Deletion of the Neuronal Transcription Factor Satb1 Induced Disturbance of the Kinome and Mechanisms of Hypoxic Preconditioning
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture Preparation
2.3. Fluorescent Ca2+ Measurements
2.4. The Technique for Short-Term Hypoxia Episode Generation
2.5. Extraction of RNA and Real-Time Polymerase Chain Reaction (RT-qPCR)
2.6. Immunocytochemistry
2.7. Statistical Analysis
3. Results
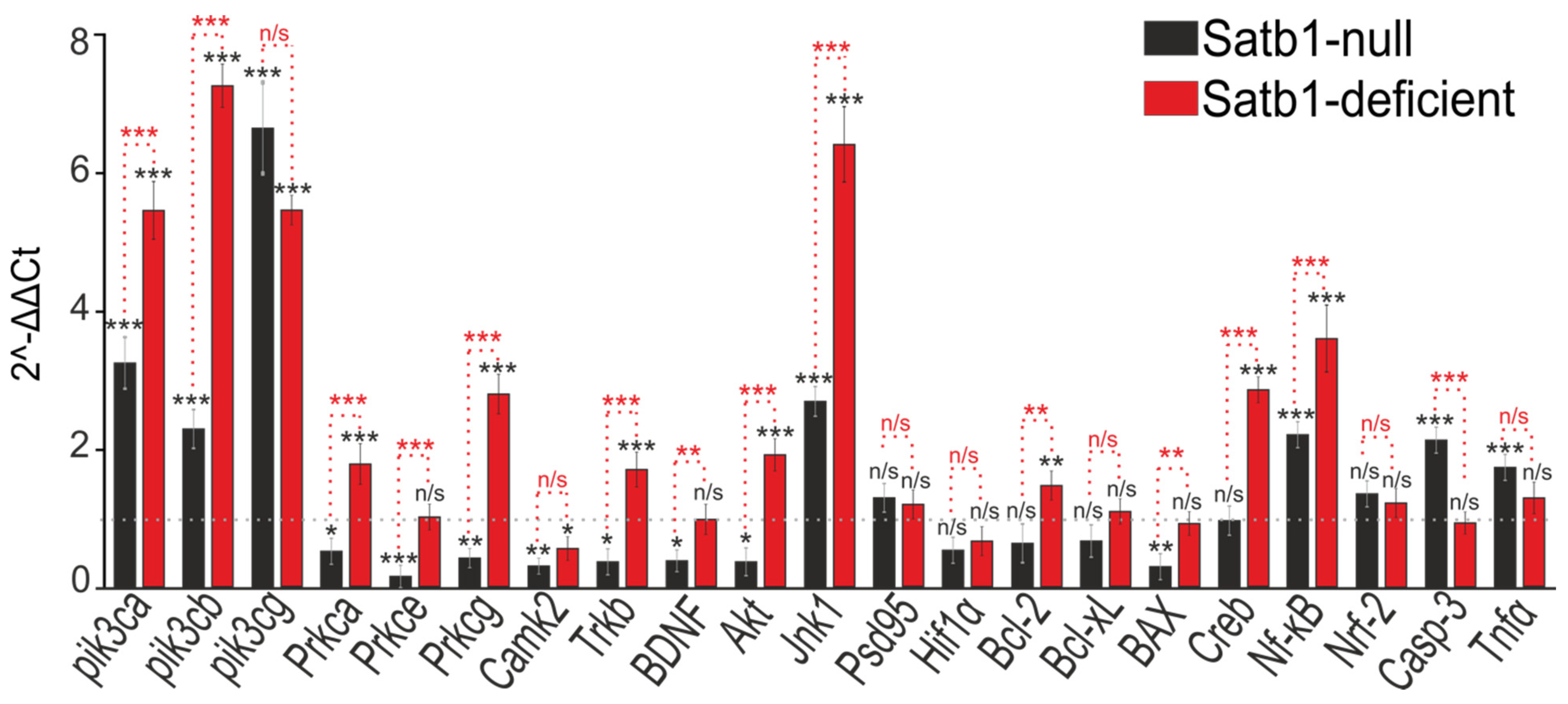
3.1. Deletion of the Transcription Factor Satb1 in Cortical Neurons Leads to Impaired Expression of Key Protein Kinases and Genes Regulating Cell Viability
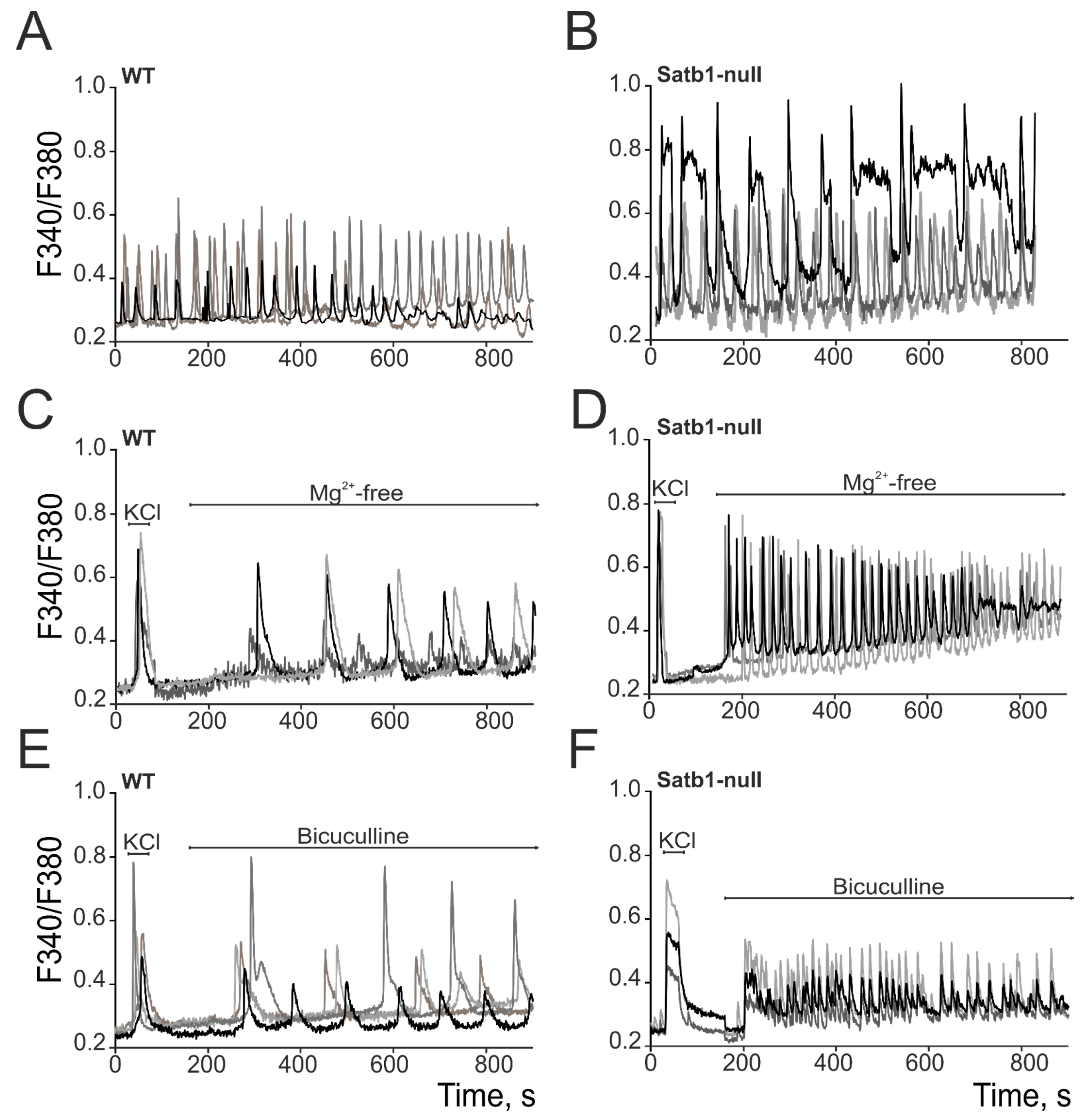
3.2. Deletion of the Transcription Factor Satb1 Correlates with Hyperexcitation of Cortical Neurons
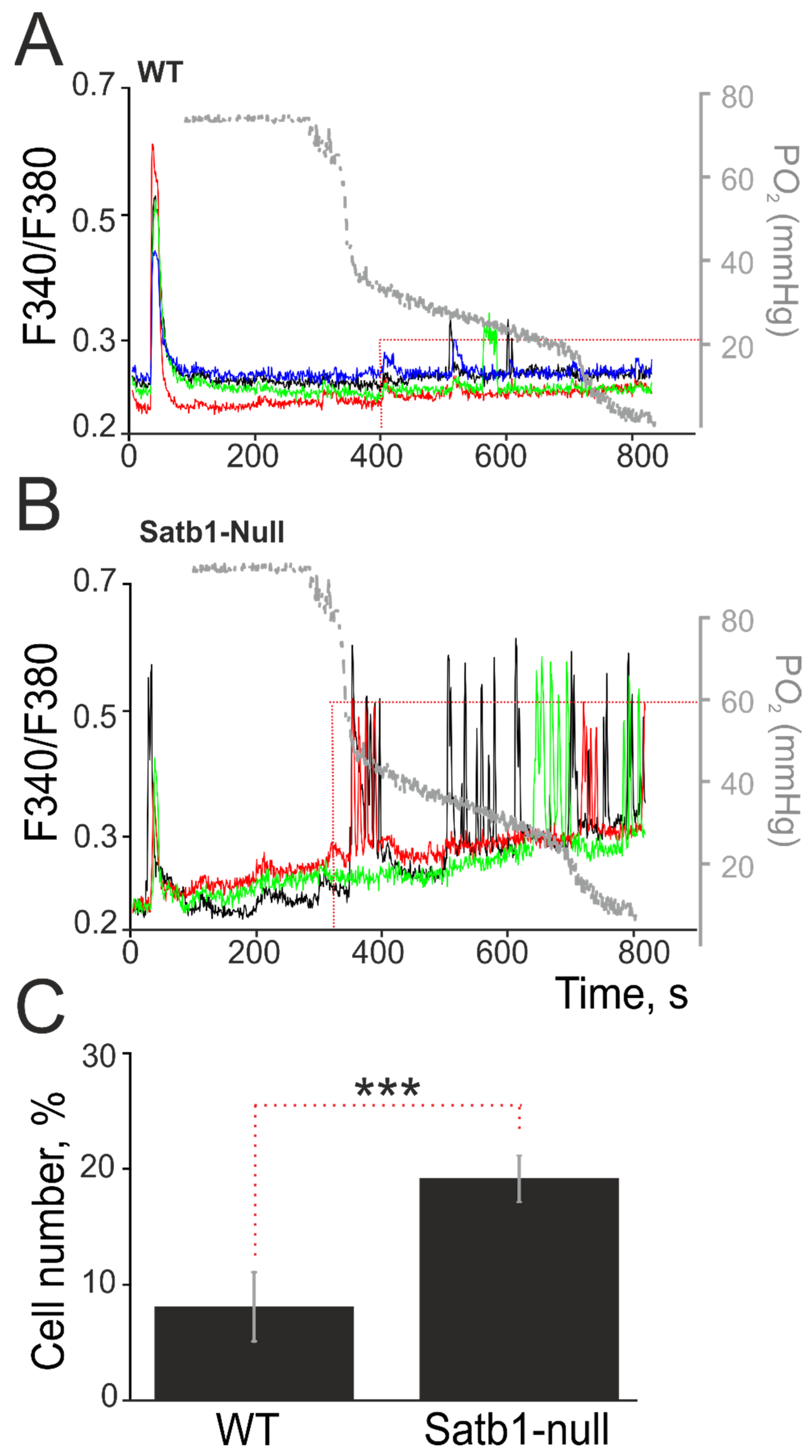
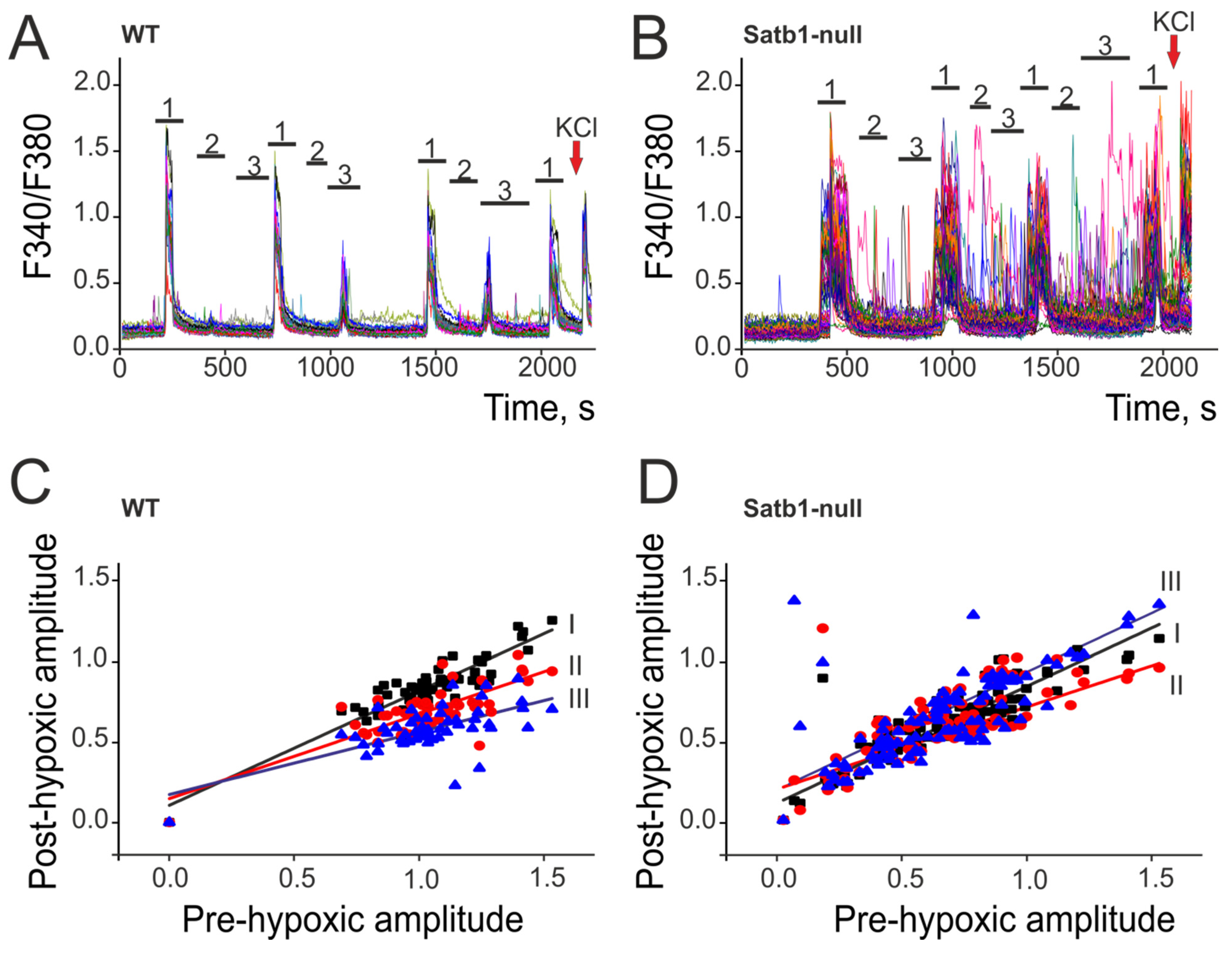
3.3. Deletion of the Transcription Factor Satb1 Affects the Sensitivity of Cortical Neurons to Hypoxia and the Induction of Hypoxic Preconditioning
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, R.S.; Walsh, C.A. Molecular insights into human brain evolution. Nature 2005, 437, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, R.; Dobyns, W.B. Malformations of cortical development: Clinical features and genetic causes. Lancet Neurol. 2014, 13, 710–726. [Google Scholar] [CrossRef] [PubMed]
- Lodato, S.; Shetty, A.S.; Arlotta, P. Cerebral cortex assembly: Generating and reprogramming projection neuron diversity. Trends Neurosci. 2015, 38, 117–125. [Google Scholar] [CrossRef]
- Denaxa, M.; Kalaitzidou, M.; Garefalaki, A.; Achimastou, A.; Lasrado, R.; Maes, T.; Pachnis, V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2012, 2, 1351–1362. [Google Scholar] [CrossRef]
