretion from the isolated perfused rat pancreas. Pancreas 1990, 5, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Otsuki, M.; Fujii, M.; Nakamura, T.; Okabayashi, Y.; Tani, S.; Fujisawa, T.; Koide, M.; Baba, S. Effects of a new proglumide analogue CR 1392 on pancreatic exocrine secretion in the rat. Digestion 1989, 42, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Mirabella, S.; Gomez-Paz, S.; Lam, E.; Gonzalez-Mosquera, L.; Fogel, J.; Rubinstein, S. Glucose dysregulation and its association with COVID-19 mortality and hospital length of stay. Diabetes Metab. Syndr. 2022, 16, 102439. [Google Scholar] [CrossRef]
- Ismail, S.I.; Naffa, R.G.; Yousef, A.M.; Ghanim, M.T. Incidence of bcr-abl fusion transcripts in healthy individuals. Mol. Med. Rep. 2014, 9, 1271–1276. [Google Scholar] [CrossRef]
- Arbore, D.R.; Galdean, S.M.; Dima, D.; Rus, I.; Kegyes, D.; Ababei, R.G.; Dragancea, D.; Tomai, R.A.; Trifa, A.P.; Tomuleasa, C. COVID-19 Impact on Chronic Myeloid Leukemia Patients. J. Pers. Med. 2022, 12, 1886. [Google Scholar] [CrossRef]
- Waliany, S.; Sainani, K.L.; Park, L.S.; Zhang, C.A.; Srinivas, S.; Witteles, R.M. Increase in Blood Pressure Associated With Tyrosine Kinase Inhibitors Targeting Vascular Endothelial Growth Factor. JACC CardioOncol. 2019, 1, 24–36. [Google Scholar] [CrossRef]
- Meinel, S.; Gekle, M.; Grossmann, C. Mineralocorticoid receptor signaling: Crosstalk with membrane receptors and other modulators. Steroids 2014, 91, 3–10. [Google Scholar] [CrossRef]
- Martinez-Beamonte, R.; Lou-Bonafonte, J.M.; Martinez-Gracia, M.V.; Osada, J. Sphingomyelin in high-density lipoproteins: Structural role and biological function. Int. J. Mol. Sci. 2013, 14, 7716–7741. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, C.; Li, W.; Martinez, D.R.; Drelich, A.; Baek, D.S.; Liu, X.; Mellors, J.W.; Tseng, C.T.; Baric, R.S.; et al. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. MAbs 2020, 12, 1778435. [Google Scholar] [CrossRef]
- Gomez-Lopez, N.; Romero, R.; Escobar, M.F.; Carvajal, J.A.; Echavarria, M.P.; Albornoz, L.L.; Nasner, D.; Miller, D.; Gallo, D.M.; Galaz, J.; et al. Pregnancy-specific responses to COVID-19 revealed by high-throughput proteomics of human plasma. Commun. Med. 2023, 3, 48. [Google Scholar] [CrossRef]
- Cobb, J.A.; Bjergbaek, L.; Gasser, S.M. RecQ helicases: At the heart of genetic stability. FEBS Lett. 2002, 529, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Pitsillou, E.; Liang, J.; Hung, A.; Karagiannis, T.C. The SARS-CoV-2 helicase as a target for antiviral therapy: Identification of potential small molecule inhibitors by in silico modelling. J. Mol. Graph. Model. 2022, 114, 108193. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Nogueira, M.L.; Vogel, J.L.; Kristie, T.M. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc. Natl. Acad. Sci. USA 2010, 107, 2461–2466. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ebata, A.; Dong, Y.; Rizki, G.; Iwata, T.; Lee, S.S. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008, 6, e233. [Google Scholar] [CrossRef]
- Vogel, J.L.; Kristie, T.M. The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection. Viruses 2013, 5, 1272–1291. [Google Scholar] [CrossRef]
- Chen, F.; Zhu, S.; Kang, R.; Tang, D.; Liu, J. ATP6V0D1 promotes alkaliptosis by blocking STAT3-mediated lysosomal pH homeostasis. Cell Rep. 2023, 42, 111911. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Chang, C.C.; Lin, K.H. Thyroid hormone upregulates LAMP2 expression and lysosome activity. Biochem. Biophys. Res. Commun. 2023, 662, 66–75. [Google Scholar] [CrossRef]
