Simple Summary
Wounded skin can naturally be repaired by a mechanism called wound healing. Human skin is a habitat of various pathogenic and commensal bacteria. While these bacteria are in balance in healthy skin, they can lose the balance by wounding, which leads to delay in the wound-healing process. Moreover, commensal and pathogenic bacteria inhabit skin tissue and have constant communication with the immune system, which can increase and decrease the healing efficiency, respectively. This indicates that cutaneous bacteria have important effects on wound healing. Herein, we discuss some important bacteria (coagulase-negative Staphylococci (CoNS), S. aureus, P. aeruginosa, and Lactobacilli) present in human skin, the effects of communication of bacteria with the immune system and epithelial cells on wound healing, and the identification techniques and manipulation strategies of the bacterial population in wounded skin tissue.
Abstract
Cutaneous wound healing is a natural and complex repair process that is implicated within four stages. However, microorganisms (e.g., bacteria) can easily penetrate through the skin tissue from the wound bed, which may lead to disbalance in the skin microbiota. Although commensal and pathogenic bacteria are in equilibrium in normal skin, their imbalance in the wound area can cause the delay or impairment of cutaneous wounds. Moreover, skin microbiota is in constant crosstalk with the immune system and epithelial cells, which has significance for the healing of a wound. Therefore, understanding the major bacteria species in the cutaneous wound as well as their communication with the immune system has gained prominence in a way that allows for the emergence of a new perspective for wound healing. In this review, the major bacteria isolated from skin wounds, the role of the crosstalk between the cutaneous microbiome and immune system to heal wounds, the identification techniques of these bacteria populations, and the applied therapies to manipulate the skin microbiota are investigated.
1. Introduction
Cutaneous wound healing is a complicated and well-organized natural repair process that comprises four stages: hemostasis, inflammation, proliferation, and remodeling [1]. Skin damage, inducing a wound, allows organisms from foreign bodies to penetrate through the wound site [2,3]. In other words, a wound procures an occasion for both commensal and pathogenic microorganisms to access underlying tissue, then grow and colonize after reaching ideal conditions [4], which may cause further impairment in wound healing.
The complex and rich ecosystem of skin microbiota (microbiome) arises from diverse microorganisms, i.e., bacteria, fungi, viruses, and yeasts [5], and has a significant role in the protection of skin tissue and ensuring hemostasis [6,7]. The bacteria inhabiting the cutaneous microbiota can be classified as commensal and pathogenic. Pathogenic bacteria are a harmful bacteria type that can directly be transmitted to the host tissue and lead to infection. In contrast, commensal bacteria can supply essential nutrients to the host tissue and benefit in fighting infection. Although bacteria are the most abundant microorganisms in skin microbiota, only about 25% of them can move through the deeper skin layers [8], being important players in skin physiology and disease processes [9]. Commensals present in skin microbiota have been determined as beneficial with their ability to originate immune response thanks to their communication with cutaneous cells such as keratinocytes and fibroblasts [10,11]. These types of bacteria have advantageous effects on wound healing by providing a barrier function for the skin and combating pathogenic microorganisms [12]. In contrast, pathogenic bacteria may give rise to delayed or impaired wound healing [13,14] by leading to infection in the wound site.
The crosstalk between the cutaneous microbiome, immune system, and epithelial cells is evaluated significantly for tissue repair and regeneration in vertebrates [15]. The harmony among all these provides an efficacious approach to wound healing and an invasive system by possible pathogens in equilibrium [16]. However, in the case of the disequilibrium between commensals and pathogens, cutaneous diseases may appear. In this perspective, understanding of the communication between the microbiome and the immune system as well as the identification and manipulation of the microbiome by several approaches has become prominent as an alternative solution for cutaneous wound treatment. In this review, we first discuss the abundant pathogenic and commensal bacteria that inhabit a wound. Moreover, we examine the effects of the crosstalk between skin microbiota and the immune system, as well as the identification and manipulation of skin microbiota, to come up with a different perspective for cutaneous wound healing mechanisms.
2. The Abundant Bacteria Implicated in Wound
Both aerobic and anaerobic microorganisms that constitute skin microbiota inhabit the skin surface soon after birth in a dynamic correlation with the host [17]. Although the bacteria protect the skin balance, they may penetrate through the underlying skin tissues when their continuity is broken by intrinsic and/or extrinsic factors. Thereafter, these penetrated bacteria can lead to the formation of colonization or contamination. Contamination is the presence of potentially pathogenic microorganisms in the wound area, whilst colonization is the existence of replicating microorganisms with no damage to the wound. However, critical colonization is the threshold that may delay the healing of the wound due to the high number of bacterial counts. Local infection with critical colonization, and proliferation of microorganisms, as well as local tissue reactions, can cause generalized host reactions, thereby an invasive infection [18]. Therefore, understanding the species and effects of the bacteria in skin tissue is important to develop solutions for infected wounds.
Even though many studies have revealed the positive effect of the skin microbiota in wound healing either by modulating immune response or preventing pathogen invasion, the precise relationship between commensal microbiota and impaired wound healing remains unclear [19]. Some studies state that regardless of the destination between friend and foe, the skin microbiota tends to play a negative role in wound healing in different ways such as the elevation of pro-inflammatory mediators [20]. For instance, the persistence of bacteria in wounds impairs the healing process by elevation of pro-inflammatory cytokines such as interleukin-1 and tumor necrosis factor-alpha that in turn cause increased levels of matrix metalloproteinases (MMPs), a decreased level of tissue inhibitors to the MMPs, and decreased production of growth factors [21]. Cytokines are important signaling proteins that modulate fundamental pathophysiological and hemostatic processes, e.g., wound healing, by inducing downstream signal transduction pathways via specific cytokine receptors [22,23]. Therefore, cytokines can regulate the function of other receptors such as sodium channels, as well as the transient vanilloid receptors, and modulate the hemostatic process. Hence, a significant reduction in the number of pathogenic microbes using an appropriate antimicrobial agent is vital in regularizing wound healing. Moreover, satisfactory wound repair is possible only when the infection is brought under control. In other words, acceleration in the wound healing process is proportional to the reduction of the number of pathogenic microbes in the wound bed [24,25,26,27].
Numerous studies have reported that the most common pathogens associated with wound infections are Staphylococcus aureus (S. aureus); Pseudomonas aeruginosa (P. aeruginosa); Escherichia coli (E. coli); coagulase-negative Staphylococci (CoNS), i.e., Staphylococcus epidermidis (S. epidermidis); Streptococcus pyogenes (S. pyogenes); Klebsiella spp.; and Proteus spp. [28,29,30,31,32]. In chronic wounds, S. aureus, followed by P. aeruginosa, is the most common isolated microorganism [2,33,34], inhibiting wound healing, and is considered dominant [35,36]. However, many other commensal species have been isolated from cutaneous wounds, such as lactobacilli, which might have a positive therapeutic effect. Herein, we focus on the effect of CoNS, S. aureus, P. aeruginosa, and Lactobacilli on the wound-healing process, as illustrated in Figure 1.
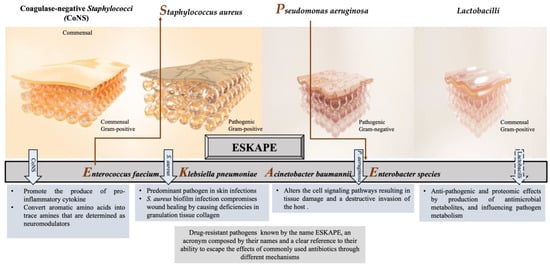
Figure 1.
Different responses of the most often isolated bacteria to the process of wound healing.