- Fogarty, M.; Grist, M.; Gelman, D.; Marín, O.; Pachnis, V.; Kessaris, N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007, 27, 10935–10946. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.; Pradhan, S.J.; Galande, S. Chromatin organizer SATB1 as a novel molecular target for cancer therapy. Curr. Drug Targets 2012, 13, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- Turovsky, E.A.; Turovskaya, M.V.; Fedotova, E.I.; Babaev, A.A.; Tarabykin, V.S.; Varlamova, E.G. Role of Satb1 and Satb2 Transcription Factors in the Glutamate Receptors Expression and Ca2+ Signaling in the Cortical Neurons In Vitro. Int. J. Mol. Sci. 2021, 22, 5968. [Google Scholar] [CrossRef]
- Alvarez, J.D.; Yasui, D.H.; Niida, H.; Joh, T.; Loh, D.Y.; mKohwi-Shigematsu, T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000, 14, 521–535. [Google Scholar] [CrossRef]
- Balamotis, M.A.; Tamberg, N.; Woo, Y.J.; Li, J.; Davy, B.; Kohwi-Shigematsu, T.; Kohwi, Y. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol. Cell. Biol. 2012, 32, 333–347. [Google Scholar] [CrossRef]
- Close, J.; Xu, H.; De Marco García, N.; Batista-Brito, R.; Rossignol, E.; Rudy, B.; Fishell, G. Satb1 is an activity-modulated transcription factor required for the terminal differentiation and connectivity of medial ganglionic eminence-derived cortical interneurons. J. Neurosci. 2012, 32, 17690–17705. [Google Scholar] [CrossRef]
- Goebbels, S.; Bormuth, I.; Bode, U.; Hermanson, O.; Schwab, M.H.; Nave, K.A. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 2006, 44, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Turovsky, E.A.; Varlamova, E.G.; Gudkov, S.V.; Plotnikov, E.Y. The Protective Mechanism of Deuterated Linoleic Acid Involves the Activation of the Ca2+ Signaling System of Astrocytes in Ischemia In Vitro. Int. J. Mol. Sci. 2021, 22, 13216. [Google Scholar] [CrossRef]