- Avari, P.; Eng, P.C.; Hu, M.; Chen, R.; Popovic, N.; Polychronakos, C.; Spalding, D.; Rutter, G.A.; Oliver, N.; Wernig, F. A Novel Somatic Mutation Implicates ATP6V0D1 in Proinsulin Processing. J. Endocr. Soc. 2023, 7, bvac196. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, T.; Ding, Y.; Yu, T.; Cui, Y.; Nie, H. Expression profiles of respiratory V-ATPase and calprotectin in SARS-CoV-2 infection. Cell Death Discov. 2022, 8, 362. [Google Scholar] [CrossRef]
- Stille, G.; Michaelis, W. Imipramine demethylation and norepinephrine storage in brain. Eur. J. Pharmacol. 1970, 10, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Armando, I.; Villar, V.A.; Jose, P.A. Dopamine and renal function and blood pressure regulation. Compr. Physiol. 2011, 1, 1075–1117. [Google Scholar] [CrossRef] [PubMed]
- Nakhaee, H.; Zangiabadian, M.; Bayati, R.; Rahmanian, M.; Ghaffari Jolfayi, A.; Rakhshanderou, S. The effect of antidepressants on the severity of COVID-19 in hospitalized patients: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0267423. [Google Scholar] [CrossRef]
- Shen, J.; Wu, Q.; Liang, T.; Zhang, J.; Bai, J.; Yuan, M.; Shen, P. TRIM40 inhibits IgA1-induced proliferation of glomerular mesangial cells by inactivating NLRP3 inflammasome through ubiquitination. Mol. Immunol. 2021, 140, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhao, C.; Zhao, W. Emerging Roles of MHC Class I Region-Encoded E3 Ubiquitin Ligases in Innate Immunity. Front. Immunol. 2021, 12, 687102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ling, J.; Barcia, G.; Jing, L.; Wu, J.; Barry, B.J.; Mochida, G.H.; Hill, R.S.; Weimer, J.M.; Stein, Q.; et al. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am. J. Hum. Genet. 2014, 94, 547–558. [Google Scholar] [CrossRef]
- Thiele, F.; Cohrs, C.M.; Flor, A.; Lisse, T.S.; Przemeck, G.K.; Horsch, M.; Schrewe, A.; Gailus-Durner, V.; Ivandic, B.; Katus, H.A.; et al. Cardiopulmonary dysfunction in the Osteogenesis imperfecta mouse model Aga2 and human patients are caused by bone-independent mechanisms. Hum. Mol. Genet. 2012, 21, 3535–3545. [Google Scholar] [CrossRef]
- Alshukairi, A.N.; Doar, H.; Al-Sagheir, A.; Bahasan, M.A.; Sultan, A.A.; Al Hroub, M.K.; Itani, D.; Khalid, I.; Saeedi, M.F.; Bakhamis, S.; et al. Outcome of COVID19 in Patients with Osteogenesis Imperfecta: A Retrospective Multicenter Study in Saudi Arabia. Front. Endocrinol. 2021, 12, 800376. [Google Scholar] [CrossRef]
- Ringden, O.; Moll, G.; Gustafsson, B.; Sadeghi, B. Mesenchymal Stromal Cells for Enhancing Hematopoietic Engraftment and Treatment of Graft-Versus-Host Disease, Hemorrhages and Acute Respiratory Distress Syndrome. Front. Immunol. 2022, 13, 839844. [Google Scholar] [CrossRef]
- Szlezak, D.; Bronowicka-Adamska, P.; Hutsch, T.; Ufnal, M.; Wrobel, M. Hypertension and Aging Affect Liver Sulfur Metabolism in Rats. Cells 2021, 10, 1238. [Google Scholar] [CrossRef]