2.1. Coagulase-Negative Staphylococci (CoNS)
Staphylococci (mainly S. epidermidis, S. haemolyticus, S. hominis) are generally abundant bacteria that inhabit normal skin flora. Some CoNS species demonstrate a promoting influence on the wound as well as preventing a chronic process. For example, commensals of the human skin microbiota, such as S. epidermidis, stimulate IL-17+ CD8+ T cells, which are able to produce the pro-inflammatory cytokine IL-17A and restrict pathogen intrusion [37]. Moreover, S. caprae inhibits the quorum sensing (QS) of S. aureus by forming an autoinducing peptide, which leads to the expression of diverse virulence genes [38]. On the other hand, the lantibiotics gallidermin and epidermin, which are produced from certain CoNSs or antimicrobial peptides, displayed the blockage of cell wall biosynthesis in Gram-positive bacteria that might cause the inhibition of S. aureus [39].
S. epidermidis has been getting attention with its triggering mechanisms that reduce harmful microbes and encourage wound healing in the acute phase among the other CoNSs [40]. Recent studies have shown that S. epidermidis plays an active role in skin immunity, protecting from the invasion of Gram-positive and Gram-negative pathogens [41]. The SadA-expressing S. epidermidis strains that commonly inhabit human skin and gut microbiota [31,36] can convert aromatic amino acids into trace amines that are determined as neuromodulators by interacting with diverse adrenergic receptors. The wound-healing process is enhanced thanks to this biological reaction that induces the increase in keratinocyte migration, re-epithelialization rate, and extracellular-signal-regulated kinase level [42,43]. On the other hand, trace-amine-producing skin commensals can contribute to the acceleration of wound healing by suppressing the adrenaline and represent a promising therapeutic option [44].
2.2. Staphylococcus aureus
S. aureus is one of the most common and predominant pathogens with approximately 65% prevalence, involved in skin infections worldwide, which can cause persistent infections in chronic wounds with adverse effects [45]. This bacterium was first described by the Scottish surgeon Alexander Ogston as an isolate from a wound, and he demonstrated its significance as a pus pathogen [46]. As a commensal or opportunistic pathogen, S. aureus has been isolated from various reservoirs with a tendency to disseminate among them, such as humans, animals, and the environment [47,48,49], revealing genetic relatedness and sometimes threatening public health [50,51].
S. aureus is a pathobiont for humans and animals, leading to the emergence of more virulence and multidrug-resistant strains. Hence, this situation makes S. aureus-caused wound infections dangerous and requires the development of new prevention models and treatment strategies [52]. Because of the invasion, this bacterium can cause a variety of diseases, ranging from minor skin and soft tissue infections such as impetigo, folliculitis, and abscesses, to life-threatening systemic infections such as sepsis, endocarditis, or toxic shock syndrome [53]. It is also worth mentioning that the biofilm formation ability of most S. aureus strains is one of the specific virulence factors enabling them to adapt to the chronic wound environment [54].
Some S. aureus-strain-infected wounds may be challenging or almost impossible to treat due to their mostly antibiotic-resistant and high virulence nature, contributing to the pathogenicity of the host [55]. This high virulence allows it to escape the immune system’s reaction and thus the antibiotic activity. For instance, the existence of a β-lactamase enzyme in S. aureus enables it to break the ring of β-lactam antibiotics, making the antibiotics inactive [56]. In particular, some strains are resistant to methicillin as well as to other antibiotics. Methicillin resistance occurs due to the acquisition of mecA or mecC genes by previously susceptible strains that are consequently called methicillin-resistant S. aureus (MRSA) [57], followed by resistance to all β-lactam antibiotics, except for the fifth-generation cephalosporins [32,58]. MRSA is responsible for nosocomial infections, which are more difficult to treat. Patients with infections caused by MRSA have a much greater mortality risk than methicillin-sensitive strains [59].
As a consequence, the emergence and the unceasing spread of multi-resistant S. aureus strains, such as methicillin or vancomycin-resistant S. aureus, complicate the treatment of Staphylococcal infections with detrimental impact on global health and the economy. Therefore, the WHO has justifiably enlisted S. aureus as one of the major health threats in the so-called “post-antibiotic era” [60]. Thus, the development of new antibiotics and alternative prophylactic or therapeutic strategies have become inevitable to combat S. aureus, as well as other five ESKAPE bacteria (Figure 1), which possess multidrug resistance and high virulence [61,62].
2.3. Pseudomonas aeruginosa
P. aeruginosa is a Gram-negative bacterium included in the ESKAPE bacteria with concerns for public health, like S. aureus, and has a place in the concept of the “one health approach” [63]. It elicits prolonged hospitalization with increased morbidity and mortality rates [64] as a common opportunistic pathogen that causes several chronic, treatment-resistant infections in humans, i.e., certain skin, respiratory, and urinary tract infections [65,66]. Moreover, it has been isolated from patients with burn wounds, cystic fibrosis, acute leukemia, organ transplants, and intravenous drug addiction [67]. This bacterium penetrates wounds, holding the ability to form intact biofilms and subsequently degrading the extracellular matrix and altering the cell signaling pathways, resulting in tissue damage and a destructive invasion of the host [68,69]. P. aeruginosa has been reported as a detrimental pathogen during the past two decades, with grounds for 10 to 20% of infections in most hospitals, being determined as one of the twelve prior pathogens that pose the greatest threat to human health according to the WHO [62]. In 2017, it was estimated that 32,600 infections were caused by multidrug-resistant P. aeruginosa among hospitalized patients, and 2700 deaths occurred in the US [70].
The emergence of multidrug-resistant P. aeruginosa resulting in the persistence and non-response to clinical treatment of infectious diseases such as infected wounds has been referred to by several studies [71,72,73]. P. aeruginosa has been evaluated as resistant to diverse antibiotics, such as β-lactams, aminoglycosides, quinolones, and sulfonamides [74,75]. This resistance is derived from its excellent ability to select chromosomal mutations and acquire resistant genes, bearing multiple antimicrobial resistance mechanisms, which led P. aeruginosa to become one of the most difficult bacteria to treat [76,77].
Different mechanisms are involved in the expression of resistance of P. aeruginosa coming from innate and acquired ways. Innate resistance is related to an overexpressed efflux pump and low permeability of the outer membrane of the bacterium [78]. Acquired resistance involves the acquisition of a resistance gene or mutation in genes encoding porins, efflux pumps, penicillin-binding proteins, and chromosomal β-lactamase, all contributing to resistance to β-lactams, carbapenems, aminoglycosides, and fluoroquinolones [79]. P. aeruginosa strains have also been found resistant to aminoglycosides carrying the mexXY genes that induce the modification of aminoglycoside enzymes [80]. On the other hand, the genes crpP and qnrVC1 have been identified in clinical isolates of P. aeruginosa, which demonstrated resistance to fluoroquinolone [81]. Hence, all these concerns become P. aeruginosa-infected wounds, making developing treatment strategies vital.
2.4. Lactobacilli
In general, Lactobacilli are defined as beneficial bacteria and constitute a valuable member of an organs’ microbiota where they are located [82]. They are endogenous inhabitants of healthy skin, contrary to inflammatory skin, which is often associated with disturbed skin microbiota. The abundance and the diversity of Lactobacilli in the skin depend on the phylum of the host as well as on the anatomical areas [83].