- Turovskaya, M.V.; Turovsky, E.A.; Zinchenko, V.P.; Levin, S.G.; Shamsutdinova, A.A.; Godukhin, O.V. Repeated brief episodes of hypoxia modulate the calcium responses of ionotropic glutamate receptors in hippocampal neurons. Neurosci. Lett. 2011, 496, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Tekin, D.; Gursoy, E.; Salloum, F.; Levasseur, J.E.; Kukreja, R.C. Evidence that NOS2 acts as a trigger and mediator of late preconditioning induced by acute systemic hypoxia. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, 5–12. [Google Scholar] [CrossRef]
- Turovsky, E.A.; Turovskaya, M.V.; Kononov, A.V.; Zinchenko, V.P. Short-term episodes of hypoxia induce posthypoxic hyperexcitability and selective death of GABAergic hippocampal neurons. Exp. Neurol. 2013, 250, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moody, W.J.; Bosma, M.M. Ion channel development, spontaneous activity, and activity-dependent development in nerve and muscle cells. Physiol. Rev. 2005, 85, 883–941. [Google Scholar] [CrossRef]
- Jiang, M.; Swann, J.W. A role for L-type calcium channels in the maturation of parvalbumin-containing hippocampal interneurons. Neuroscience 2005, 135, 839–850. [Google Scholar] [CrossRef]
- Batista-Brito, R.; Rossignol, E.; Hjerling-Leffler, J.; Denaxa, M.; Wegner, M.; Lefebvre, V.; Pachnis, V.; Fishell, G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron 2009, 63, 466–481. [Google Scholar] [CrossRef]
- Vasilopoulos, N.; Kaplanian, A.; Vinos, M.; Katsaiti, Y.; Christodoulou, O.; Denaxa, M.; Skaliora, I. The role of selective SATB1 deletion in somatostatin expressing interneurons on endogenous network activity and the transition to epilepsy. J. Neurosci. Res. 2023, 101, 424–447. [Google Scholar] [CrossRef]
- Turovskaya, M.V.; Gaidin, S.G.; Vedunova, M.V.; Babaev, A.A.; Turovsky, E.A. BDNF Overexpression Enhances the Preconditioning Effect of Brief Episodes of Hypoxia, Promoting Survival of GABAergic Neurons. Neurosci. Bull. 2020, 36, 733–760. [Google Scholar] [CrossRef]