- Luo, L.; Liu, D.; Tang, C.; Du, J.; Liu, A.D.; Holmberg, L.; Jin, H. Sulfur dioxide upregulates the inhibited endogenous hydrogen sulfide pathway in rats with pulmonary hypertension induced by high pulmonary blood flow. Biochem. Biophys. Res. Commun. 2013, 433, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Carballo, I.; Bakola, M.; Stuckler, D. The impact of air pollution on COVID-19 incidence, severity, and mortality: A systematic review of studies in Europe and North America. Environ. Res. 2022, 215, 114155. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.; Tian, R.; Zarrabi, I.; Hill, G.; Williams, E.; Sanchez-Duffhues, G.; Thorikay, M.; Ramachandran, P.; Siddaway, R.; Wong, J.F.; et al. Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas. Cancer Cell 2020, 37, 308–323.e12. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-John, T. VPS54 and the wobbler mouse. Front. Neurosci. 2015, 9, 381. [Google Scholar] [CrossRef] [PubMed]
- Realegeno, S.; Puschnik, A.S.; Kumar, A.; Goldsmith, C.; Burgado, J.; Sambhara, S.; Olson, V.A.; Carroll, D.; Damon, I.; Hirata, T.; et al. Monkeypox Virus Host Factor Screen Using Haploid Cells Identifies Essential Role of GARP Complex in Extracellular Virus Formation. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Meng, L.B.; Shan, M.J.; Qiu, Y.; Qi, R.; Yu, Z.M.; Guo, P.; Di, C.Y.; Gong, T. TPM2 as a potential predictive biomarker for atherosclerosis. Aging 2019, 11, 6960–6982. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, J.; Li, J.; Zhao, W. Identification and Validation of Immune Markers in Coronary Heart Disease. Comput. Math. Methods Med. 2022, 2022, 2877679. [Google Scholar] [CrossRef]
- Maiese, A.; Manetti, A.C.; La Russa, R.; Di Paolo, M.; Turillazzi, E.; Frati, P.; Fineschi, V. Autopsy findings in COVID-19-related deaths: A literature review. Forensic Sci. Med. Pathol. 2021, 17, 279–296. [Google Scholar] [CrossRef]
- Moll, T.; Odon, V.; Harvey, C.; Collins, M.O.; Peden, A.; Franklin, J.; Graves, E.; Marshall, J.N.G.; Souza, C.D.S.; Zhang, S.; et al. Low expression of EXOSC2 protects against clinical COVID-19 and impedes SARS-CoV-2 replication. bioRxiv 2022. [Google Scholar] [CrossRef]
- Calame, D.G.; Herman, I.; Fatih, J.M.; Du, H.; Akay, G.; Jhangiani, S.N.; Coban-Akdemir, Z.; Milewicz, D.M.; Gibbs, R.A.; Posey, J.E.; et al. Risk of sudden cardiac death in EXOSC5-related disease. Am. J. Med. Genet. A 2021, 185, 2532–2540. [Google Scholar] [CrossRef]
- Sato, T.; Uchiumi, T.; Arakawa, M.; Kominami, R. Serological association of lupus autoantibodies to a limited functional domain of 28S ribosomal RNA and to the ribosomal proteins bound to the domain. Clin. Exp. Immunol. 1994, 98, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, L.; Dontaraju, V.S.; Troyer, J.; Sahota, J. New onset systemic lupus erythematosus after COVID-19 infection: A case report. AME Case Rep. 2022, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.L.; Qian, Y.; Jin, X.H.; Yu, H.R.; Du, L.; Wu, H.; Chen, H.L.; Shi, Y.Q. COVID-19 in patients with systemic lupus erythematosus: A systematic review. Lupus 2022, 31, 684–696. [Google Scholar] [CrossRef]
- Raza, H.A.; Tariq, J.; Agarwal, V.; Gupta, L. COVID-19, hydroxychloroquine and sudden cardiac death: Implications for clinical practice in patients with rheumatic diseases. Rheumatol. Int. 2021, 41, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Rajput, A.; Thakur, A.; Mukhopadhyay, A.; Kamboj, S.; Rastogi, A.; Gautam, S.; Jassal, H.; Kumar, M. Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning. Comput. Struct. Biotechnol. J. 2021, 19, 3133–3148. [Google Scholar] [CrossRef] [PubMed]
- Yano, Y.; Nakazato, M.; Toshinai, K.; Inokuchi, T.; Matsuda, S.; Hidaka, T.; Hayakawa, M.; Kangawa, K.; Shimada, K.; Kario, K. Circulating des-acyl ghrelin improves cardiovascular risk prediction in older hypertensive patients. Am. J. Hypertens. 2014, 27, 727–733. [Google Scholar] [CrossRef]