The potential benefit of Lactobacilli as a skin habitat is based on the competition that exerts against skin pathogens through adhesion inhibition, production of antimicrobial metabolites, and influencing pathogen metabolism. Furthermore, their metabolites have proven to be immunomodulators, reducing excessive skin inflammation. In accordance with these effects, the functions of Lactobacilli as a skin barrier have already been tested in several clinical trials, making them a primarily promising alternative agent for promoting skin health [84]. Beyond the typical skin commensals, other bacterial species such as Lactobacilli might be beneficial for wound healing, either by their lysate that increases the migration and proliferation of keratinocytes or by the production of organic acids that act against pathogens and inhibit biofilm formation on wounds [85,86,87]. Several studies have shown that the application of probiotics causes a reduction in wound infections [86,88], especially when used as an adjuvant to antibiotic therapy. Lactobacillus plantarum, Lactobacillus casei, Lactobacillus acidophilus, and Lactobacillus rhamnosus are the most commonly used probiotics for most studies, which are well-known strains of the species with their positive effects [85]. All in vitro studies with these probiotics revealed successful inhibition of chosen skin or wound pathogens. Moreover, within the scope of in vivo studies on mice, rats, and rabbits, probiotics exhibited strong opportunities for counteracting wound infections caused mainly by S. aureus and P. aeruginosa. In regards, clinical studies generally showed a slight or statistically significant lower incidence of infections for patients using probiotics [85]. In particular, antimicrobial, quorum sensing, anti-biofilm, and adhesion assays were carried out with several probiotic strains and tested against selected skin pathogens. For instance, the tested Lactobacilli except for Lactobacillus delbrueckii exerted antimicrobial activity against skin pathogens, mainly due to organic acid production. On the other hand, most of them could prevent biofilm formation by selected pathogens [87].
Although probiotic Lactobacilli have a ‘generally regarded as safe’ (GRAS) status for food, the potential risks of live probiotics entering the bloodstream through breached skin has not been assessed. Hence, as opposed to previously mentioned studies, some investigations were conducted with the lysates of these probiotic strains as they could represent a safer alternative to the use of live bacteria in a wound situation. Moreover, the use of lysates may be of more utility to potential wound care manufacturers than live bacteria by overcoming the logistical requirements of maintaining viable bacteria within a formulation or wound dressing [89].
3. Communication between Skin Microbiota, Immune System, and Epithelial Cells
In vertebrates, regularly, all anatomical surfaces that communicate with the environment are colonized by microbes that compose the microbiome. Therefore, the study of the physiological and metabolic effects of the human microbiome on multicellular organisms regarding both health and disease concerns has become prominent.
The largest organ of the human body, the skin, is home to approximately 1012 bacterial cells [90,91] that comprise the skin microbiota, thanks to its immediate interface with the environment [17]. The skin microbiome, as mentioned, comprises a diverse population of fungi, bacteria, archaea, viruses, and sometimes parasites in close interaction with vertebrate hosts [92], and it contributes to the barrier function and hemostasis of skin tissue in various ways. For instance, the secretion of protease and lipase enzymes are involved in the desquamation and lipid surface degradation processes, respectively. Likewise, free fatty acid and sebum formation take place in the pH regulation of the skin tissue [93].
The skin microbiota has significant roles, such as the production of biofilms, bacteriocins, and quorum sensing [94,95]. It protects the skin tissue against pathogenic microorganisms by competition [96,97] and leads the production of antimicrobial peptides by virtue of commensal bacteria [39,98], which are in crosstalk with the immune system continuously that may be beneficial for the healing of the wound [99].
The internal communication between skin microbiota, epithelial cells, and the immune system is a primary mechanism to combat pathogenic invasion, as well as to ensure the maintenance of the skin commensals [14], which is schematically illustrated in Figure 2. This primary interaction has been initiated by keratinocytes toward the binding of pathogen-associated molecular patterns to pattern recognition receptors, resulting in the release of antimicrobial peptides as an inhibitory agent to diverse pathogens in the infection area. Moreover, the efficacy of various impacts of different microorganisms on the immune system has been determined as an important phenomenon in wound healing. For instance, the colony formation in wound bed by S. epidermidis has led to an increase in the expression of the cytokine, interleukin 1α (IL-1α). This cytokine is responsible for contributing to skin inflammation and host defense, which directly promotes wound healing [100]. In a study, the communication among commensal organisms, pathogens, and keratinocytes was investigated using polymicrobial biofilms formed by a mixture of commensal strains (S. epidermidis and M. luteus) and pathogens (S. aureus and P. aeruginosa). The commensals demonstrated the reduction of the damage caused by pathogens on the keratinocyte monolayer via degrading biofilm thickness and forming a layer between the keratinocytes and pathogens [101].
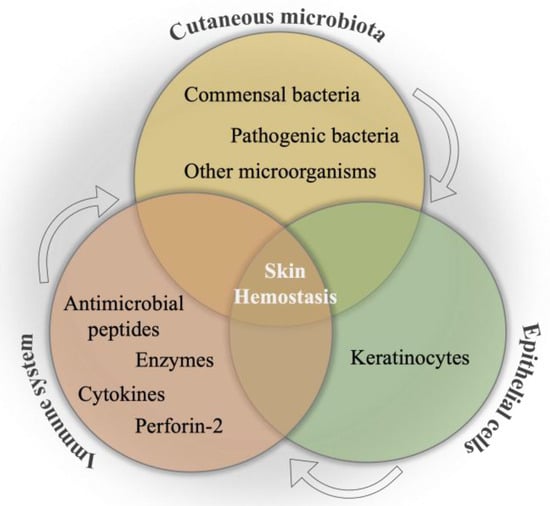
Figure 2.
Schematical illustration of the relation between cutaneous microbiota, immune system, and epithelial cells for skin hemostasis.
The innate immune protein Perforin-2, which is expressed by keratinocytes, is a key element for the inhibition of several Gram-positive and Gram-negative bacteria in the wound area. Even though it has proven inhibition activity on bacterial cells, in a study, S. aureus caused infection by inhibiting the expression of the Perforin-2 protein, which caused the delay of wound closure [102]. On the other hand, in vitro MRSA and P. aeruginosa caused polymicrobial wounds, demonstrating a delay in the reepithelization of the wound due to the downregulation of keratinocyte growth factor-1 expression, which is vital to promoting the proliferation and migration of keratinocytes [103]. In contrast to those negative impacts, skin commensals such as S. epidermidis were evaluated with their promoting effect on wound recovery, owing to their ability to induce diverse antimicrobials [11]. Alongside the inhibitory action of S. aureus on Perforin-2 expression, skin commensals are able to inhibit bacterial wound infections by altering the wound environment as well as skin tissue.
The microbiome composition of a wound may vary according to the wound type and influence its healing potential directly. Several studies have proved that most nonhealing wounds have a polymicrobial nature [30]. Both intracellular and extracellular bacteria exist in a biofilm, indicating several effects on the wound. Therefore, interactions between skin commensals and pathogens have gained importance in understanding wound healing regulation. As previously mentioned, S. aureus is one of the most abundant bacteria on wounds that mostly shows antimicrobial resistance, hence leading to delayed or impaired wound healing. Some species of CoNS are able to inhibit the S. aureus colonization or S. aureus-related biofilm formation. S. lugdunensis is able to prevent S. aureus colonization by producing the antibiotic lugdunin [104]. Likewise, S. hominis demonstrates the synergetic effects with antimicrobial peptide LL-37, owing to the lantibiotics they synthesize, hence prohibiting the colony formation of S. aureus [39]. It was also shown that serine protease glutamyl endopeptidase expressing S. epidermidis killed the S. aureus cells by promoting keratinocytes for the production of antimicrobial peptides [105]. Although some species of skin bacteria commensals seem beneficial and prominent for fighting S. aureus-caused infections, coproporphyrin III produced by Propionibacterium acnes led to the biofilm formation of S. aureus [17].