- Lukyanova, L.D.; Germanova, E.L.; Kopaladze, R.A. Development of resistance of an organism under various conditions of hypoxic preconditioning: Role of the hypoxic period and reoxygenation. Bull. Exp. Biol. Med. 2009, 147, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Slevin, M.; Krupinski, J.; Kumar, P.; Gaffney, J.; Kumar, S.J. Gene activation and protein expression following ischaemic stroke: Strategies towards neuroprotection. Cell Mol Med. 2005, 9, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Shirai, Y. Protein kinase C gamma (PKC gamma): Function of neuron specific isotype. J. Biochem. 2002, 132, 683–687. [Google Scholar] [CrossRef]
- Oehrlein, S.A.; Maelicke, A.; Herget, T. Expression of protein kinase C gene family members is temporally and spatially regulated during neural development in vitro. Eur. J. Cell. Biol. 1998, 77, 323–337. [Google Scholar] [CrossRef]
- Cao, J.; Semenova, M.M.; Solovyan, V.T.; Han, J.; Coffey, E.T.; Courtney, M.J. Distinct requirements for p38alpha and c-Jun N-terminal kinase stress-activated protein kinases in different forms of apoptotic neuronal death. J. Biol. Chem. 2004, 279, 35903–35913. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Kuan, C.Y.; Whitmarsh, A.J.; Rincon, M.; Zheng, T.S.; Davis, R.J.; Rakic, P.; Flavell, R.A. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 1997, 389, 865–870. [Google Scholar] [CrossRef]
- Whitmarsh, A.J.; Kuan, C.Y.; Kennedy, N.J.; Kelkar, N.; Haydar, T.F.; Mordes, J.P.; Appel, M.; Rossini, A.A.; Jones, S.N.; Flavell, R.A.; et al. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev. 2001, 15, 2421–2432. [Google Scholar] [CrossRef]