- Forman, L.J.; Estilow, S.; Hock, C.E. Localization of beta-endorphin in the rat heart and modulation by testosterone. Proc. Soc. Exp. Biol. Med. 1989, 190, 240–245. [Google Scholar] [CrossRef]
- Genazzani, A.R.; Petraglia, F.; Facchinetti, F.; Golinelli, S.; Oltramari, P.; Santoro, V.; Volpe, A. Evidences for a dopamine-regulated peripheral source of circulating beta-endorphin. J. Clin. Endocrinol. Metab. 1988, 66, 279–282. [Google Scholar] [CrossRef]
- Vitolo, E.; Castini, D.; Colombo, A.; Collini, P.; Gueli Alletti, D.; Bianchi, M.; Panerai, A.E. Plasma beta-endorphin and beta-lipotropin in congestive heart failure in man. Acta Cardiol. 1990, 45, 65–74. [Google Scholar]
- Collins, A.B.; Zhao, L.; Zhu, Z.; Givens, N.T.; Bai, Q.; Wakefield, M.R.; Fang, Y. Impact of COVID-19 on Male Fertility. Urology 2022, 164, 33–39. [Google Scholar] [CrossRef]
- Luay Kamil, A.; Al-Kawaz, U.M.; Al-Essawe, E.M. Gonadotropin and Sex Steroid Hormones in Males with Post COVID-19 Infection. Wiad Lek. 2022, 75, 2222–2225. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.A.; Tobler, P.H.; Henke, H.; Tschopp, F.A. Salmon and human calcitonin-like peptides coexist in the human thyroid and brain. J. Clin. Endocrinol. Metab. 1983, 57, 1314–1316. [Google Scholar] [CrossRef] [PubMed]
- Wells, G.; Chernoff, J.; Gilligan, J.P.; Krause, D.S. Does salmon calcitonin cause cancer? A review and meta-analysis. Osteoporos. Int. 2016, 27, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Najafi, R.; Mamizadeh, N.; Hosseini, S.H.; Roushenas, S.; Bazhdan, L. A challenging case of COVID-19: A COVID-19 positive adolescent presented with severe diabetic ketoacidosis, resistant hypertension. BMC Endocr. Disord. 2022, 22, 90. [Google Scholar] [CrossRef]
- Sun, X.; Tang, S.; Hou, B.; Duan, Z.; Liu, Z.; Li, Y.; He, S.; Wang, Q.; Chang, Q. Overexpression of P-glycoprotein, MRP2, and CYP3A4 impairs intestinal absorption of octreotide in rats with portal hypertension. BMC Gastroenterol. 2021, 21, 2. [Google Scholar] [CrossRef]
- Luty, J.; Hayward, L.; Jackson, M.; Duell, P.B. Severe respiratory failure in a patient with COVID-19 and acromegaly: Rapid improvement after adding octreotide. BMJ Case Rep. 2021, 14, e243900. [Google Scholar] [CrossRef]
- Mittal, L.; Kumari, A.; Srivastava, M.; Singh, M.; Asthana, S. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J. Biomol. Struct. Dyn. 2021, 39, 3662–3680. [Google Scholar] [CrossRef]
- Mercuri, V.; D’Amico, T.; Gargiulo, P. Letter to the Editor: “COVID-19 and the endocrine system: Exploring the unexplored”. Focus on acromegaly. J. Endocrinol. Investig. 2021, 44, 637–638. [Google Scholar] [CrossRef]
- Baykan, E.K.; Baykan, A.R.; Utlu, M.; Deve, E.; Yildiz, F.; Birdal, C.; Ozdemir, Y.; Aslan, M.H.; Altinkaynak, K. Growth hormone level in COVID-19 patients. North. Clin. Istanb. 2022, 9, 470–475. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhao, Z.; Chen, X.; Chu, Z.; He, Y.; Tan, Y.; Zhou, J.; Tang, C. Effects of growth hormone/estrogen/androgen on COVID-19-type proinflammatory responses in normal human lung epithelial BEAS-2B cells. BMC Mol. Cell. Biol. 2022, 23, 42. [Google Scholar] [CrossRef]
- Palmeiro, C.R.; Anand, R.; Dardi, I.K.; Balasubramaniyam, N.; Schwarcz, M.D.; Weiss, I.A. Growth hormone and the cardiovascular system. Cardiol. Rev. 2012, 20, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Itoh, H. Hypertension as a Metabolic Disorder and the Novel Role of the Gut. Curr. Hypertens. Rep. 2019, 21, 63. [Google Scholar] [CrossRef] [PubMed]