4. Identification of the Skin Microbiota
The identification of patients’ ‘core’ microbiome can help to understand its role in the pathogenesis of disease and subsequently discover new therapeutic strategies through microbial intervention [106]. The culture-dependent, denatured, and temperature gradient gel electrophoresis (DGGE and TGGE), metagenomic, and culturomic are techniques commonly used for identifying the composition of the skin microbiome (Table 1).

Table 1.
The identification techniques for skin microbiota.
The bacterial diversity of wounds has traditionally been detected by culture-dependent methods immediately after the collection of samples from the wound area. The samples are exposed to specific growth media to let the unknown bacteria grow. This cost-friendly and easy-to-study technique depends on the growth conditions and media according to the specific microorganisms [107]. Even though culture methods tend to select microorganisms that can grow under certain laboratory conditions, the detection of non-culturable microorganisms becomes limited.
The advanced sequencing methodologies, also called metagenomic techniques, provide more clarity as well as unbiased profiling on wounded skin microbiomes compared to culture techniques with higher taxonomical determination and functional investigations of bacteria, e.g., antibiotic resistance. On the other hand, these techniques are beneficial for the detection of bacterial cells in the formed biofilms on the wound area, which are highly difficult to reveal by culturing techniques [110,111]. These amplicon-based methods demonstrate more accuracy for bacterial diversity in collected samples than bacteria that grow in an artificial environment. Hence, culture-independent techniques have revealed many recent strains that previously have not been detected via culturing methods. In other words, wound microbiomes have started to show more diversity than previously suggested. For example, in a study, researchers compared culture and advanced sequencing diagnostic methods on bacterial diversity in wound microbiota by sampling 168 different wounds. The molecular diagnosis technique identified up to 33 different bacteria in the individual wound, whilst up to 3 bacteria were identified by culture-dependent methods [112].
Next-generation sequencing-based techniques can characterize the wound bioburden, which provides an important understanding of the role of the microbiota in regulating the wound healing process, as well as clinical outcomes including infection-caused complications [18]. These amplicon-based sequencing techniques with the advantage of 16s rRNA, which exists in all prokaryotes, contrary to eukaryotes, and is conserved among different bacterial species, greatly expand the knowledge of wound microbiota. Moreover, analyses focusing on fungal ITS gene sequencing provide additional insights into the impacts of non-bacterial microorganisms in chronic wounds.
Whole-genome sequencing (WGS) is a versatile analysis to determine a microbiome that represents not only the wound bioburden but also discriminates the microbial diversity of healthy and wounded skin with the beneficial effects to prevent and/or treat the wound. In comparison with published sequencing analyses of normal human skin microbiota, chronic wounds have more anaerobes, Gram-positive cocci, and Gram-negative rods, and fewer commensals, e.g., Propionibacterium [113]. In particular, Staphylococcus, Pseudomonas, and Corynebacterium are mostly isolated species from chronic wounds. The WGS, also called shotgun metagenomics sequencing, also contributes to the understanding of clinically relevant insights into the effects of strain level diversity, mechanisms of virulence, and responses to therapeutics as well as specific essential genes [114,115].
5. Manipulation of the Skin Microbiota
Most skin conditions such as wound infections are related to the instability in skin microbiota [114]. Microorganisms in wounded skin microbiota may delay or block the natural healing process as well as inflammation, even if the bacterial infection has not initiated wounding. Therefore, the dynamic skin microbiota presents an adequate platform for wound-healing treatment [116,117]. The manipulation of the skin microbiota has been studied by antimicrobial and prebiotic/probiotic therapies to accelerate the healing of the wound (Figure 3).
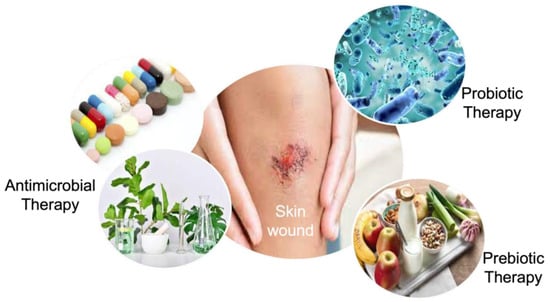
Figure 3.
The manipulation strategies of the wounded skin microbiome.
Antimicrobial therapy is the most studied manipulation method for wound healing by aiming for the inhibition and/or reduction of a considerable number of bacteria in the wounded skin microbiota. Antibiotics are a well-known and extensively used antimicrobial agent for this purpose. For instance, in a retrospective study that identified wounds frequently co-infected by S. aureus and P. aeruginosa, ampicillin indicated the higher resistance on 134 isolates, corresponding to 56.1%, followed by gentamicin, penicillin, trimethoprim/sulfamethoxazole, and piperacillin, in that order [118]. However, the misuse and overuse of antibiotics have led to the generation of drug-resistant pathogens [119,120], and antimicrobial resistance concerns were reported as one of the top global public health threats facing humanity by the World Health Organization [121]. Therefore, exploring alternative and cost-effective antimicrobials based on traditional plant-based medicines has become prominent [122]. Plant-based antimicrobials (e.g., essential oils, herbal extracts) have been determined as effective alternatives to accelerate the natural wound healing process, as well as tissue regeneration in the wound area due to the broad range of natural bioactive compounds contained in herbs [123,124,125]. Moreover, approximately 73% of pharmaceutical products include ingredients derived from natural products [126]. Recent studies have shown that medicinal plants improve wound healing in diabetic, infected, and opened wounds. For example, Moringa oleifera seeds loaded with n-hexane hydrogels inhibited the growth of S. aureus, E. coli, and P. aeruginosa, showing almost complete closure of excision wounds in a male Swiss albino mice model [127]. In another study, composite polymeric hydrogels, including Didymocarpus pedicellatus plant extract, gave rise to complete wound closure in diabetic rats [128]. However, plant-based antimicrobials are generally incorporated into a biomaterial in order to improve their stability, sensitivity, and on-target inability.
Probiotic and prebiotic therapies are another manipulation method for wounded skin microbiota. This manipulation technique aims to increase the number of beneficial microorganisms in the wound area by using living organisms (probiotics) [129], specific nutrients (prebiotics), or a special concept that favors the idea of the utilization of collage-type material infused with probiotics/prebiotics as a means of prevention and/or treatment options in skin disease [130,131]. Probiotics have demonstrated a reduction in bacterial load and promote wound healing [132]. Contrary to antibiotics, probiotic and prebiotic therapies are generally studied in vivo. In a clinical study with 80 patients who had second- or third-degree burn wounds, topical administration of Lactobacillus plantarum significantly accelerated the healing rate to 17% higher than the standard silver sulfadiazine for 3–7 days post-burn [133]. In comparison with probiotics, the prebiotic-based approach is an indirect method with the advantage of the reduced chance of immune response of skin tissue. However, they may also affect the low abundance of non-targeted bacteria in the skin microbiome [134].
6. Conclusions
Bacteria inhabit cutaneous microbiota, which is in balance under normal conditions, and have an important role in the natural wound healing process. The skin microbiota can easily become imbalanced after wounding of the skin tissue and enhance or diminish the healing process due to several reasons such as microbial load, dominant bacteria species, and communication between the immune system. Therefore, the understanding of the major bacteria present in the wound bed, their communication between the immune system and epithelial cells, their identification process, and their manipulation strategies to re-balance the cutaneous microbiota have come to the forefront. We believe that this review study, investigating various perspectives about the role of the skin microbiota under wounding conditions, can provide a way to develop new strategies for wound healing.