- Fojtík, P.; Beckerová, D.; Holomková, K.; Šenfluk, M.; Rotrekl, V. Both Hypoxia-Inducible Factor 1 and MAPK Signaling Pathway Attenuate PI3K/AKT via Suppression of Reactive Oxygen Species in Human Pluripotent Stem Cells. Front. Cell Dev. Biol. 2021, 8, 607444. [Google Scholar] [CrossRef]
- Della-Morte, D.; Raval, A.P.; Dave, K.R.; Lin, H.W.; Perez-Pinzon, M.A. Post-ischemic activation of protein kinase C epsilon protects the hippocampus from cerebral ischemic injury via alterations in cerebral blood flow. Neurosci. Lett. 2011, 487, 158–162. [Google Scholar] [CrossRef]
- Yu, H.; Yang, Z.; Pan, S.; Yang, Y.; Tian, J.; Wang, L.; Sun, W. Hypoxic preconditioning promotes the translocation of protein kinase C ε binding with caveolin-3 at cell membrane not mitochondrial in rat heart. Cell Cycle 2015, 14, 3557–3565. [Google Scholar] [CrossRef][Green Version]
- Feng, Y.; Rhodes, P.G.; Bhatt, A.J. Hypoxic preconditioning provides neuroprotection and increases vascular endothelial growth factor A, preserves the phosphorylation of Akt-Ser-473 and diminishes the increase in caspase-3 activity in neonatal rat hypoxic-ischemic model. Brain Res. 2010, 1325, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Wang, T.; Li, W.; Xu, Z.C.; Sun, W.; Xu, E. Activation of Akt/FoxO signaling pathway contributes to induction of neuroprotection against transient global cerebral ischemia by hypoxic pre-conditioning in adult rats. J. Neurochem. 2010, 114, 897–908. [Google Scholar] [CrossRef]
- Wu, Q.J.; Tymianski, M. Targeting NMDA receptors in stroke: New hope in neuroprotection. Mol. Brain 2018, 11, 15. [Google Scholar] [CrossRef]
- Gross, C.; Bassell, G.J. Neuron-specific regulation of class I PI3K catalytic subunits and their dysfunction in brain disorders. Front. Mol. Neurosci. 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Shim, D.; Kang, S.S.; Chang, S.I.; Kim, H.Y. Protein kinase B inhibits endostatin-induced apoptosis in HUVECs. J. Biochem. Mol. Biol. 2006, 39, 97–104. [Google Scholar] [CrossRef]