- Achilonu, I.; Iwuchukwu, E.A.; Achilonu, O.J.; Fernandes, M.A.; Sayed, Y. Targeting the SARS-CoV-2 main protease using FDA-approved Isavuconazonium, a P2-P3 alpha-ketoamide derivative and Pentagastrin: An in-silico drug discovery approach. J. Mol. Graph. Model. 2020, 101, 107730. [Google Scholar] [CrossRef] [PubMed]
- Petronilho, F.; Danielski, L.G.; Roesler, R.; Schwartsmann, G.; Dal-Pizzol, F. Gastrin-releasing peptide as a molecular target for inflammatory diseases: An update. Inflamm. Allergy Drug Targets 2013, 12, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Clausen, C.L.; Rasmussen, A.K.; Johannsen, T.H.; Hilsted, L.M.; Skakkebaek, N.E.; Szecsi, P.B.; Pedersen, L.; Benfield, T.; Juul, A. Thyroid function in COVID-19 and the association with cytokine levels and mortality. Endocr. Connect. 2021, 10, 1234–1242. [Google Scholar] [CrossRef]
- Kilicoglu, H.; Shin, D.; Fiszman, M.; Rosemblat, G.; Rindflesch, T.C. SemMedDB: A PubMed-scale repository of biomedical semantic predications. Bioinformatics 2012, 28, 3158–3160. [Google Scholar] [CrossRef]
| ID | Analysis Deliverable | Target Node(s) with CUI |
|---|---|---|
| A6 | Source nodes related to COVID-19 | COVID-19 (C5203670) |
| A5 | Source nodes related to resistant hypertension | Resistant hypertension (C0745130) |
| A4 | Intersection of high-ranking A5 and A6 source nodes to determine hub nodes for cross-domain analysis in A1, A2, A3 | COVID-19 (C5203670); resistant hypertension (C0745130) |
| A3 | Relationship of AT1 receptor (a RAAS hub node) with COVID-19 | AT1 receptor (C0529330); COVID-19 (C5203670) |
| A2 | Relationships of epithelial sodium channel (a RAAS hub node) and COVID-19 | Epithelial sodium channel (C0384156); COVID-19 (C5203670) |
| A1 | Relationships of mineralocorticoid receptor (a RAAS hub node) with COVID-19 | Mineralocorticoid receptor (C0066563); COVID-19 (C5203670) |
| ID | Target Node(s) | Source Node Count | Metapath Count | Average HeteSim | Minimum HeteSim | Maximum HeteSim |
|---|---|---|---|---|---|---|
| A6 | COVID-19 | 75,731 | 41,932 | 0.2198 | 0.0001 | 1.0000 |
| A5 | Resistant hypertension | 24,699 | 2883 | 0.3493 | 0.0003 | 1.0000 |
| A3 | AT1 receptor, COVID-19 | 67,779 | 69,722 | 0.2264 | 0.0002 | 1.0000 |
| A2 | Epithelial sodium channel, COVID-19 | 61,104 | 55,338 | 0.2091 | 0.0005 | 0.8292 |
| A1 | Mineralocorticoid receptor, COVID-19 | 70,038 | 77,486 | 0.2263 | 0.0002 | 1.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kartchner, D.; McCoy, K.; Dubey, J.; Zhang, D.; Zheng, K.; Umrani, R.; Kim, J.J.; Mitchell, C.S. Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19. Biology 2023, 12, 1269. https://doi.org/10.3390/biology12091269
Kartchner D, McCoy K, Dubey J, Zhang D, Zheng K, Umrani R, Kim JJ, Mitchell CS. Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19. Biology. 2023; 12(9):1269. https://doi.org/10.3390/biology12091269
Chicago/Turabian StyleKartchner, David, Kevin McCoy, Janhvi Dubey, Dongyu Zhang, Kevin Zheng, Rushda Umrani, James J. Kim, and Cassie S. Mitchell. 2023. "Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19" Biology 12, no. 9: 1269. https://doi.org/10.3390/biology12091269
APA StyleKartchner, D., McCoy, K., Dubey, J., Zhang, D., Zheng, K., Umrani, R., Kim, J. J., & Mitchell, C. S. (2023). Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19. Biology, 12(9), 1269. https://doi.org/10.3390/biology12091269