Author Contributions
Conceptualization: C.E. and A.T.; resources: C.E. and C.V.; literature review and writing—original draft preparation: C.E.; writing—review and editing: C.E., A.T., I.S., S.S., C.V. and D.I.Z.; supervision: I.S., A.T. and D.I.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union, EuroNanoMed3, project nAngioDerm, through the Greek General Secretariat for Research and Innovation ERA-NETS (code number T9EPA3-00022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pugliese, E.; Coentro, J.Q.; Raghunath, M.; Zeugolis, D.I. Wound Healing and Scar Wars. Adv. Drug Deliv. Rev. 2018, 129, 1–3. [Google Scholar] [CrossRef]
- Rahim, K.; Saleha, S.; Zhu, X.; Huo, L.; Basit, A.; Franco, O.L. Bacterial contribution in chronicity of wounds. Microb. Ecol. 2017, 73, 710–721. [Google Scholar] [CrossRef]
- Nasser, S.; Mabrouk, A.; Maher, A. Colonization of burn wounds in Ain Shams University burn unit. Burns 2003, 29, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Zeeuwen, P.L.; Boekhorst, J.; van den Bogaard, E.H.; de Koning, H.D.; van de Kerkhof, P.; Saulnier, D.M.; van Swam, I.I.; van Hijum, S.A.; Kleerebezem, M.; Schalkwijk, J. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fournière, M.; Latire, T.; Souak, D.; Feuilloley, M.G.; Bedoux, G. Staphylococcus epidermidis and Cutibacterium acnes: Two major sentinels of skin microbiota and the influence of cosmetics. Microorganisms 2020, 8, 1752. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.; King, T.; Aminov, R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat. Res. Fund. Mol. M. 2007, 622, 58–69. [Google Scholar] [CrossRef]
- Macia, L.; Thorburn, A.N.; Binge, L.C.; Marino, E.; Rogers, K.E.; Maslowski, K.M.; Vieira, A.T.; Kranich, J.; Mackay, C.R. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol. Rev. 2012, 245, 164–176. [Google Scholar] [CrossRef]
- Lange-Asschenfeldt, B.; Marenbach, D.; Lang, C.; Patzelt, A.; Ulrich, M.; Maltusch, A.; Terhorst, D.; Stockfleth, E.; Sterry, W.; Lademann, J. Distribution of bacteria in the epidermal layers and hair follicles of the human skin. Ski. Pharmacol. Physiol. 2011, 24, 305–311. [Google Scholar] [CrossRef]
- Rosenthal, M.; Goldberg, D.; Aiello, A.; Larson, E.; Foxman, B. Skin microbiota: Microbial community structure and its potential association with health and disease. Infect. Genet. Evol. 2011, 11, 839–848. [Google Scholar] [CrossRef]
- Pistone, D.; Meroni, G.; Panelli, S.; D’Auria, E.; Acunzo, M.; Pasala, A.R.; Zuccotti, G.V.; Bandi, C.; Drago, L. A journey on the skin microbiome: Pitfalls and opportunities. Int. J. Mol. Sci. 2021, 22, 9846. [Google Scholar] [CrossRef]
- Harrison, O.J.; Linehan, J.L.; Shih, H.-Y.; Bouladoux, N.; Han, S.-J.; Smelkinson, M.; Sen, S.K.; Byrd, A.L.; Enamorado, M.; Yao, C. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 2019, 363, eaat6280. [Google Scholar] [CrossRef] [PubMed]
- Balato, A.; Cacciapuoti, S.; Di Caprio, R.; Marasca, C.; Masarà, A.; Raimondo, A.; Fabbrocini, G. Human microbiome: Composition and role in inflammatory skin diseases. Arch. Immunol. Ther. Exp. 2019, 67, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.R.; Meisel, J.S.; Loesche, M.A.; Horwinski, J.; Soaita, I.; Chen, X.; Uberoi, A.; Gardner, S.E.; Grice, E.A. Strain-and species-level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe 2019, 25, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Tomic-Canic, M.; Burgess, J.L.; O’Neill, K.E.; Strbo, N.; Pastar, I. Skin microbiota and its interplay with wound healing. Am. J. Clin. Dermatol. 2020, 21, 36–43. [Google Scholar] [CrossRef]
- Wang, G.; Sweren, E.; Liu, H.; Wier, E.; Alphonse, M.P.; Chen, R.; Islam, N.; Li, A.; Xue, Y.; Chen, J. Bacteria induce skin regeneration via IL-1β signaling. Cell Host Microbe 2021, 29, 777–791.e776. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.R.; Gómez, B.I.; McIntyre, M.K.; Dubick, M.A.; Christy, R.J.; Nicholson, S.E.; Burmeister, D.M. The cutaneous microbiome and wounds: New molecular targets to promote wound healing. Int. J. Mol. Sci. 2018, 19, 2699. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Minasyan, H. Sepsis: Mechanisms of bacterial injury to the patient. Scand. J. Trauma Resusc. Emerg. Med. 2019, 27, 1–22. [Google Scholar] [CrossRef]
- Flowers, L.; Grice, E.A. The skin microbiota: Balancing risk and reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- Canesso, M.C.; Vieira, A.T.; Castro, T.B.; Schirmer, B.G.; Cisalpino, D.; Martins, F.S.; Rachid, M.A.; Nicoli, J.R.; Teixeira, M.M.; Barcelos, L.S. Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J. Immunol. 2014, 193, 5171–5180. [Google Scholar] [CrossRef]
- Tarnuzzer, R.W.; Schultz, G.S. Biochemical analysis of acute and chronic wound environments. Wound Repair Regen. 1996, 4, 321–325. [Google Scholar] [CrossRef]
- Zgheib, C.; Xu, J.; Liechty, K.W. Targeting inflammatory cytokines and extracellular matrix composition to promote wound regeneration. Adv. Wound Care 2014, 3, 344–355. [Google Scholar] [CrossRef]
- Xiao, T.; Yan, Z.; Xiao, S.; Xia, Y. Proinflammatory cytokines regulate epidermal stem cells in wound epithelialization. Stem Cell Res. Ther. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Burke, J.F. The effective period of preventive antibiotic action in experimental incisions and dermal lesions. Surgery 1961, 50, 161–168. [Google Scholar]
- Kumar, M.S.; Sripriya, R.; Raghavan, H.V.; Sehgal, P.K. Wound healing potential of Cassia fistula on infected albino rat model. J. Surg. Res. 2006, 131, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, G.S.; Cooper, D.M.; Knighton, D.R.; Margolis, D.J.; Percoraro, R.E.; Rodeheaver, G.; Robson, M.C. Definitions and guidelines for assessment of wounds and evaluation of healing. Wound Repair Regen. 1994, 2, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.C. Wound infection: A failure of wound healing caused by an imbalance of bacteria. Surg. Clin. N. Am. 1997, 77, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.; Harding, K.G. Bacteria and wound healing. Curr. Opin. Infect. Dis. 2004, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Collier, M. Recognition and management of wound infections. World Wide Wounds 2004, 7, 8–14. [Google Scholar]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef]