- Yeo, E.J. Hypoxia and aging. Exp. Mol. Med. 2019, 51, 67. [Google Scholar] [CrossRef]
- Bopassa, J.C.; Ferrera, R.; Gateau-Roesch, O.; Couture-Lepetit, E.; Ovize, M. PI3-kinase regulates the mitochondrial transition pore in controlled reperfusion and postconditioning. Cardiovasc. Res. 2006, 69, 178–185. [Google Scholar] [CrossRef]
- Rellos, P.; Pike, A.C.; Niesen, F.H.; Salah, E.; Lee, W.H.; von Delft, F.; Knapp, S. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010, 8, 1000426. [Google Scholar] [CrossRef]
- Giese, K.P.; Fedorov, N.B.; Filipkowski, R.K.; Silva, A.J. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science 1998, 279, 870–873. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Takahashi, E.; Li, W.; Halt, A.; Wiltgen, B.; Ehninger, D.; Li, G.D.; Hell, J.W.; Kennedy, M.B.; Silva, A.J. Interactions between the NR2B receptor and CaMKII modulate synaptic plasticity and spatial learning. J. Neurosci. 2007, 27, 13843–13853. [Google Scholar] [CrossRef]
- Turovskaya, M.V.; Epifanova, E.A.; Tarabykin, V.S.; Babaev, A.A.; Turovsky, E.A. Interleukin-10 restores glutamate receptor-mediated Ca2+-signaling in brain circuits under loss of Sip1 transcription factor. Int. J. Neurosci. 2022, 132, 114–125. [Google Scholar] [CrossRef]
- Fink, C.C.; Meyer, T. Molecular mechanisms of CaMKII activation in neuronal plasticity. Curr. Opin. Neurobiol. 2002, 12, 293–299. [Google Scholar] [CrossRef]
- Johns, L.; Sinclair, A.J.; Davies, J.A. Hypoxia/hypoglycemia-induced amino acid release is decreased in vitro by preconditioning. Biochem. Biophys. Res. Commun. 2000, 276, 134–136. [Google Scholar] [CrossRef]