- Bessa, L.J.; Fazii, P.; Di Giulio, M.; Cellini, L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: Some remarks about wound infection. Int. Wound J. 2015, 12, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Coelho, C.; Santos-Silva, I.; de Lencastre, H.; Aires-de-Sousa, M. Epidemiology of methicillin-resistant and-susceptible Staphylococcus aureus in Luanda, Angola: First description of the spread of the MRSA ST5-IVa clone in the African continent. Microb. Drug Resist. 2014, 20, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Dong, W.; Lu, Y.; Jiang, M.; Zhang, D.; Aobuliaximu, Y.; Dong, J.; Niu, Y.; Liu, Y.; Guan, B. Distribution and antibiotic resistance patterns of pathogenic bacteria in patients with chronic cutaneous wounds in China. Front. Med. 2021, 8, 609584. [Google Scholar] [CrossRef] [PubMed]
- Kirketerp-Møller, K.; Jensen, P.Ø.; Fazli, M.; Madsen, K.G.; Pedersen, J.; Moser, C.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008, 46, 2717–2722. [Google Scholar] [CrossRef]
- Serra, R.; Grande, R.; Butrico, L.; Rossi, A.; Settimio, U.F.; Caroleo, B.; Amato, B.; Gallelli, L.; De Franciscis, S. Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti Infect. Ther. 2015, 13, 605–613. [Google Scholar] [CrossRef]
- Wong, S.Y.; Manikam, R.; Muniandy, S. Prevalence and antibiotic susceptibility of bacteria from acute and chronic wounds in Malaysian subjects. J. Infect. Dev. Ctries. 2015, 9, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.-J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015, 520, 104–108. [Google Scholar] [CrossRef]
- Paharik, A.E.; Parlet, C.P.; Chung, N.; Todd, D.A.; Rodriguez, E.I.; Van Dyke, M.J.; Cech, N.B.; Horswill, A.R. Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 2017, 22, 746–756.e745. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- Claudel, J.-P.; Auffret, N.; Leccia, M.-T.; Poli, F.; Corvec, S.; Dréno, B. Staphylococcus epidermidis: A potential new player in the physiopathology of acne? Dermatology 2019, 235, 287–294. [Google Scholar] [CrossRef]
- Stacy, A.; Belkaid, Y. Microbial guardians of skin health. Science 2019, 363, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Pullar, C.E.; Isseroff, R.R. The β2-adrenergic receptor activates pro-migratory and pro-proliferative pathways in dermal fibroblasts via divergent mechanisms. J. Cell Sci. 2006, 119, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Steenhuis, P.; Huntley, R.; Gurenko, Z.; Yin, L.; Dale, B.; Fazel, N.; Isseroff, R. Adrenergic signaling in human oral keratinocytes and wound repair. J. Dent. Res. 2011, 90, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Luqman, A.; Muttaqin, M.Z.; Yulaipi, S.; Ebner, P.; Matsuo, M.; Zabel, S.; Tribelli, P.M.; Nieselt, K.; Hidayati, D.; Götz, F. Trace amines produced by skin bacteria accelerate wound healing in mice. Commun. Biol. 2020, 3, 277. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Santra, S.; Das, A.; Dixith, S.; Sinha, M.; Ghatak, S.; Ghosh, N.; Banerjee, P.; Khanna, S.; Mathew-Steiner, S. Staphylococcus aureus biofilm infection compromises wound healing by causing deficiencies in granulation tissue collagen. Ann. Surg. 2020, 271, 1174. [Google Scholar] [CrossRef]
- Alexander, O. Classics in infectious diseases. On abscesses. Rev. Infect. Dis 1984, 6, 122–128. [Google Scholar]
- Heaton, C.J.; Gerbig, G.R.; Sensius, L.D.; Patel, V.; Smith, T.C. Staphylococcus aureus epidemiology in wildlife: A systematic review. Antibiotics 2020, 9, 89. [Google Scholar] [CrossRef]
- Pirolo, M.; Visaggio, D.; Gioffrè, A.; Artuso, I.; Gherardi, M.; Pavia, G.; Samele, P.; Ciambrone, L.; Di Natale, R.; Spatari, G. Unidirectional animal-to-human transmission of methicillin-resistant Staphylococcus aureus ST398 in pig farming; evidence from a surveillance study in southern Italy. Antimicrob. Resist. Infect. Control. 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Voidarou, C.; Tzora, A.; Skoufos, I.; Vassos, D.; Galogiannis, G.; Alexopoulos, A.; Bezirtzoglou, E. Experimental effect of ozone upon some indicator bacteria for preservation of an ecologically protected watery system. Water Air Soil Pollut. 2007, 181, 161–171. [Google Scholar] [CrossRef]
- Patrick, S. Bacteroides. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 917–944. [Google Scholar]
- Salgueiro, V.; Manageiro, V.; Bandarra, N.M.; Ferreira, E.; Clemente, L.; Caniça, M. Genetic relatedness and diversity of Staphylococcus aureus from different reservoirs: Humans and animals of livestock, poultry, zoo, and aquaculture. Microorganisms 2020, 8, 1345. [Google Scholar] [CrossRef]
- Mrochen, D.M.; Fernandes de Oliveira, L.M.; Raafat, D.; Holtfreter, S. Staphylococcus aureus host tropism and its implications for murine infection models. Int. J. Mol. Sci. 2020, 21, 7061. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Paleczny, J.; Junka, A.; Brożyna, M.; Dydak, K.; Oleksy-Wawrzyniak, M.; Ciecholewska-Juśko, D.; Dziedzic, E.; Bartoszewicz, M. The high impact of Staphylococcus aureus biofilm culture medium on in vitro outcomes of antimicrobial activity of wound antiseptics and antibiotic. Pathogens 2021, 10, 1385. [Google Scholar] [CrossRef]
- Alavi, S.M.; Khosravi, A.D.; Sarami, A.; Dashtebozorg, A.; Montazeri, E.A. Bacteriologic study of diabetic foot ulcer. Pak. J. Med. Sci. 2007, 23, 684. [Google Scholar] [CrossRef]
- Fatimah, S.; Nadifah, F.; Burhanudin, I. Uji daya hambat ekstrak etanol kubis (Brassica oleracea var. capitata f. alba) terhadap bakteri Staphylococcus aureus secara in vitro. Biogenesis J. Ilm. Biol. 2016, 4, 102–106. [Google Scholar] [CrossRef][Green Version]
- Boswihi, S.S.; Udo, E.E. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pract. 2018, 8, 18–24. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Tacconelli, E.; Pop-Vicas, A.; D’Agata, E. Increased mortality among elderly patients with meticillin-resistant Staphylococcus aureus bacteraemia. J. Hosp. Infect. 2006, 64, 251–256. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Dalton, K.R.; Rock, C.; Carroll, K.C.; Davis, M.F. One Health in hospitals: How understanding the dynamics of people, animals, and the hospital built-environment can be used to better inform interventions for antimicrobial-resistant gram-positive infections. Antimicrob. Resist. Infect. Control. 2020, 9, 1–17. [Google Scholar] [CrossRef]
- Alhussain, F.A.; Yenugadhati, N.; Al Eidan, F.A.; Al Johani, S.; Badri, M. Risk factors, antimicrobial susceptibility pattern and patient outcomes of Pseudomonas aeruginosa infection: A matched case-control study. J. Infect. Public Health 2021, 14, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Raizman, R.; Little, W.; Smith, A.C. Rapid diagnosis of Pseudomonas aeruginosa in wounds with point-of-care fluorescence Imaing. Diagnostics 2021, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Vanderwoude, J.; Fleming, D.; Azimi, S.; Trivedi, U.; Rumbaugh, K.P.; Diggle, S.P. The evolution of virulence in Pseudomonas aeruginosa during chronic wound infection. Proc. R. Soc. B 2020, 287, 20202272. [Google Scholar] [CrossRef]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.B.; Shruptha, P.; Prabhu, V.; Srujan, C.; Nayak, U.Y.; Anuradha, C.K.R.; Ramachandra, L.; Keerthana, P.; Joshi, M.B.; Murali, T.S. Pseudomonas aeruginosa virulence proteins pseudolysin and protease IV impede cutaneous wound healing. Lab. Investig. 2020, 100, 1532–1550. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Holst, E.; Tapper, H.; Björck, L. Elastase-producing Pseudomonas aeruginosa degrade plasma proteins and extracellular products of human skin and fibroblasts, and inhibit fibroblast growth. Microb. Pathog. 2003, 34, 47–55. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United State; Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob. Agents Chemother. 2006, 50, 43–48. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s not easy being green: A narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- Miyoshi-Akiyama, T.; Tada, T.; Ohmagari, N.; Viet Hung, N.; Tharavichitkul, P.; Pokhrel, B.M.; Gniadkowski, M.; Shimojima, M.; Kirikae, T. Emergence and spread of epidemic multidrug-resistant Pseudomonas aeruginosa. Genome Biol. Evol. 2017, 9, 3238–3245. [Google Scholar] [CrossRef] [PubMed]
- Sonmezer, M.C.; Ertem, G.; Erdinc, F.S.; Kaya Kilic, E.; Tulek, N.; Adiloglu, A.; Hatipoglu, C. Evaluation of risk factors for antibiotic resistance in patients with nosocomial infections caused by Pseudomonas aeruginosa. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 1321487. [Google Scholar] [CrossRef] [PubMed]
- Montero, M.M.; López Montesinos, I.; Knobel, H.; Molas, E.; Sorlí, L.; Siverio-Parés, A.; Prim, N.; Segura, C.; Duran-Jordà, X.; Grau, S. Risk factors for mortality among patients with Pseudomonas aeruginosa bloodstream infections: What is the influence of XDR phenotype on outcomes? J. Clin. Med. 2020, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Krebes, K.; McNally, C.; Neshat, S. Multiple antibiotic resistance in Pseudomonas aeruginosa: Evidence for involvement of an efflux operon. J. Bacteriol. Res. 1993, 175, 7363–7372. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Oie, S.; Fukui, Y.; Yamamoto, M.; Masuda, Y.; Kamiya, A. In vitro antimicrobial effects of aztreonam, colistin, and the 3-drug combination of aztreonam, ceftazidime and amikacin on metallo-β-lactamase-producing Pseudomonas aeruginosa. BMC Infect. Dis. 2009, 9, 1–5. [Google Scholar] [CrossRef]
- Atassi, G.; Scheetz, M.; Nozick, S.; Rhodes, N.J.; Murphy-Belcaster, M.; Murphy, K.R.; Ozer, E.A.; Hauser, A.R. Genomics of aminoglycoside resistance in pseudomonas aeruginosa bloodstream infections at a United States Academic Hospital. Medrxiv 2021, 11, e05087-22. [Google Scholar] [CrossRef]
- Khan, M.; Summers, S.; Rice, S.A.; Stapleton, F.; Willcox, M.D.; Subedi, D. Acquired fluoroquinolone resistance genes in corneal isolates of Pseudomonas aeruginosa. Infect. Genet. Evol. 2020, 85, 104574. [Google Scholar] [CrossRef]
- Ishaq, M.; Khan, A.; Bacha, A.S.; Shah, T.; Hanif, A.; Ahmad, A.A.; Ke, W.; Li, F.; Ud Din, A.; Ding, Z. Microbiota targeted interventions of probiotic lactobacillus as an anti-ageing approach: A review. Antioxidants 2021, 10, 1930. [Google Scholar] [CrossRef]
- Kim, J.-H.; Son, S.-M.; Park, H.; Kim, B.K.; Choi, I.S.; Kim, H.; Huh, C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021, 11, 16269. [Google Scholar] [CrossRef]
- Delanghe, L.; Spacova, I.; Van Malderen, J.; Oerlemans, E.; Claes, I.; Lebeer, S. The role of lactobacilli in inhibiting skin pathogens. Biochem. Soc. Trans. 2021, 49, 617–627. [Google Scholar] [PubMed]
- Fijan, S.; Frauwallner, A.; Langerholc, T.; Krebs, B.; ter Haar née Younes, J.A.; Heschl, A.; Mičetić Turk, D.; Rogelj, I. Efficacy of using probiotics with antagonistic activity against pathogens of wound infections: An integrative review of literature. BioMed Res. Int. 2019, 2019, 7585486. [Google Scholar] [CrossRef] [PubMed]
- Mohammedsaeed, W.; Cruickshank, S.; McBain, A.J.; O’Neill, C.A. Lactobacillus rhamnosus GG lysate increases re-epithelialization of keratinocyte scratch assays by promoting migration. Sci. Rep. 2015, 5, 16147. [Google Scholar] [CrossRef]
- Lopes, E.G.; Moreira, D.A.; Gullón, P.; Gullón, B.; Cardelle-Cobas, A.; Tavaria, F.K. Topical application of probiotics in skin: Adhesion, antimicrobial and antibiofilm in vitro assays. J. Appl. Microbiol. 2017, 122, 450–461. [Google Scholar] [CrossRef]
- Heydari Nasrabadi, M.; Tajabadi Ebrahimi, M.; Dehghan Banadaki, S. Study of cutaneous wound healing in rats treated with Lactobacillus plantarum on days 1, 3, 7, 14 and 21. Afr. J. Pharm. Pharmacol. 2011, 5, 2395–2401. [Google Scholar] [CrossRef]
- Mohammedsaeed, W.; McBain, A.J.; Cruickshank, S.M.; O’Neill, C.A. Lactobacillus rhamnosus GG inhibits the toxic effects of Staphylococcus aureus on epidermal keratinocytes. Appl. Environ. Microbiol. 2014, 80, 5773–5781. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996, 4, 430–435. [Google Scholar] [CrossRef]
- Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977, 31, 107–133. [Google Scholar] [CrossRef]
- Ross, A.A.; Rodrigues Hoffmann, A.; Neufeld, J.D. The skin microbiome of vertebrates. Microbiome 2019, 7, 1–14. [Google Scholar]
- Meisel, J.S.; Sfyroera, G.; Bartow-McKenney, C.; Gimblet, C.; Bugayev, J.; Horwinski, J.; Kim, B.; Brestoff, J.R.; Tyldsley, A.S.; Zheng, Q. Commensal microbiota modulate gene expression in the skin. Microbiome 2018, 6, 1–15. [Google Scholar]
- Baldwin, H.E.; Bhatia, N.; Friedman, A.; Prunty, T.; Martin, R.; Seite, S. The role of cutaneous microbiota harmony in maintaining a functional skin barrier. Skin 2017, 1, s139. [Google Scholar]
- Williams, M.R.; Costa, S.K.; Zaramela, L.S.; Khalil, S.; Todd, D.A.; Winter, H.L.; Sanford, J.A.; O’Neill, A.M.; Liggins, M.C.; Nakatsuji, T. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 2019, 11, eaat8329. [Google Scholar] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing skin microbiome research: A method to the madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [PubMed]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95, fiy241. [Google Scholar] [PubMed]
- Coates, M.; Lee, M.J.; Norton, D.; MacLeod, A.S. The skin and intestinal microbiota and their specific innate immune systems. Front. Immunol. 2019, 10, 2950. [Google Scholar]
- Maheswary, T.; Nurul, A.A.; Fauzi, M.B. The insights of microbes’ roles in wound healing: A comprehensive review. Pharmaceutics 2021, 13, 981. [Google Scholar]