- Semenov, D.G.; Samoilov, M.O.; Lazarewicz, J.W. Calcium transients in the model of rapidly induced anoxic tolerance in rat cortical slices: Involvement of NMDA receptors. Neurosignals 2002, 11, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Turovskaya, M.V.; Zinchenko, V.P.; Babaev, A.A.; Epifanova, E.A.; Tarabykin, V.S.; Turovsky, E.A. Mutation in the Sip1 transcription factor leads to a disturbance of the preconditioning of AMPA receptors by episodes of hypoxia in neurons of the cerebral cortex due to changes in their activity and subunit composition. The protective effects of interleukin-10. Arch. Biochem. Biophys. 2018, 654, 126–135. [Google Scholar]
- Mitroshina, E.V.; Mishchenko, T.A.; Shishkina, T.V.; Vedunova, M.V. Role of Neurotrophic Factors BDNF and GDNF in Nervous System Adaptation to the Influence of Ischemic Factors. Bull. Exp. Biol. Med. 2019, 167, 574–579. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turovsky, E.A.; Tarabykin, V.S.; Varlamova, E.G. Deletion of the Neuronal Transcription Factor Satb1 Induced Disturbance of the Kinome and Mechanisms of Hypoxic Preconditioning. Biology 2023, 12, 1207. https://doi.org/10.3390/biology12091207
Turovsky EA, Tarabykin VS, Varlamova EG. Deletion of the Neuronal Transcription Factor Satb1 Induced Disturbance of the Kinome and Mechanisms of Hypoxic Preconditioning. Biology. 2023; 12(9):1207. https://doi.org/10.3390/biology12091207
Chicago/Turabian StyleTurovsky, Egor A., Viktor S. Tarabykin, and Elena G. Varlamova. 2023. "Deletion of the Neuronal Transcription Factor Satb1 Induced Disturbance of the Kinome and Mechanisms of Hypoxic Preconditioning" Biology 12, no. 9: 1207. https://doi.org/10.3390/biology12091207
APA StyleTurovsky, E. A., Tarabykin, V. S., & Varlamova, E. G. (2023). Deletion of the Neuronal Transcription Factor Satb1 Induced Disturbance of the Kinome and Mechanisms of Hypoxic Preconditioning. Biology, 12(9), 1207. https://doi.org/10.3390/biology12091207