- Jordana-Lluch, E.; Garcia, V.; Kingdon, A.D.; Singh, N.; Alexander, C.; Williams, P.; Hardie, K.R. A simple polymicrobial biofilm keratinocyte colonization model for exploring interactions between commensals, pathogens and antimicrobials. Front. Microbiol. 2020, 11, 291. [Google Scholar]
- Strbo, N.; Pastar, I.; Romero, L.; Chen, V.; Vujanac, M.; Sawaya, A.P.; Jozic, I.; Ferreira, A.D.; Wong, L.L.; Head, C. Single cell analyses reveal specific distribution of anti-bacterial molecule Perforin-2 in human skin and its modulation by wounding and Staphylococcus aureus infection. Exp. Dermatol. 2019, 28, 225–232. [Google Scholar]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013, 8, e56846. [Google Scholar]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [PubMed]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and gut microbiome in psoriasis: Gaining insight into the pathophysiology of it and finding novel therapeutic strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [PubMed]
- Grogan, M.D.; Bartow-McKenney, C.; Flowers, L.; Knight, S.A.; Uberoi, A.; Grice, E.A. Research techniques made simple: Profiling the skin microbiota. J. Investig. Dermatol. 2019, 139, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, J.-P.; Sotto, A.; Dunyach-Remy, C.; Lipsky, B.A. New molecular techniques to study the skin microbiota of diabetic foot ulcers. Adv. Wound Care 2015, 4, 38–49. [Google Scholar]
- Sandhu, S.S.; Pourang, A.; Sivamani, R.K. A review of next generation sequencing technologies used in the evaluation of the skin microbiome: What a time to be alive. Dermatol. Online J. 2019, 25, 13030. [Google Scholar]
- Brandwein, M.; Steinberg, D.; Meshner, S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes 2016, 2, 3. [Google Scholar]
- Xu, Z.; Hsia, H.C. The impact of microbial communities on wound healing: A review. Ann. Plast. Surg. 2018, 81, 113–123. [Google Scholar]
- Rhoads, D.D.; Wolcott, R.D.; Sun, Y.; Dowd, S.E. Comparison of culture and molecular identification of bacteria in chronic wounds. Int. J. Mol. Sci. 2012, 13, 2535–2550. [Google Scholar]
- Han, A.; Zenilman, J.M.; Melendez, J.H.; Shirtliff, M.E.; Agostinho, A.; James, G.; Stewart, P.S.; Mongodin, E.F.; Rao, D.; Rickard, A.H. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen. 2011, 19, 532–541. [Google Scholar]
- Kalan, L.; Grice, E.A. Fungi in the wound microbiome. Adv. Wound Care 2018, 7, 247–255. [Google Scholar] [CrossRef]
- Lipof, J.S.; Jones, C.M.C.; Daiss, J.; Oh, I. Comparative study of culture, next-generation sequencing, and immunoassay for identification of pathogen in diabetic foot ulcer. J. Orthop. Res. 2021, 39, 2638–2645. [Google Scholar] [CrossRef]
- Smythe, P.; Wilkinson, H.N. The Skin Microbiome: Current Landscape and Future Opportunities. Int. J. Mol. Sci. 2023, 24, 3950. [Google Scholar] [CrossRef]
- Callewaert, C.; Knödlseder, N.; Karoglan, A.; Güell, M.; Paetzold, B. Skin microbiome transplantation and manipulation: Current state of the art. Comput. Struct. Biotechnol. J. 2021, 19, 624–631. [Google Scholar] [CrossRef]
- Puca, V.; Marulli, R.Z.; Grande, R.; Vitale, I.; Niro, A.; Molinaro, G.; Prezioso, S.; Muraro, R.; Di Giovanni, P. Microbial species isolated from infected wounds and antimicrobial resistance analysis: Data emerging from a three-years retrospective study. Antibiotics 2021, 10, 1162. [Google Scholar] [CrossRef] [PubMed]
- Ersanli, C.; Tzora, A.; Skoufos, I.; Fotou, K.; Maloupa, E.; Gridoriadou, K.; Voidarou, C.; Zeugolis, D.I. The Assessment of Antimicrobial and Anti-Biofilm Activity of Essential Oils against Staphylococcus aureus Strains. Antibiotics 2023, 12, 384. [Google Scholar] [CrossRef]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 10, 2041. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Mitropoulou, G.; Stavropoulou, E.; Vaou, N.; Tsakris, Z.; Voidarou, C.; Tsiotsias, A.; Tsigalou, C.; Taban, B.M.; Kourkoutas, Y.; Bezirtzoglou, E. Insights into Antimicrobial and Anti-Inflammatory Applications of Plant Bioactive Compounds. Microorganisms 2023, 11, 1156. [Google Scholar] [CrossRef]
- Gorain, B.; Pandey, M.; Leng, N.H.; Yan, C.W.; Nie, K.W.; Kaur, S.J.; Marshall, V.; Sisinthy, S.P.; Panneerselvam, J.; Molugulu, N. Advanced drug delivery systems containing herbal components for wound healing. Int. J. Pharm. 2022, 617, 121617. [Google Scholar] [CrossRef] [PubMed]
- Hajialyani, M.; Tewari, D.; Sobarzo-Sánchez, E.; Nabavi, S.M.; Farzaei, M.H.; Abdollahi, M. Natural product-based nanomedicines for wound healing purposes: Therapeutic targets and drug delivery systems. Int. J. Nanomed. 2018, 13, 5023. [Google Scholar] [CrossRef]
- Yazarlu, O.; Iranshahi, M.; Kashani, H.R.K.; Reshadat, S.; Habtemariam, S.; Iranshahy, M.; Hasanpour, M. Perspective on the application of medicinal plants and natural products in wound healing: A mechanistic review. Pharmacol. Res. 2021, 174, 105841. [Google Scholar] [CrossRef] [PubMed]
- Wangchuk, P. Therapeutic applications of natural products in herbal medicines, biodiscovery programs, and biomedicine. J. Biol. Act. Prod. Nat. 2018, 8, 1–20. [Google Scholar] [CrossRef]
- Ali, A.; Garg, P.; Goyal, R.; Kaur, G.; Li, X.; Negi, P.; Valis, M.; Kuca, K.; Kulshrestha, S. A novel herbal hydrogel formulation of moringa oleifera for wound healing. Plants 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.K.; Bhardwaj, R.; Arora, R.; Singh, A.; Mukherjee, M.; Rajput, S.K. Acceleration of wound healing in diabetic rats through poly dimethylaminoethyl acrylate–hyaluronic acid polymeric hydrogel impregnated with a Didymocarpus pedicellatus plant extract. ACS Omega 2020, 5, 24239–24246. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Vitamin and Mineral Requirements in Human Nutrition; World Health Organization: Geneva, Switzerland, 2004; pp. 17–299. [Google Scholar]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Sorushanova, A.; Skoufos, I.; Tzora, A.; Mullen, A.M.; Zeugolis, D.I. The influence of animal species, gender and tissue on the structural, biophysical, biochemical and biological properties of collagen sponges. J. Mater. Sci. Mater. Med. 2021, 32, 12. [Google Scholar] [CrossRef]
- Nole, K.L.B.; Yim, E.; Keri, J.E. Probiotics and prebiotics in dermatology. J. Am. Acad. Dermatol. 2014, 71, 814–821. [Google Scholar] [CrossRef]
- Peral, M.C.; Huaman Martinez, M.A.; Valdez, J.C. Bacteriotherapy with Lactobacillus plantarum in burns. Int. Wound J. 2009, 6, 73–81. [Google Scholar] [CrossRef]
- Roberfroid, M. Prebiotics: The concept revisited. J. Nutr. 2007, 137, 830S–837S. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).